Abstract
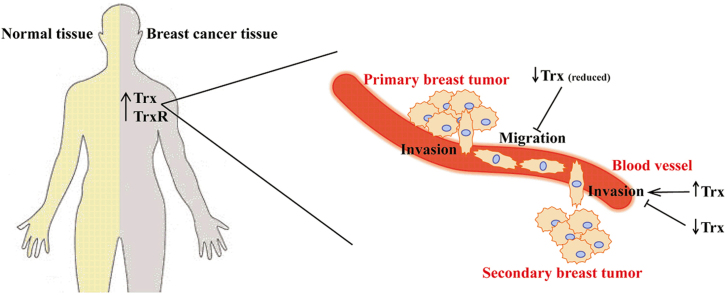
Metastasis is the most life threatening aspect of breast cancer. It is a multi-step process involving invasion and migration of primary tumor cells with a subsequent colonization of these cells at a secondary location. The aim of the present study was to investigate the role of thioredoxin (Trx1) in the invasion and migration of breast cancer cells and to assess the strength of the association between high levels of Trx1 and thioredoxin reductase (TrxR1) expression with breast cancer patient survival. Our results indicate that the expression of both Trx1 and TrxR1 are statistically significantly increased in breast cancer patient cells compared with paired normal breast tissue from the same patient. Over-expression of Trx1 in MDA-MB-231 breast cancer cell lines enhanced cell invasion in in vitro assays while expression of a redox inactive mutant form of Trx1 (designated 1SS) or the antisense mRNA inhibited cell invasion. Addition of exogenous Trx1 also enhanced cell invasion, while addition of a specific monoclonal antibody that inhibits Trx1 redox function decreased cell invasion. Over-expression of intracellular Trx1 did not increase cell migration but expression of intracellular 1SS inhibited migration. Addition of exogenous Trx1 enhanced cell migration while 1SS had no effect. Treatment with auranofin inhibited TrxR activity, cell migration and clonogenic activity of MDA-MB-231 cells, while increasing reactive oxygen species (ROS) levels. Analysis of 25 independent cohorts with 5910 patients showed that Trx1 and TrxR1 were both associated with a poor patient prognosis in terms of overall survival, distant metastasis free survival and disease free survival. Therefore, targeting the Trx system with auranofin or other specific inhibitors may provide improved breast cancer patient outcomes through inhibition of cancer invasion and migration.
Abbreviations: ATL, adult T-cell leukemia; CI, confidence interval; FBS, fetal bovine serum; DFS, disease free survival; DMFS, distant-metastasis free survival; DTNB, 5,5′-Dithio-Bis (2-Nitrobenzoic Acid); DTT, dithiothreitol; GFP, green fluorescent protein; H2DCF-DA, 2′,7′-dichlorodihydrofluorescein diacetate; HR, hazard ratio; HUVEC, human umbilical vein endothelial cells; MMP, matrix metalloproteinase; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OS, overall survival; RFU, relative fluorescence units; ROS, reactive oxygen species; TCGA, the cancer genome atlas; TIMP, tissue inhibitor of metalloproteinase; TNB, 2-nitro-5-thiobenzoate anion; TPM, transcripts per million; Trx, thioredoxin; TrxR, thioredoxin reductase; TXNIP, thioredoxin interacting protein; VEGF, vascular endothelial growth factor
Keywords: Thioredoxin, Breast cancer, Cell invasion, Cell migration, Patient survival, Auranofin
Graphical abstract
Highlights
-
•
Over expression of thioredoxin in MDA-MB-231 cells enhanced cell invasion in vitro.
-
•
Thioredoxin inhibition reduced cell invasion and migration of MDA-MB-231 cells.
-
•
Addition of thioredoxin enhanced migration of MDA-MB-231 cells in vitro.
-
•
Auranofin treatment inhibited MDA-MB-231 cell migration and clonogenic activity.
-
•
High Trx1 and TrxR1 expression is associated with a poor breast cancer prognosis.
1. Introduction
Oxidative stress is a characteristic feature of many cancers, including breast cancer, and has been implicated in cancer development and progression [1], [2], [3], [4]. Higher levels of reactive oxygen species (ROS) are generated through the increased metabolic activity of cancer cells, enhanced signaling pathways or mitochondrial dysfunction [5]. As a consequence many antioxidants and redox control systems are up-regulated, suggesting that a balance of ROS levels is important to maintain cancer cell function [6]. One of the most important cellular cytoplasmic redox systems is the thioredoxin (Trx) system, comprised of Trx1, thioredoxin reductase (TrxR1) and NADPH [7] and it is highly expressed in many cancer cells, including breast tumors [8].
Thioredoxins are small (12 KDa) redox active proteins with a highly conserved active dithiol containing site of –Cys–Gly–Pro–Cys–, which participates in redox reactions through a reversible oxidation cycle [7]. The reduced Trx protein is a very effective protein disulfide reductase and through this action it is able to effect disulfide-dependent structural changes in other molecules, causing the activation or inhibition of target proteins. The oxidized Trx is then reduced by the NADPH dependent TrxR [7].
Multiple protein substrates have been identified for Trx1, including ribonucleotide reductase [7], various transcription factors [9], [10], peroxiredoxins [11] and the methionine sulfoxide reductases [12]. Furthermore Trx1 can also act as a direct scavenger of ROS [13] and reduced Trx1 inhibits apoptosis by binding to ASK-1 [14]. Involvement of Trx1 in protecting cells from apoptosis has many consequences for cancer, in terms of maintaining cell viability and protection from various chemotherapeutic treatments [15], [16].
Over recent years the Trx1 system has been linked with tumor development and cancer. Both Trx1 and TrxR1 are highly expressed in many cancer cell lines, [17], [18], [19], [20] and in patient tumors compared to corresponding normal tissue [8], [21], [22], [23]. Significant evidence links the expression of the Trx1 system with the proliferative capacity and the apoptotic resistance of different cancers. An in vivo study observed a positive correlation between Trx1 over-expression and cancer cell proliferation and decreased apoptosis in primary gastric carcinomas, which correlated with a poor clinical outcome [21]. Other studies have shown similar correlations with high Trx1 levels and decreased patient survival levels in non-small cell lung carcinoma [22] and colorectal cancer [23]. An association between high levels of Trx1 expression and the aggressiveness of tumors in human lung cancer [24], prostate carcinoma [25] and in skin cancers and mammary tumors [8] has also been observed.
An increase in Trx1 levels does not occur just as a consequence of cancer growth but rather, Trx1 has an active functional role in cancer metastasis and progression [6]. Several studies have been conducted using breast cancer cells to assess the role of Trx1 in cancer development and invasive processes. An early study showed that transfection of MCF-7 breast cancer cells with the gene encoding a redox inactive mutated Trx1 protein (with both active site cysteines mutated) reversed the transformed phenotype of cancer cells with the expressed protein acting in a dominant negative manner. When the transfected MCF-7 cells were inoculated into scid mice, tumor formation was almost completely suppressed with only microscopic tumor cell deposits being observed and no evidence of metastasis to other organs [26].
Trx1 expression has also been associated with the regulation of other molecules that are necessary for breast cancer cell invasion, including vascular endothelial growth factor (VEGF) and matrix metalloproteinase-9 (MMP-9). Transfection of MCF-7 breast cancer cells with a construct that over-expresses Trx1 increased VEGF production and secretion in vitro while transfection of constructs expressing the redox-inactive Trx1 protein resulted in a decrease of VEGF expression [27]. Transfection of the MDA-MB-231 breast cancer cell line with a Trx1 expressing construct resulted in an increase of MMP-9 expression and also enhanced cell invasion of these transfected cells in vitro, while transfection with a construct expressing the redox inactive protein resulted in decreased MMP-9 expression and decreased cancer cell invasion [28]. A previous study had shown that Trx1 preferentially inhibits the tissue inhibitor of metalloproteinases (TIMPs) so that MMP activity is favored, leading to extracellular matrix degradation and an increase in cancer cell invasion [29]. These results suggest that Trx1 plays an active role in regulating processes that lead to increased cancer cell invasion.
Trx1 also has extracellular functions. Trx1 can be secreted by cells by a non-classical pathway [19] and can act as an autocrine growth factor as well as displaying co-cytokine activity augmenting the effects of other cytokines [30]. Extracellular Trx1 may be bound to the external cell membrane where it may participate in and regulate cell to cell contact and contribute to redox regulation in the extracellular space [31]. Redox processes in the extracellular environment that involve Trx1 have also been linked to regulation of cellular processes that encompass cell differentiation, proliferation and death [32], [33]. This leads to the possibility that Trx1 may also control the progression or metastatic pathways of cancer cells through an extracellular activity [34]. A recent study showed that Trx1 is present at high levels in the serum of breast cancer patients [35] and it is therefore feasible for Trx1 to have extracellular roles in breast cancer.
This study defines the active site of Trx1 as required for stimulating cancer cell invasion and migration. Inhibition of the Trx1 system through the addition of auranofin, an inhibitor of TrxR1, also decreased the cell migration capacity of the breast cancer cells. We also show that tumor tissue isolated from breast cancer patients has higher Trx1 and TrxR1 expression than from the paired normal tissue. Patients with high Trx1 and TrxR1 expression levels were also shown to have poor outcomes in terms of overall survival, distant-metastasis free survival, and disease free survival.
