Abstract
Background
Biomarker-assisted diagnosis of acute aortic dissection (AAD) is important for diagnosis and treatment. However, identification of biomarkers for AAD in blood is a challenging task. The aim of this study is to search for new potentially microRNA (miRNAs) biomarkers in AAD.
Methods
The miRNAs expression profiles in ascending aortic tissue and plasma were examined by microarray analysis in two sets or groups. The tissue group was composed of four patients with AAD and four controls of healthy male organ donors. The plasma group included 20 patients with AAD and 20 controls without cardiovascular disease. Bioinformatics was used to analyze the potential targets of the differentially expressed miRNAs.
Results
Our study revealed that in AAD patients, the aortic tissue had 30 differentially expressed miRNAs with 13 up-regulated and 17 down-regulated, and plasma had 93 differentially expressed miRNAs, of which 33 were up-regulated and 60 were down-regulated. Four miRNAs were found to be up-regulated in both aortic tissue and plasma in AAD patients. The predicted miRNA targets indicated the four dysregulated miRNAs mainly targeted genes that were associated with cell-cell adhesion, extracellular matrix metabolism, cytoskeleton organization, inflammation, and multiple signaling pathways related to cellular cycles.
Conclusions
Four miRNAs, which are up-regulated both in aortic tissue and in plasma in AAD patients, have been identified in this study. These miRNAs might be potential diagnostic biomarkers for AAD. Larger sample investigations are needed for further verification.
Keywords: Acute aortic dissection, Biomarker, Diagnosis, MicroRNAs
1. Introduction
Acute aortic dissection (AAD) is the most frequent and catastrophic manifestation of the acute aortic syndrome characterized by acute onset and rapid progress. Mechanically, circulating blood flows into the media of the aorta through the rupture of the intima and forms true and false lumens. Since AAD is highly lethal and requires prompt diagnosis and treatment and despite advances in imaging methods to identify the disease, misdiagnosis occurs in 25%–50% of patients on initial evaluation with symptoms mimicking acute myocardial infarction and other cardiovascular disorders.[1]–[4] Currently, computed tomography, magnetic resonance imaging, and transthoracic or transesophageal echocardiography are the commonly used modalities for diagnosing AAD. However, they are usually time-consuming or limited by unavailability at bedside. Blood testing is widely used to diagnose various diseases in clinical practice due to the rapid, easy and convenient operation at bedside. Several plasma markers such as D-di-mer,[5] smooth muscle myosin heavy chain,[6],[7] BB-isozyme of creatine kinase,[8] calponin,[9] and elastin[10] were studied as potential candidates for use as biomarkers of AAD. But, except D-dimer,[11] none of the other biomarkers has been adopted into routine clinical practice because they have not been able to meet the requirements of a ‘gold standard’ biomarker including adequate sensitivity and specificity, in addition to a favorable time course of release that covers a time window necessary for non-ambiguity in the clinical setting.[12] Therefore, novel biomarkers characterized by high sensitivity and specificity, as well as suitable for early diagnosis, are highly desirable for the diagnosis of AAD.
MicroRNAs (miRNAs) are endogenous non-coding RNA molecules that regulate gene expression on the post-transcriptional, or transcriptional level by targeting mRNAs for cleavage or translational repression.[13] The circulating miRNAs have been reported to be the biomarkers for diagnosis and treatment of diseases.[14] In recent years, growing evidence suggest that miRNAs not only play crucial roles in physiological processes of cardiovascular development, but also in pathologic processes of cardiovascular diseases.[15],[16] However, there are also limited studies focused on miRNAs in AAD and the results were inconsistent.[17],[18] Here, we analyzed the differential expression profile of miRNAs in aortic tissue and plasma between AAD patients and healthy control subjects by microarray experiments and our results may provide useful evidence for identifying novel biomarkers of AAD.
2. Methods
Study protocols were approved by the ethical committees of Fuwai hospital and complied with the declaration of Helsinki. All persons gave their informed consent prior to their inclusion in the study.
2.1. Aortic tissue samples
The dissecting tissue samples of ascending aorta were collected from four male patients (mean age 49 years) with type A AAD, who were identified without Marfan syndrome, Loeys-Dietz Syndrome, and familial aortic dissection. All patients received surgical operation within 24 h in the Department of Vascular Surgery of Fuwai Hospital (Beijing, China). Four normal aortic tissue samples were collected from organ donors (mean age 48 years) without aortic disease. The aortic tissue samples were placed in freezing tubes and frozen immediately in liquid nitrogen and then stored at −80°C until RNA extraction.
2.2. RNA extraction and miRNA microarray analysis
Total RNA was extracted and purified using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, US) following the manufacturer's instructions and checked for a RIN number to inspect RNA integration by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). Then miRNA in the total RNA was labeled by miRNA Complete Labeling and Hyb Kit (Agilent technologies, Santa Clara, CA, US) followed the manufacturer's instruc-tions. Each slide was hybridized with 100 ng Cy3-labeled RNA using miRNA Complete Labeling and Hyb Kit (Agilent technologies, Santa Clara, CA, US) in a hybridization Oven (Agilent technologies, Santa Clara, CA, US) at 55°C, 20 r/min for 20 h according to the manufacturer' s instructions. After hybridization, slides were washed in staining dishes (Thermo Shandon, Waltham, MA, US) with Gene Expression Wash Buffer Kit (Agilent technologies, Santa Clara, CA, US). Slides were then scanned by Agilent Microarray Scanner (Agilent technologies, Santa Clara, CA, US) and Feature Extraction software 10.7 (Agilent technologies, Santa Clara, CA, US) with default settings. Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent technologies, Santa Clara, CA, US).