2. Materials and methods
2.1. Cells and reagents
The human breast cancer cell line, MDA-MB-231 was obtained from Prof. Erik Thompson (Queensland University of Technology, Brisbane Australia). Cells were cultured in RPMI-1640 medium containing 10% (v/v) fetal bovine serum (FBS), 200 mM l-glutamine, and 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, VIC, Australia). Auranofin was purchased from ICN Biomedicals (NSW, Australia). The anti-human Trx1 monoclonal antibodies, 9G9 and 2B1, were made and characterized for their Trx1 binding properties as described previously [32]. Recombinant human Trx1 and the redox inactive mutated form of human Trx1 (designated 1SS) was expressed and purified as previously described [36]. All chemicals were purchased from Sigma (NSW, Australia) unless otherwise stated.
2.2. Gene expression data and survival analysis
We used 25 gene expression datasets obtained by microarray analysis of tumor specimens from a total of 4839 patients with primary breast cancers (Table 1). The Cancer Genome Atlas (TCGA) Data Portal was queried for RNA-seq expression and clinical information (n=1071) of breast invasive carcinoma patients (last update May 2015). Details on data generation are available in the original TCGA study [37]. The normalized gene expression results analyzed according to the RNASeqV2 protocol were obtained from the TCGA portal. Analysis was restricted to patients with matched tumor and normal samples (n=114). The statistical significance (p-value) of the difference in expression levels for a gene was calculated using a two-sample paired t test.
Table 1.
Datasets used for gene expression analysis. Total n= 5910
| Dataset | Microarray technology | Survival data | No. of patients | PMID |
|---|---|---|---|---|
| TCGA | RNA-seq | OS | 1071 | 23000897 |
| E-TABM-158 | Affymetrix HGU | OS, DMFS, DFS | 117 | 17157792 |
| GSE11121 | Affymetrix HGU | DMFS | 200 | 18593943 |
| GSE12093 | Affymetrix HGU | DMFS | 136 | 18821012 |
| GSE12276 | Affymetrix HGU | DMFS | 204 | 19421193 |
| GSE17705 | Affymetrix HGU | DMFS | 298 | 20697068 |
| GSE19615 | Affymetrix HGU | DMFS | 155 | 20098429 |
| GSE25066 | Affymetrix HGU | OS | 508 | 21558518 |
| GSE2603 | Affymetrix HGU | DMFS | 82 | 16049480 |
| GSE2990 | Affymetrix HGU | DMFS, DFS | 187 | 16478745 |
| GSE3143 | Affymetrix HGU | OS | 171 | 16273092 |
| GSE5327 | Affymetrix HGU | DMFS | 58 | 17420468 |
| GSE6532 | Affymetrix HGU | DMFS | 114 | 17401012 |
| GSE7390 | Affymetrix HGU | OS, DMFS, DFS | 198 | 17545524 |
| GSE9195 | Affymetrix HGU | DMFS, DFS | 77 | 18498629 |
| GSE18229 | Agilent | OS, DFS | 252 | 20813035 |
| Van de Vijver | Agilent | OS, DFS | 296 | 12490681 |
| E-UCON-1 | Agilent | DFS | 305 | 22366898 |
| GSE1379 | Arcturus 22k human oligonucleotide microarray | DFS | 60 | 15193263 |
| GSE1456 | Affymetrix HGU | DFS | 159 | 16280042 |
| GSE2034 | Affymetrix HGU | DFS | 286 | 15721472 |
| GSE3494 | Affymetrix HGU | DFS | 236 | 16141321 |
| GSE4922 | Affymetrix HGU | DFS | 249 | 17079448 |
| GSE6532 | Affymetrix HGU | DFS | 241 | 17401012 |
| Vant'Veer | Agilent | DFS | 95 | 11823860 |
| GSE9893 | MLRG Human 21K V12.0 | OS | 155 | 18347175 |
For survival analysis patients were dichotomized based on gene expression levels. The optimal cut-points were searched within the inner 80% selection interval and chosen based on a minimal corrected p-value as proposed by Altman and colleagues [38] and based on maximum Harrell's C indices [39]. Kaplan Meier estimator of survival was used to visualize the survival curves. Hazard ratio and the logrank test were used to compare overall survival (OS), distant metastasis-free survival (DMFS) and disease free survival (DFS) between patients in different groups. All analyses were performed using the statistical software environment R (package survival).
2.3. Plasmid constructs
The constructs used for the cell invasion studies that expressed either wild-type Trx1 (pTrx-wt) or the 1SS redox inactive mutated form of Trx1 (pTrx-1SS) from the pcDNA3.1 vector were described previously [40]. To generate the antisense construct the wild-type Trx1 coding sequences were inserted into the pcDNA3.1 vector in the opposite direction with respect to the CMV promoter (pTrx-as). The constructs used for the cell migration work were made by excising the wild type Trx1 coding sequences or the 1SS redox inactive mutant sequences from the pTrx-wt and pTrx-1SS constructs and inserting into pIRES-EGFP (Clontech, CA, USA) to generate pGFP-Trx and pGFP-1SS.
2.4. Production and analysis of stable cell lines
For the cancer cell invasion studies MDA-MB-231 breast cancer cells were transfected with pTrx-wt, pTrx-1SS, pTrx-as and pcDNA3.1 using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Cells were grown for 3 weeks in 1000 μg/ml geneticin (Sigma). At least three clones from each transfection were randomly selected for further investigation and the data presented for the invasion and migration studies using the stable cell lines is combined from individual analysis of each clone. Confirmation of the construct was obtained by PCR of genomic DNA of each clone and subsequent sequencing of the amplified product. Specific expression from each construct in each clone was confirmed by quantitative RT-PCR using primers specific for the construct. For cell migration studies MDA-MB-231 breast cancer cells were transfected with pIRES-GFP, pGFP-Trx and pGFP-1SS as described above except that fluorescence was also used to confirm expression of the constructs.
2.5. Cell invasion assay
Assays were performed based on the Boyden chamber invasion method described previously [41]. Essentially, a 13 mm diameter polycarbonate filter with 12 μm pores (Neuro Probe, MD, USA) was coated with a 1:30 dilution of Matrigel (BD Biosciences, NSW, Australia) (17 μg/50 μL) and placed over the lower well of the Boyden chamber that had been filled with RPMI containing 10% FBS. The upper compartment was then created by screwing in the retainer with a rubber washer placed above the Matrigel-coated membrane. Then 2.5×105 MDA-MB-231 cells were added to the upper compartment. Cells were allowed to invade for 9 h, and then the cells on the lower surface of the filter were stained using the Quick-Dip dye system (Histolabs, NSW, Australia). The filter was mounted for microscopy, and the cells present in five low-power fields (100×total magnification) on each filter were counted. The experiments were done in triplicate and repeated 3 times. When adding reduced recombinant human Trx1, samples were treated with 0.5 mM DTT for 15 min at 37 °C prior to addition to the bottom well of the chamber.
2.6. Cell proliferation assays
The MDA-MB-231 cells were seeded in triplicate in sterile 96-well plates at a density of 3×104 cells/well and cultured in a CO2 incubator for 24 h before the medium was removed and replaced with 0.1% serum containing media. Plates were then incubated for a further 24 h, after which the media was removed and replaced with 200 µl of media containing auranofin. Cell growth was measured 24 h after auranofin addition. 5 mg/ml MTT (ICN Biochemicals), prepared using sterile filtered 1×PBS, was added to each well and incubated for 3 h. 50 µl of 20% (w/v) SDS in 0.01 M HCl was then added to each well. The plates were incubated overnight and then read on a Spectramax M3 Plate Reader (Molecular Devices, Vic, Australia) at a wavelength of 570 nm. All assays were measured in triplicate from experiments performed 3 times.
2.7. TrxR activity assays
The activity assays were based on previously described procedures [42]. Cells were lysed in 200 µl NP-40 extraction buffer (150 mM NaCl; 50 mM Tris-Cl, pH 8; 0.5% Nonidet P-40; 0.5 mM EDTA; 2 mM PMSF; 1 µl/ml Protease Inhibitor Cocktail VI; 1× PBS). Samples were spun at 14,000 rpm for 10 minutes and the supernatant retained for activity analysis. The TrxR activity was measured in 0.5 M potassium phosphate, 200 mM EDTA, 20 mM NADPH, with 125 mM DTNB. TNB production was measured by following the increase in absorbance at 412 nm for 10 minutes. The DC protein assay kit (BioRad, NSW, Australia) was used to determine the concentration of proteins extracted from cells according to manufacturer's instructions. Units of TrxR activity (µmoles TNB produced/min) were calculated using an extinction coefficient of 13.6×103 M−1 for TNB at 412 nm. The specific activity of thioredoxin reductase was determined using the following equation: Specific activity (U/mg)=U/total protein.
2.8. Monolayer scratch assays
This method was adapted from a previously described protocol [43]. Before seeding out the cells, a scalpel was used to etch horizontal and vertical lines, resulting in a cross at the center of each well, on the underside of a 24-well plate. These lines were used as reference points. Cells were seeded out and incubated at 37 °C until they were 85-90% confluent. The cells were then scratched using a P1000 pipette tip to create a scratch passing through the reference mark. The media and displaced cells were then removed and replaced with 10 mM thymidine containing media to act as an anti-proliferative agent. Photos were taken of the scratches, including the reference points, at 0, 24, and 48 h to monitor closure of the scratch. At the start of each experiment, the distance migrated by cells for each scratch-closure was designated as 0. The distance migrated was calculated for 24 and 48 h based on the reduction of the scratch width with respect to the 0 hour time point. When recombinant Trx1 or the 1SS redox inactive mutated form of human Trx1 was added to migration assays the samples were first reduced with DTT such that the wells contained a final concentration of 0.626 μM DTT. When applicable, the appropriate concentration of auranofin was added along with thymidine-containing media.
2.9. ROS assays
Cells (1.5×106) were grown in 6-well plates, after which 5 μM H2DCF-DA (2′,7′–dichlorodihydrofluorescein diacetate) (Molecular Probes, Vic, Australia) was added to the cells and incubated for 30 mins at 37°C. Cells were then lysed in 200 µL of ice-cold NP-40 lysis buffer (150 mM NaCl; 50 mM Tris-Cl, pH 8; 0.5% Nonidet P-40; 0.5 mM EDTA; 2 mM PMSF; 1 µl/ml Protease Inhibitor Cocktail VI; 1× PBS). The fluorescence was measured using 495 nm excitation and 515 nm emission wavelengths with a 495 nm cut-off on the SpectraMax M3 plate reader (Molecular Devices). The relative fluorescence units (RFUs) were calculated per mg of the total cellular protein (quantified using the DC protein assay kit from BioRad according to manufacturer’s instructions). Cells were also treated with the appropriate auranofin concentration for 24 h prior to incubation with H2DCF-DA.
2.10. Clonogenic assays
Cells (2×106) were grown in 25 cm2 flasks and treated with appropriate concentrations of auranofin for 24 hours. Cells were then harvested and 1000 cells were seeded into 35 mm petri dishes. Cells were then allowed to form colonies for 5 days at the end of which, cells were washed with 1× PBS and fixed with ice-cold methanol. Crystal violet solution (0.5% (w/v) crystal violet and 25% (v/v) methanol) was then used to stain the colonies. The colonies were washed with mqH2O three times and then counted.
2.11. Statistical analysis
Experimental data were analyzed by using the software GraphPad Prism 6 (GraphPad Software, CA, USA). Values are presented as mean±SEM. Statistical significance was determined by the statistical tests specified in the. p<0.05 was considered significant.
3. Results
3.1. Over-expression of the Trx system in breast cancer patients
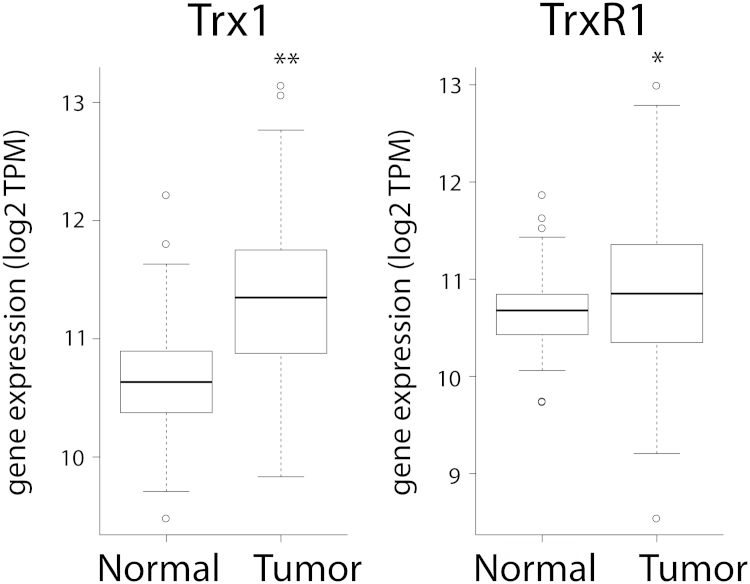
We first investigated the expression of the Trx system in patient breast cancer samples. Using RNAseq data obtained from The Cancer Genome Atlas (TCGA) portal we found that the expression of both Trx1 and TrxR1 are statistically significantly increased in breast cancer patient cells compared with paired normal breast tissue from the same patients (Fig. 1).
Fig. 1.
Expression of Trx1 and TrxR1 in patient breast cancer samples. The normalized gene expression levels (transcripts per million (TPM)) encoding Trx1 and TrxR1 were analyzed according to the RNASeqV2 protocol using data obtained from the TCGA portal. Analysis was restricted to patients with matched tumor and normal samples (n=114). Both Trx1 and TrxR1 expression was shown to be statistically significantly different in tumor samples compared to normal tissue, using a two-sample paired t test, *p<0.001, **p<0.0001.
3.2. Thioredoxin in breast cancer cell invasion
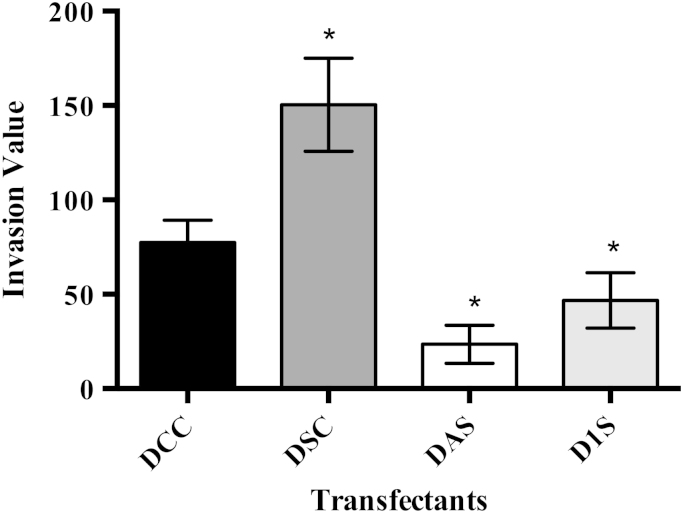
Using an in vitro cell culture system we investigated the role that Trx1 may play in cancer cell invasion. Stable transfectants were made of MDA-MB-231 breast cancer cells using constructs that over-expressed wild type Trx1, a mutated form of Trx1 with both active site cysteines converted to serines (Trx-1SS), an antisense Trx1 mRNA (Trx-as) and the vector (pcDNA3.1) as a control. At least 4 clones for each transfection were selected and tested to ensure they were expressing the correct construct. Transfectants over-expressing wild type Trx1 showed a significant increase in invasion compared to the control clones, while transfectants expressing either the redox inactive form of Trx1 (Trx-1SS) or the antisense form of Trx1 (Trx-as) showed a statistically significant decrease in invasion compared to control cells (Fig. 2). These results confirm the role of Trx1 in the invasion of breast cancer cells [28]. There was no significant difference between clones transfected with the antisense Trx1 construct or the redox inactive Trx-1SS construct, suggesting each strategy was effective at disrupting Trx1 function.
Fig. 2.
Invasion of MDA-MB-231 transfectants in the Boyden chamber invasion assay. DCC – Clones transfected with pcDNA3.1 vector only; DSC – Sense Trx1 clones; DAS – Antisense Trx1 clones; D1S – 1SS redox inactive Trx1 clones (n=3). Results from three independent experiments performed in triplicate are presented as mean±SEM. A one-way ANOVA followed by Tukey's post test was employed and a significant difference in invasion values compared to the control cells is indicated by *p<0.05.
3.3. Exogenous Trx1 and breast cancer cell invasion
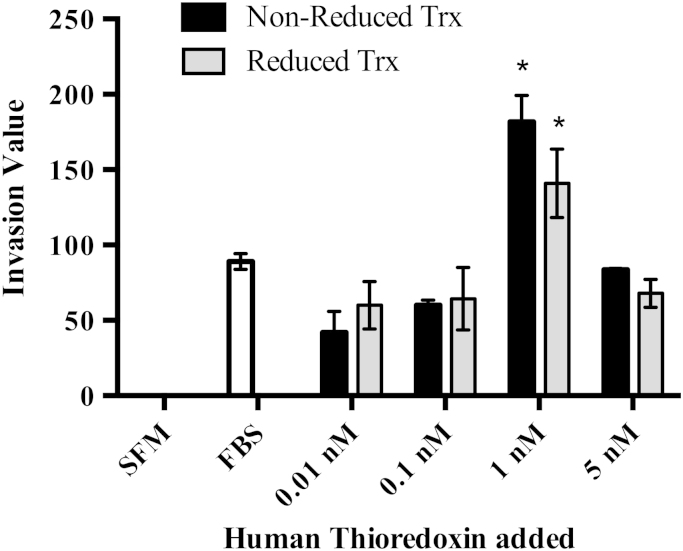
Since Trx1 is secreted from cancer cells [19], [30], [31], [44], we also assessed if addition of Trx1 into the extracellular environment had an effect on the invasion of MDA-MB-231 cells. Addition of either reduced or non-reduced Trx1 to the bottom well of the Boyden chamber was shown to increase cell invasion of MDA-MB-231 cells in a bell-shaped dose response (Fig. 3), similar to that observed in studies showing Trx1 is chemotactic for neutrophils, monocytes and T-cells [45]. The bell-shaped curve is characteristic of the response exhibited by other chemokines [45]. There was no statistical difference on cell invasion when comparing the addition of reduced or non-reduced Trx1 at the same concentration.
Fig. 3.
Effect of exogenously added recombinant human Trx1 on MDA-MB-231 cell invasion. Trx1 concentration is as indicated. SFM is serum free media (negative control) and FBS (fetal bovine serum) in the bottom well was used as a positive control. Results from three independent experiments performed in triplicate are presented as mean±SEM. A two-way ANOVA followed by Tukey’s post test was employed and a significant difference in invasion values compared to the FBS treated control cells and to other concentrations of Trx1 is indicated by *p<0.05.
3.4. Targeting the redox site of extracellular Trx1
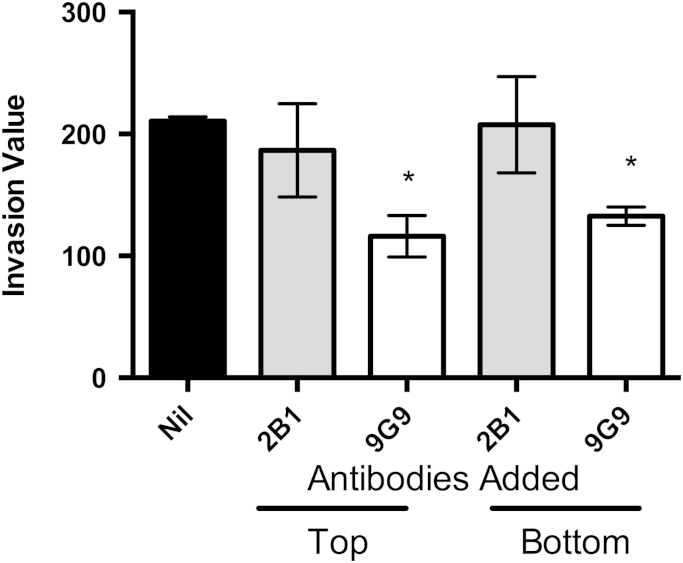
Two specific anti-human Trx1 monoclonal antibodies were used to determine if the redox function of Trx1 is required to stimulate cell invasion. The monoclonal antibody 2B1 specifically recognizes Trx1, but does not neutralize the active site, while the antibody 9G9, binds to the active site region and inhibits the redox activity of Trx1 [32]. These antibodies were added to either the bottom or top well of the chamber and the invasion of the MDA-MB-231 cells was assessed. The addition of the 2B1 monoclonal antibody to either the top or bottom well did not result in any statistically significant change in cell invasion compared to cells with no added antibody (Fig. 4). In contrast, the addition of the 9G9 monoclonal antibody to either the top or bottom well significantly decreased the level of invasion. These results indicate that the redox activity of Trx1 is important for its role in invasion.
Fig. 4.
Effect of adding anti-Trx1 human monoclonal antibodies on the invasion of MDA-MB-231 cells. Antibodies were added to either the top or bottom well of the Boyden chamber. Results are from three independent experiments performed in triplicate presented as mean±SEM. The invasion value is the cells/field in the test assay less the cells/field in the negative control. A one-way ANOVA followed by Tukey's post test was employed and a significant difference in invasion values compared to the control cells (Nil antibody added) is indicated by *p<0.05.
3.5. Effect of thioredoxin on migration
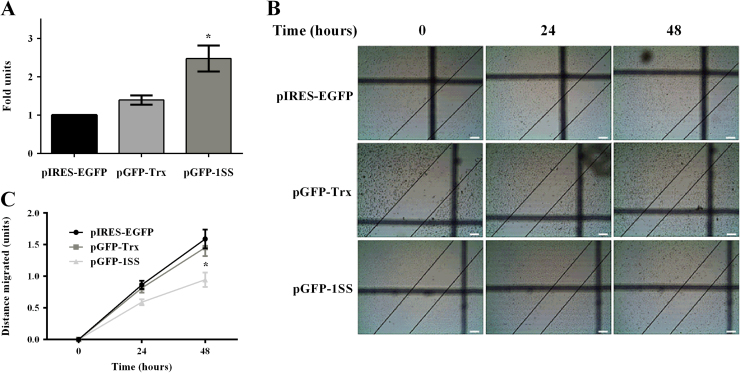
We also investigated the effect of Trx1 on cancer cell migration through the use of in vitro monolayer scratch assays [43]. Stable transfectants of the MDA-MB-231 cell line over-expressing either wild type Trx1 or the 1SS redox inactive mutated form of Trx1 were used in these experiments. The wild type and 1SS constructs, as used previously, were cloned into a GFP-expressing vector (pIRES-EGFP) so that fluorescence could be monitored to confirm ongoing expression from the construct. Stable transfectants of the MDA-MB-231 cells were made using the pIRES-EGFP, pGFP-Trx and pGFP-1SS constructs. At least 3 clones for each plasmid were selected and tested to ensure they contained the correct construct. In addition ROS levels were also determined in the 3 groups of stable transfected cell lines to further characterize the intracellular redox state of the cells. Cells expressing the 1SS construct had significantly higher ROS levels than the cells expressing either wild type Trx1 or the empty vector (Fig. 5A).
Fig. 5.
Effect of Trx1 on the migration of MDA-MB-231 cells. (A) ROS levels in stable transfected MDA-MB-231 cells analyzed using H2DCF-DA and represented as a fold change over pIRES-EGFP transfected cells. A one-way ANOVA followed by Sidak’s post test was employed and showed that cells over-expressing the 1SS redox inactive Trx1 protein exhibited significantly higher levels of ROS compared to the control cells, *p<0.05. Results are presented as mean±SEM of three independent experiments performed in triplicate. (B) Representative images of the transfected cells used in the monolayer scratch assays over a period of 48 h. The images were taken at 4× using a Tucsen TCA 3MP camera and the scale bar is 50 µm. (C) Migration of stable transfected cells in response to over-expression of wild-type Trx1 and the 1SS redox inactive Trx1 protein. A repeated-measures two-way ANOVA followed by Sidak’s post test was employed and showed a statistical difference at 48 hour time point between 1SS over-expressing cells and control cells, *p<0.0001. Results are presented as mean±SEM of at least three independent experiments performed in quadruplicate.
The stable transfectants were then tested in monolayer scratch assays. The representative images of the scratches monitored over a period of 48 h are shown in Fig. 5B. As shown in Fig. 5B and C, Trx1 over-expression (pGFP-Trx) did not cause any increase in the migration of cells as compared to the control cells transfected with the vector only (pIRES-EGFP). However, the cells over-expressing the 1SS redox inactive form of Trx1 (pGFP-1SS) migrated slower than the vector-only controls, with a significant difference at the 48 hour time point when analyzed by a repeated measures two-way ANOVA.
3.6. Effect of extracellular Trx1 on migration
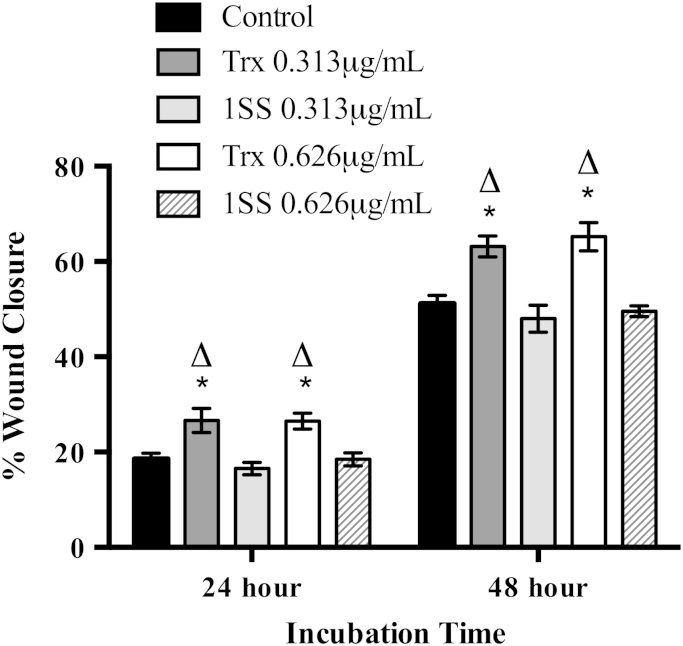
The effect of extracellular Trx1 on the migration of MDA-MB-231 breast cancer cells was tested. Wild type recombinant human Trx1 and the 1SS redox inactive mutated form of human Trx1 were reduced with DTT and then incubated with MDA-MB-231 cells during monolayer scratch assays. Untreated cells were used as the control. At both the 24 h and 48 h time points Trx1 exhibited a significant effect on cell migration compared to both the control cells and to the 1SS treated cells (Fig. 6). Control cells and 1SS treated cells showed no significant difference in migration. Other controls were also conducted, using DTT at the same concentration as used to reduce Trx1, and recombinant Trx1 that had been boiled, to ensure no other non-protein factors in the sample were stimulating the migration. No change in cell migration was observed in either case (data not shown).
Fig. 6.
Differential effect of wild-type Trx1 and the 1SS redox inactive Trx1 form on MDA-MB-231 breast cancer cell migration. Two different concentrations of wild-type Trx1 or the 1SS redox inactive Trx1 form were used to treat MDA-MB-231 cells. Untreated cells were used as the control. Scratch closure was expressed as the percentage scratch closure calculated at the 24 or 48 h time points using 0 hour measurements as 0%. A two-way ANOVA employing Tukey's post test showed a significant difference between untreated cells and both the 0.313 μg/mL and 0.626 μg/mL Trx1 treated cells at 24 and 48 h incubation time, *p<0.05. There was also a difference between the Trx1 and 1SS redox inactive Trx1 treated cells at 24 and 48 h incubation time, ∆p<0.05. Trx: Wild-type Trx1, 1SS: 1SS redox inactive Trx1 form. Results are presented as mean±SEM from three independent experiments conducted in quadruplicate.
3.7. Inhibition of the thioredoxin system by auranofin and effects on cell migration
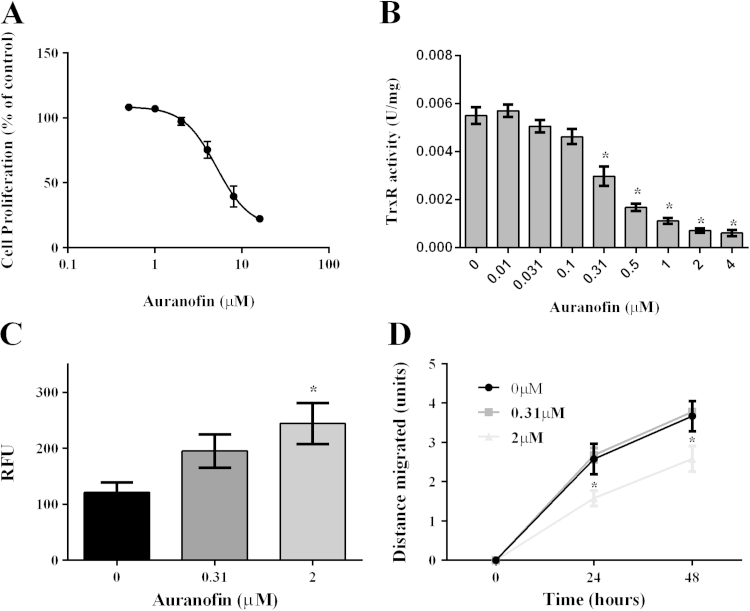
Since Trx1 had been implicated in cancer cell migration we used auranofin, a potent anti-cancer drug, as a tool to inhibit the Trx1 system. Auranofin is a gold-based drug that irreversibly inhibits the activity of TrxR, resulting in a shift in the cellular redox balance towards a less reduced state [46]. We first assessed the effect of a range of auranofin concentrations on MDA-MB-231 cell proliferation (Fig. 7A) and on TrxR activity (Fig. 7B). The IC50 for cell proliferation was determined to be 5.1 µM and for TrxR activity was 0.31 µM. For subsequent experiments we used two different concentrations of auranofin (0.31 µM and 2 µM), at which the TrxR activity was inhibited without an effect on cell proliferation. An increase in ROS levels was observed for auranofin treated cells compared to the untreated controls with a statistically significant difference observed when 2 µM auranofin was used (Fig. 7C).
Fig. 7.
Effect of auranofin induced inhibition of the TrxR system. (A) Proliferation of MDA-MB-231 cells in response to auranofin treatment. The IC50 for auranofin-induced inhibition of MDA-MB-231 cell proliferation was determined to be 5.1 µM. At least three independent experiments were performed in triplicate. (B) TrxR1 specific activity in MDA-MB-231 cells upon auranofin treatment. Auranofin significantly inhibited the TrxR1 activity at 0.31 µM and higher concentrations, *p<0.0001. A one-way ANOVA followed by Sidak's post test was employed and results are presented as mean±SEM of at least three independent experiments performed in triplicate. (C) ROS levels in auranofin treated MDA-MB-231 cells analyzed using H2DCF-DA. A one-way ANOVA followed by Sidak’s post test was employed. Cells treated with 2 µM auranofin had significantly higher levels of ROS compared to the control cells, *p<0.05. (D) Migration of MDA-MB-231 cells in response to auranofin treatment. Cells treated with 2µM auranofin showed a significant decrease in the distance migrated as compared to the control cells, at both 24 and 48 h time points, *p<0.0001. Data was analyzed using a repeated measures two-way ANOVA followed by Sidak’s post test and results are presented as mean±SEM of three independent experiments.
Next we examined the effect of auranofin on the migration of MDA-MB-231 cells using monolayer scratch assays. As shown in Fig. 7D, at the higher concentration of 2 µM, auranofin significantly inhibited the migration of the cells at both 24 and 48 h time points compared to the untreated controls when analyzed by a repeated measures two-way ANOVA. However, the lower concentration of 0.31 µM auranofin did not affect cell migration.
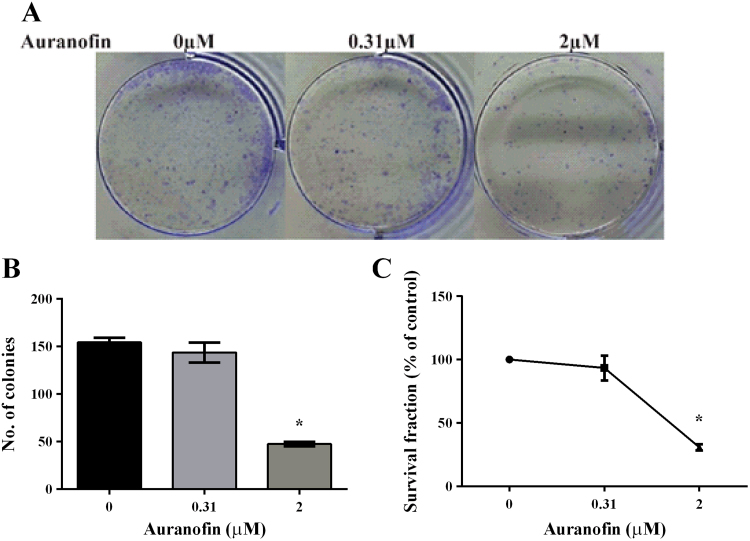
3.8. Auranofin and clonogenic capacity of MDA-MB-231 cells
We also examined the effect of auranofin treatment on the clonogenic activity of MDA-MB-231 cells. Cells were treated with 0.31 µM and 2 µM auranofin for 24 h and then seeded out for colony formation over a period of 5 days. The representative images are shown in Fig. 8A. As compared to the untreated cells, no difference was observed in the number of colonies obtained with cells treated with 0.31 µM auranofin. However, treatment with 2 µM auranofin significantly lowered the clonogenic capacity of MDA-MB-231 cells with around 30% survival (Fig. 8B and C).
Fig. 8.
Effect of auranofin pre-treatment on the clonogenic capacity of MDA-MB-231 cells. (A) Representative images taken after 5 days of colony formation. (B) Number of colonies obtained after 5 days. Cells that were pre-treated with 2 µM auranofin had significantly less number of colonies compared to the untreated cells, *p<0.05. A one-way ANOVA followed by Sidak's post test was employed and results are presented as mean±SEM of two independent experiments performed in duplicate. (C) Survival data is presented as percentage of the untreated control cells.
3.9. Thioredoxin system in the survival of breast cancer patients
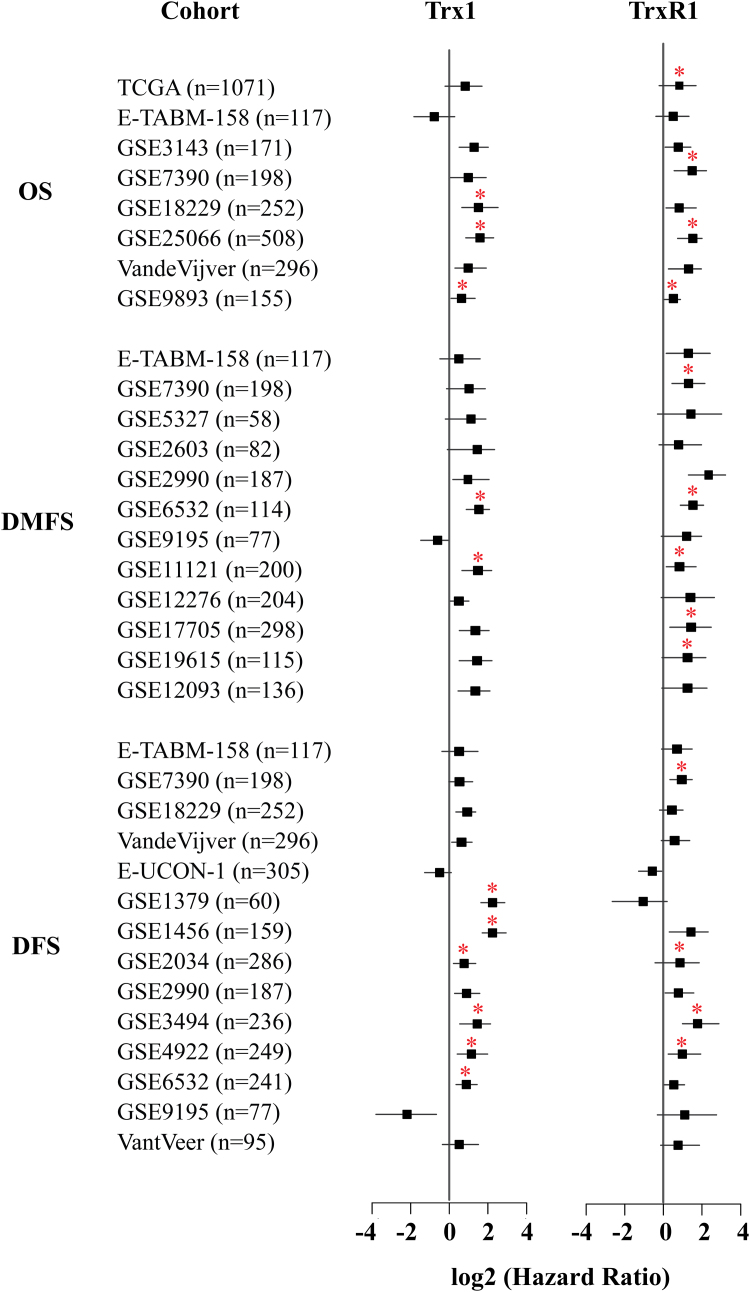
We used a meta-analytical approach to investigate the evidence linking the prognostic markers (Trx1 and TrxR1) with survival of breast cancer patients. We analyzed 25 independent cohorts (the TCGA cohort and 24 microarray cohorts) containing a total of 5910 patient samples. We prepared a forest plot (Fig. 9) to identify any association between the Trx1 system and the overall survival (OS) of patients, distant-metastasis free survival (DMFS) and disease free survival (DFS) in each of these independent cohorts. Overall survival refers to the time interval until breast cancer associated death whereas disease free survival refers to time before recurrence of breast cancers. DMFS refers to the time before any distant metastases are detected. In general, high levels of Trx1 and TrxR1 are associated with poor outcomes. TrxR1 is consistently associated with poor prognosis in all cohorts for OS and DMFS, with the hazard ratio (HR)>1 (Fig. 9). Trx1 is also associated with poor patient outcomes in many of the cohorts.
Fig. 9.
Breast cancer patient survival analysis. Survival analysis for Trx1 and TrxR1 using RNA sequencing data (TCGA cohort) as well as microarray data from 25 independent cohorts, as indicated in the figure with associated number of patients. OS, overall survival; DMFS, distant metastasis-free survival; DFS, disease free survival. A forest plot with the corresponding hazard ratios is shown on the right with the associated confidence intervals indicated. Asterisks mark the results that are statistically significant. A hazard ratio (HR) >1 is indicative of a poor outcome.
4. Discussion
Metastasis is the most life threatening aspect of breast cancer. It is the process by which tumor cells detach from their primary tissue, migrate and establish secondary tumors [47], which can be far from the original tumor site and thus difficult to detect. The first stage of metastasis is the detachment of cells from the primary tumor and attachment to the extracellular matrix (ECM), which requires chemotactic migration of cancer cells [48], [49]. Subsequently there is degradation of the ECM and entry of the cells into the bloodstream [48]. The next step is the extravasation of the cells from the circulation, degradation of the ECM of the secondary tissue and establishment of a tumor colony with subsequent proliferation [48], [50]. In this paper we have shown that the Trx1 system plays a role in cancer cell invasion, migration and colony formation.
The Trx1 system is highly expressed in breast tumors [8], [51] and Trx1 has previously been suggested as a suitable serum marker and diagnostic marker for breast cancer [35]. Our paper extends these previous studies by using data obtained from a RNAseq database to show that both Trx1 and TrxR1 are expressed at statistically significant higher levels in breast tumor tissue compared to the paired normal tissue from the same patient (Fig. 1). By limiting our analysis to the 114 patient samples that have paired data, a stronger association can be made between the higher expression of both Trx1 and TrxR1 in tumor tissue compared to normal tissue. This data suggests a role for the Trx1 system in breast cancer carcinogenesis. Since high expression levels of Trx1 protein had been previously correlated with the most aggressive and metastatic breast tumors [8] this current study was focused on refining the role of Trx1 in cancer cell invasion and migration.
Intracellular Trx1 had previously been shown to have a role in breast cancer cell invasion using in vitro invasion assays [28]. Our data confirms the role of intracellular Trx1 in invasion since decreasing the levels of Trx1 in MDA-MB-231 breast cancer cells using either an antisense approach or through expressing a redox inactive Trx1 protein (1SS) resulted in decreased cell invasion. This current study also extended previous results by showing that there is a role for extracellular Trx1 in breast cancer cell invasion. Addition of wild type recombinant human Trx1 to the bottom well of the invasion chamber resulted in increased cell invasion in a bell-shaped dose response. This shows that Trx1 enhances the motility of breast cancer cells.
Our results also suggest that the redox active site of Trx1 is important for its extracellular role in cancer cell invasion. A specific monoclonal antibody (9G9) that binds to Trx1, and inhibits its redox function, resulted in significantly decreased cell invasion of MDA-MB-231 cells. A specific monoclonal antibody (2B1) that binds to Trx1, but does not interfere with its redox function did not have any effect on cell invasion. The 9G9 antibody was able to inhibit cell invasion when added to either the top or the bottom well of the chamber, with no exogenous Trx1 added (Fig. 4). When added to the bottom chamber it is likely that the antibody is inhibiting the action of secreted Trx1, thereby reducing breast cancer cell motility. When added to the top well the antibody may be acting on either secreted Trx1 or on any cell surface bound Trx1. Extracellular Trx1 has also been shown to destabilize the basement membrane by reducing laminin and affecting its interaction with galectin-3 in Matrigel based assays, thereby promoting endothelial cell proliferation and spreading [52]. This effect was found to be dependent on the Trx1 redox activity and was not observed when the 1SS redox inactive Trx1 mutant was added [52]. In the extracellular environment Trx1 was also shown to tip the balance towards MMP function [29]. This suggests that extracellular Trx1 could be a therapeutic target to reduce the invasion and therefore metastasis of breast cancer cells.
Cell migration is an important part of the metastatic process and since Trx1 was shown to have a role in invasion, we therefore investigated whether Trx1 could be implicated in the migration of breast cancer cells. A low dose of a Trx1 inhibitor, PX-12, resulted in decreased cell migration of colorectal cancer cells [53], raising the possibility that Trx1 may have a role in regulating cancer cell migration. Whether this role involves either intracellular or extracellular Trx1 in cancer cell migration was not established in the previously published study [53]. However, PX-12 is not a suitable drug for testing against MDA-MB-231 cells [54]. We therefore modulated the levels of intracellular Trx1 through the transfection of constructs that over-expressed wild type Trx1 and also the redox inactive 1SS. Over-expression of intracellular Trx1 had no statistically significant effect on cell migration, but intracellular expression of 1SS resulted in statistically significantly decreased cell migration. It is possible that the endogenous Trx1 levels were sufficient to stimulate the migration of cells and therefore the Trx1 protein over-expressed from the transgene did not exhibit any additional effect on the cell migration. This may be the reason why the distance migrated by pGFP-Trx cells as well as the ROS levels in these cells were similar to those observed for the pIRES-EGFP control cells. Furthermore, 1SS over-expression may be blocking the endogenous Trx1 by acting as a competitive inhibitor for TrxR1 in the intracellular environment. Therefore, a decrease in cell migration was observed compared to cells transfected with only the vector.
Extracellular Trx1 levels were modified by adding either recombinant human wild type Trx1 or 1SS. Addition of wild type Trx1 enhanced migration while addition of the equivalent concentration of 1SS had no effect on migration of cells. This result correlates with the chemotactic properties exhibited by Trx1, but not by 1SS, towards monocytes and polymorphonuclear neutrophils [55]. Exogenously added 1SS has previously been shown to be incapable of binding to the cell surface of adult T-cell leukemia (ATL) cells, whereas wild type Trx1 was able to bind, suggesting that a disulfide bond is involved in cell surface binding [56]. Therefore, exogenously added 1SS may have failed to enhance cell migration because it lacks the ability to interact with Trx1 cell surface targets. Overall these data suggest that the redox site of Trx1 is required for both its intracellular and extracellular functions with respect to cell migration.
Auranofin is a gold-containing compound known to inhibit TrxR activity [46] and has previously been shown to inhibit proliferation of cancer cell lines, including the MDA-MB-231 cell line [57]. Currently auranofin is being tested in phase I/II clinical trials for the treatment of chronic lymphocytic leukemia, small lymphocytic leukemia and prolymphocytic lymphoma [58]. Auranofin was reported to decrease VEGF mRNA levels of human umbilical vein endothelial cells (HUVEC) and decreased endothelial cell migration in monolayer scratch assays. In a separate study, auranofin was shown to down-regulate the VEGFR3-mediated migration of endothelial cells [59]. We therefore tested auranofin on the migration of breast cancer cells. Addition of auranofin was shown to inhibit the migration of MDA-MB-231 cells, compared to untreated cells, at the 2 μM concentration, at which TrxR activity was inhibited without an effect on cell proliferation. This result suggests that inhibiting TrxR activity is a possible therapeutic strategy for targeting the cell migration component of the cancer metastatic process.
Clonogenic assays were also performed to assess if pre-treatment with auranofin has any effect on the ability of breast cancer cells to form secondary colonies in a drug-free environment. Pre-treatment with auranofin resulted in an inhibition of colony formation at the higher concentration (2 μM), which correlated with the concentration of auranofin that inhibited cell migration. Therefore, auranofin can target two components of the metastatic pathway, cell migration and colonization.
We showed that high levels of Trx1 and TrxR1 expression are correlated with poor patient outcomes in terms of survival and metastases formation. A previous study showed that TrxR1 and thioredoxin interacting protein (TXNIP) are associated with prognosis in breast cancer [60]. High levels of TrxR1 were associated with worse metastasis free survival, disease free survival and overall survival. In contrast high levels of TXNIP were associated with better metastasis-free survival [60]. Our results were based on the analysis of data from a much larger sample size comprising 25 independent cohorts containing a total of 5910 patient samples, including 1071 RNA seq samples from the TCGA database. We found similar results to the previous study in that high levels of TrxR1 are associated with a poor patient prognosis. TrxR1 is strongly associated with poor outcomes in terms of overall survival, disease free survival and distant metastasis free survival in all of the independent cohorts. We also assessed the link between Trx and breast cancer prognosis. Our results support the previous smaller study in showing that Trx was associated with worse metastasis free survival [60], although not all independent cohorts were statistically significant in our study. In addition to the metastasis free survival reported in the previous study, we also assessed disease free survival and overall survival, with high levels of Trx again associated with a poor prognosis. However statistical significance associating Trx 1 with a poor prognosis was obtained for a lower number of cohorts than that observed for TrxR1, indicating that TrxR1 may be a better prognostic indicator.
The association of high levels of Trx1 and TrxR1 expression with both survival and formation of distant metastases correlates with its roles in in vitro cancer cell invasion and migration as shown in this current study. Therefore, targeting the function of TrxR by drugs such as auranofin, or using other Trx system inhibitors, in combination with other therapeutic agents may offer an opportunity to reduce metastases and enhance survival for breast cancer patients.
Acknowledgments
The authors wish to thank Cancer Council Queensland for their financial support. We also wish to thank Griffith University for supporting this research through scholarships and project support to Maneet Bhatia, Kelly McGrath and Fenil Shah. The results shown here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov).
References
- 1.Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 2.Kumar B., Koul S., Khandrika L., Meacham R.B., Koul H.K. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–1785. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- 3.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues P., de Marco G., Furriol J., Mansego M.L., Pineda-Alonso M., Gonzalez-Neira A., Martin-Escudero J.C., Benitez J., Lluch A., Chaves F.J., Eroles P. Oxidative stress in susceptibility to breast cancer: study in Spanish population. BMC cancer. 2014;14:861. doi: 10.1186/1471-2407-14-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic. Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris I.S., Treloar A.E., Inoue S., Sasaki M., Gorrini C., Lee K.C., Yung K.Y., Brenner D., Knobbe-Thomsen C.B., Cox M.A., Elia A., Berger T., Cescon D.W., Adeoye A., Brustle A., Molyneux S.D., Mason J.M., Li W.Y., Yamamoto K., Wakeham A., Berman H.K., Khokha R., Done S.J., Kavanagh T.J., Lam C.W., Mak T.W. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Holmgren A. Thioredoxin. Annu. Rev. Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 8.Lincoln D.T., Ali Emadi E.M., Tonissen K.F., Clarke F.M. The thioredoxin–thioredoxin reductase system: over-expression in human cancer. Anticancer Res. 2003;23:2425–2433. [PubMed] [Google Scholar]
- 9.Arrigo A.P. Gene expression and the thiol redox state. Free Radic. Biol. Med. 1999;27:936–944. doi: 10.1016/s0891-5849(99)00175-6. [DOI] [PubMed] [Google Scholar]
- 10.Hawkes H.J., Karlenius T.C., Tonissen K.F. Regulation of the human thioredoxin gene promoter and its key substrates: a study of functional and putative regulatory elements. Biochim. Biophys. Acta. 2014;1840:303–314. doi: 10.1016/j.bbagen.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Rhee S.G., Chae H.Z., Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Lowther W.T., Brot N., Weissbach H., Honek J.F., Matthews B.W. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6463–6468. doi: 10.1073/pnas.97.12.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das K.C., Das C.K. Thioredoxin, a singlet oxygen quencher and hydroxyl radical scavenger: redox independent functions. Biochem. Biophys. Res. Commun. 2000;277:443–447. doi: 10.1006/bbrc.2000.3689. [DOI] [PubMed] [Google Scholar]
- 14.Ueda S., Masutani H., Nakamura H., Tanaka T., Ueno M., Yodoi J. Redox control of cell death. Antioxid. Redox Signal. 2002;4:405–414. doi: 10.1089/15230860260196209. [DOI] [PubMed] [Google Scholar]
- 15.Kim S.J., Miyoshi Y., Taguchi T., Tamaki Y., Nakamura H., Yodoi J., Kato K., Noguchi S. High thioredoxin expression is associated with resistance to docetaxel in primary breast cancer. Clin. Cancer Res. 2005;11:8425–8430. doi: 10.1158/1078-0432.CCR-05-0449. [DOI] [PubMed] [Google Scholar]
- 16.Ravi D., Muniyappa H., Das K.C. Endogenous thioredoxin is required for redox cycling of anthracyclines and p53-dependent apoptosis in cancer cells. J. Biol. Chem. 2005;280:40084–40096. doi: 10.1074/jbc.M507192200. [DOI] [PubMed] [Google Scholar]
- 17.Berggren M., Gallegos A., Gasdaska J.R., Gasdaska P.Y., Warneke J., Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996;16:3459–3466. [PubMed] [Google Scholar]
- 18.Raninga P.V., Di Trapani G., Vuckovic S., Bhatia M., Tonissen K.F. Inhibition of thioredoxin 1 leads to apoptosis in drug-resistant multiple myeloma. Oncotarget. 2015;6:15410–15424. doi: 10.18632/oncotarget.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubartelli A., Bajetto A., Allavena G., Wollman E., Sitia R. Secretion of thioredoxin by normal and neoplastic cells through a leaderless secretory pathway. J. Biol. Chem. 1992;267:24161–24164. [PubMed] [Google Scholar]
- 20.Soderberg A., Sahaf B., Rosen A. Thioredoxin reductase, a redox-active selenoprotein, is secreted by normal and neoplastic cells: presence in human plasma. Cancer Res. 2000;60:2281–2289. [PubMed] [Google Scholar]
- 21.Grogan T.M., Fenoglio-Prieser C., Zeheb R., Bellamy W., Frutiger Y., Vela E., Stemmerman G., Macdonald J., Richter L., Gallegos A., Powis G. Thioredoxin, a putative oncogene product, is overexpressed in gastric carcinoma and associated with increased proliferation and increased cell survival. Hum. Pathol. 2000;31:475–481. doi: 10.1053/hp.2000.6546. [DOI] [PubMed] [Google Scholar]
- 22.Kakolyris S., Giatromanolaki A., Koukourakis M., Powis G., Souglakos J., Sivridis E., Georgoulias V., Gatter K.C., Harris A.L. Thioredoxin expression is associated with lymph node status and prognosis in early operable non-small cell lung cancer. Clin. Cancer Res. 2001;7:3087–3091. [PubMed] [Google Scholar]
- 23.Raffel J., Bhattacharyya A.K., Gallegos A., Cui H., Einspahr J.G., Alberts D.S., Powis G. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J. Lab. Clin. Med. 2003;142:46–51. doi: 10.1016/S0022-2143(03)00068-4. [DOI] [PubMed] [Google Scholar]
- 24.Ceccarelli J., Delfino L., Zappia E., Castellani P., Borghi M., Ferrini S., Tosetti F., Rubartelli A. The redox state of the lung cancer microenvironment depends on the levels of thioredoxin expressed by tumor cells and affects tumor progression and response to prooxidants. Int. J. Cancer. 2008;123:1770–1778. doi: 10.1002/ijc.23709. [DOI] [PubMed] [Google Scholar]
- 25.Chaiswing L., Bourdeau-Heller J.M., Zhong W., Oberley T.D. Characterization of redox state of two human prostate carcinoma cell lines with different degrees of aggressiveness. Free Radic. Biol. Med. 2007;43:202–215. doi: 10.1016/j.freeradbiomed.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 26.Gallegos A., Gasdaska J.R., Taylor C.W., Paine-Murrieta G.D., Goodman D., Gasdaska P.Y., Berggren M., Briehl M.M., Powis G. Transfection with human thioredoxin increases cell proliferation and a dominant-negative mutant thioredoxin reverses the transformed phenotype of human breast cancer cells. Cancer Res. 1996;56:5765–5770. [PubMed] [Google Scholar]
- 27.Welsh S.J., Bellamy W.T., Briehl M.M., Powis G. The redox protein thioredoxin-1 (Trx-1) increases hypoxia-inducible factor 1alpha protein expression: Trx-1 overexpression results in increased vascular endothelial growth factor production and enhanced tumor angiogenesis. Cancer Res. 2002;62:5089–5095. [PubMed] [Google Scholar]
- 28.Farina A.R., Cappabianca L., Desantis G., Ianni N.D., Ruggieri P.D., Ragone M., Merolle S., Tonissen K., Gulino A., Mackay A.R. Thioredoxin stimulates MMP-9 expression, de-regulates the MMP-9/TIMP-1 equilibrium and promotes MMP-9 dependent invasion in human MDA-MB-231 breast cancer cells. FEBS Lett. 2011 doi: 10.1016/j.febslet.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Farina A.R., Tacconelli A., Cappabianca L., Masciulli M.P., Holmgren A., Beckett G.J., Gulino A., Mackay A.R. Thioredoxin alters the matrix metalloproteinase/tissue inhibitors of metalloproteinase balance and stimulates human SK-N-SH neuroblastoma cell invasion. Eur. J. Biochem. 2001;268:405–413. doi: 10.1046/j.1432-1033.2001.01892.x. [DOI] [PubMed] [Google Scholar]
- 30.Schenk H., Vogt M., Droge W., Schulze-Osthoff K. Thioredoxin as a potent costimulus of cytokine expression. J. Immunol. 1996;156:765–771. [PubMed] [Google Scholar]
- 31.Sahaf B., Soderberg A., Spyrou G., Barral A.M., Pekkari K., Holmgren A., Rosen A. Thioredoxin expression and localization in human cell lines: detection of full-length and truncated species. Exp. Cell. Res. 1997;236:181–192. doi: 10.1006/excr.1997.3699. [DOI] [PubMed] [Google Scholar]
- 32.Angelini G., Gardella S., Ardy M., Ciriolo M.R., Filomeni G., Di Trapani G., Clarke F., Sitia R., Rubartelli A. Antigen-presenting dendritic cells provide the reducing extracellular microenvironment required for T lymphocyte activation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1491–1496. doi: 10.1073/pnas.022630299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee R. Redox outside the box: linking extracellular redox remodeling with intracellular redox metabolism. J. Biol. Chem. 2012;287:4397–4402. doi: 10.1074/jbc.R111.287995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura H., Masutani H., Yodoi J. Extracellular thioredoxin and thioredoxin-binding protein 2 in control of cancer. Semin. Cancer. Biol. 2006;16:444–451. doi: 10.1016/j.semcancer.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Park B.J., Cha M.K., Kim I.H. Thioredoxin 1 as a serum marker for breast cancer and its use in combination with CEA or CA15-3 for improving the sensitivity of breast cancer diagnoses. BMC Res. Notes. 2014;7:7. doi: 10.1186/1756-0500-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonissen K., Wells J., Cock I., Perkins A., Orozco C., Clarke F. Site-directed mutagenesis of human thioredoxin. Identification of cysteine 74 as critical to its function in the “early pregnancy factor” system. J. Biol. Chem. 1993;268:22485–22489. [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altman D.G., Lausen B., Sauerbrei W., Schumacher M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J. Natl. Cancer Inst. 1994;86:829–835. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- 39.Harrell F.E., Jr., Lee K.L., Mark D.B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Osborne S.A., Hawkes H.J., Baldwin B.L., Alexander K.A., Svingen T., Clarke F.M., Tonissen K.F. The tert-butylhydroquinone-mediated activation of the human thioredoxin gene reveals a novel promoter structure. Biochem. J. 2006;398:269–277. doi: 10.1042/BJ20060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bloomfield K.L., Baldwin B.L., Harkin D.G., Tonissen K.F. Modification of the Boyden chamber to improve uniformity of cell invasion of matrigel-coated membranes. Biotechniques. 2001;31 doi: 10.2144/01316bm02. 1242, 1244, 1246. [DOI] [PubMed] [Google Scholar]
- 42.Smith A.D., Levander O.A. High-throughput 96-well microplate assays for determining specific activities of glutathione peroxidase and thioredoxin reductase. Methods Enzym. 2002;347:113–121. doi: 10.1016/s0076-6879(02)47012-7. [DOI] [PubMed] [Google Scholar]
- 43.Dhanesuan N., Sharp J.A., Blick T., Price J.T., Thompson E.W. Doxycycline-inducible expression of SPARC/Osteonectin/BM40 in MDA-MB-231 human breast cancer cells results in growth inhibition. Breast Cancer Res. Treat. 2002;75:73–85. doi: 10.1023/a:1016536725958. [DOI] [PubMed] [Google Scholar]
- 44.Di Trapani G., Perkins A., Clarke F. Production and secretion of thioredoxin from transformed human trophoblast cells. Mol. Hum. Reprod. 1998;4:369–375. doi: 10.1093/molehr/4.4.369. [DOI] [PubMed] [Google Scholar]
- 45.Bertini R., Howard O.M., Dong H.F., Oppenheim J.J., Bizzarri C., Sergi R., Caselli G., Pagliei S., Romines B., Wilshire J.A., Mengozzi M., Nakamura H., Yodoi J., Pekkari K., Gurunath R., Holmgren A., Herzenberg L.A., Ghezzi P. Thioredoxin, a redox enzyme released in infection and inflammation, is a unique chemoattractant for neutrophils, monocytes, and T cells. J. Exp. Med. 1999;189:1783–1789. doi: 10.1084/jem.189.11.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rigobello M.P., Gandin V., Folda A., Rundlof A.K., Fernandes A.P., Bindoli A., Marzano C., Bjornstedt M. Treatment of human cancer cells with selenite or tellurite in combination with auranofin enhances cell death due to redox shift. Free Radic. Biol. Med. 2009;47:710–721. doi: 10.1016/j.freeradbiomed.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 47.Fidler I.J. Critical factors in the biology of human cancer metastasis: twenty-eighth G.H.A. Clowes memorial award lecture. Cancer Res. 1990;50:6130–6138. [PubMed] [Google Scholar]
- 48.Saiki I. Cell adhesion molecules and cancer metastasis. Jpn. J. Pharmacol. 1997;75:215–242. doi: 10.1254/jjp.75.215. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi H., Wyckoff J., Condeelis J. Cell migration in tumors. Curr. Opin. Cell. Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Lorusso G., Ruegg C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin. Cancer Biol. 2012;22:226–233. doi: 10.1016/j.semcancer.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Cha M.K., Suh K.H., Kim I.H. Overexpression of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J. Exp. Clin. Cancer. Res. 2009;28:93. doi: 10.1186/1756-9966-28-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farina A.R., Tacconelli A., Cappabianca L., DeSantis G., Gulino A., Mackay A.R. Thioredoxin inhibits microvascular endothelial capillary tubule formation. Exp. Cell. Res. 2003;291:474–483. doi: 10.1016/j.yexcr.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Wang F., Lin F., Zhang P., Ni W., Bi L., Wu J., Jiang L. Thioredoxin-1 inhibitor, 1-methylpropyl 2-imidazolyl disulfide, inhibits the growth, migration and invasion of colorectal cancer cell lines. Oncol. Rep. 2015;33:967–973. doi: 10.3892/or.2014.3652. [DOI] [PubMed] [Google Scholar]
- 54.Qu Y., Wang J., Ray P.S., Guo H., Huang J., Shin-Sim M., Bukoye B.A., Liu B., Lee A.V., Lin X., Huang P., Martens J.W., Giuliano A.E., Zhang N., Cheng N.H., Cui X. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-kappaB signaling. J. Clin. Invest. 2011;121:212–225. doi: 10.1172/JCI43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bizzarri C., Holmgren A., Pekkari K., Chang G., Colotta F., Ghezzi P., Bertini R. Requirements for the different cysteines in the chemotactic and desensitizing activity of human thioredoxin. Antioxid. Redox Signal. 2005;7:1189–1194. doi: 10.1089/ars.2005.7.1189. [DOI] [PubMed] [Google Scholar]
- 56.Kondo N., Ishii Y., Kwon Y.W., Tanito M., Sakakura-Nishiyama J., Mochizuki M., Maeda M., Suzuki S., Kojima M., Kim Y.C., Son A., Nakamura H., Yodoi J. Lipid raft-mediated uptake of cysteine-modified thioredoxin-1: apoptosis enhancement by inhibiting the endogenous thioredoxin-1. Antioxid. Redox Signal. 2007;9:1439–1448. doi: 10.1089/ars.2007.1665. [DOI] [PubMed] [Google Scholar]
- 57.Kim N.H., Park H.J., Oh M.K., Kim I.S. Antiproliferative effect of gold(I) compound auranofin through inhibition of STAT3 and telomerase activity in MDA-MB 231 human breast cancer cells. BMB Rep. 2013;46:59–64. doi: 10.5483/BMBRep.2013.46.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roder C., Thomson M.J. Auranofin: repurposing an old drug for a golden new age. Drugs R&D. 2015;15:13–20. doi: 10.1007/s40268-015-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X., Zhou H.J., Huang Q., Lu L., Min W. Novel action and mechanism of auranofin in inhibition of vascular endothelial growth factor receptor-3-dependent lymphangiogenesis. Anticancer Agents Med. Chem. 2014;14:946–954. doi: 10.2174/1871520614666140610102651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cadenas C., Franckenstein D., Schmidt M., Gehrmann M., Hermes M., Geppert B., Schormann W., Maccoux L.J., Schug M., Schumann A., Wilhelm C., Freis E., Ickstadt K., Rahnenfuhrer J., Baumbach J.I., Sickmann A., Hengstler J.G. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010;12:R44. doi: 10.1186/bcr2599. [DOI] [PMC free article] [PubMed] [Google Scholar]