2.3. Plasma miRNAs analysis
The blood samples were collected from 20 patients with type A AAD and 20 age- and gender-matched healthy controls. A venous EDTA-treated blood sample was taken from each patient within 24 h after symptom onset before any surgical procedure. The blood samples were collected from each healthy control after overnight fasting. After centrifuging at 2,000 g for 10 min at 4°C, the plasma was aliquoted and stored at −80°C until detection. The forty plasma of the two groups were mixed, then divided, to obtain two pooled samples. The processes of RNA extraction and miRNA microarray analysis were identical to that described above.
2.4. Target gene prediction
Prediction of miRNA target gene was performed by computational algorithms according to their base-pairing rules between miRNA and mRNA target sites, location of binding sequences within the target's 3′UTR, and conservation of target binding sequences within genomes. The target genes of differentially expressed miRNAs were predicted by online tools including TargetScan v5.1 (http://www.target-scan.org/), Sanger (http://www.sanger.ac.uk/), Pictar (http://pictar.bio.nyu.edu/), and Miranda v5 (http://miRNA.sanger.ac.uk/).
2.5. Statistical analysis
Continuous variables are presented as mean ± SD and categorical data are presented as numbers and proportion. The statistically significance of the microarray result was analyzed by fold change and the Student t test. The threshold value we used to screen differentially expressed miRNAs is a fold change ≥ 2.0 or ≤ 0.5 (P < 0.01).
3. Results
3.1. Baseline characteristics of participants for aortic tissue miRNAs microarray analysis and the differentially expressed miRNAs
Table 1 shows the baseline characteristics of participators for aortic tissue miRNAs microarray analysis. Participators in the two groups were comparable for age and gender. One AAD patient had concomitant hypertension and none of the patients in the two groups had atherosclerosis.
Table 1. Baseline characteristics of participators for aortic tissue miRNAs microarray analysis.
| AAD (n = 4) | Control (n = 4) | |
| Tissue sample | Ascending aorta | Ascending aorta |
| Age, yrs | 49.1 ± 4.9 | 47.9 ± 6.7* |
| Male | 4 (100%) | 4 (100%) |
| Hypertension | 1 (25%) | 0* |
| Atherosclerosis | 0 | 0 |
Data are presented as mean ± SD or n (%). *P > 0.05; AAD: acute aortic dissection.
Figure 1 shows the main differentially expressed miRNAs and 30 miRNAs were found to be dysregulated, of which 13 were up-regulated and 17 were down-regulated in AAD patients compared with healthy control subjects.
Figure 1. Volcano plots showing the differentially expressed miRNAs in AAD patients.
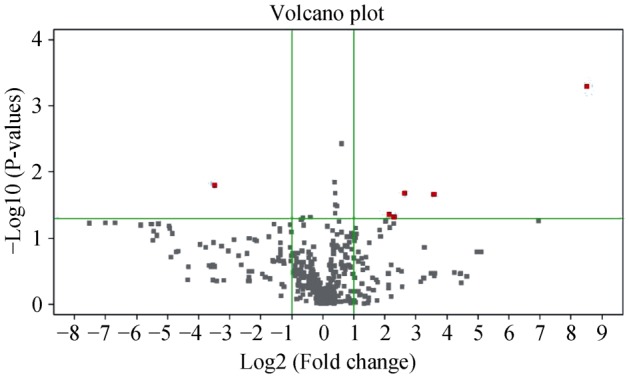
The X axis represents the log 2 fold change compared with healthy control subjects and the Y axis represents −log10 P-values obtained from the t test comparing the mean normalized miRNA signal in AAD patients to that in healthy control subjects. Top righted dots represent up-regulated miRNAs with a fold change >2, while left righted dots represent down-regulated miRNAs with fold change >2 in dissection patients compared with that of healthy control subjects. AAD: acute aortic dissection.
Table 2 displays the details of differentially expressed miRNAs. Has-miR-31 was the most up-regulated miRNA with nearly 500-fold increase in AAD patients, while the most down-regulated miRNA was has-miR-936 with over a thousand-fold decrease compared with healthy control subjects.
Table 2. Differentially expressed miRNAs in AAD patients by aortic tissue microarray analysis.
| Differential miRNAs | Fold change | P value |
| Upregulated miRNAs | ||
| has-miR-31 | 496.48 | 0.034 |
| has-miR-513b | 46.81 | 0.011 |
| has-miR-221* | 16.73 | 0.016 |
| has-miR-7 | 12.87 | 0.001 |
| has-miR-933 | 10.35 | 0.02 |
| has-miR-139-5p | 8.07 | 0.049 |
| has-miR-224 | 7.50 | 0.028 |
| has-miR-30c-1* | 5.99 | 0.026 |
| has-miR-718 | 2.16 | 0.02 |
| has-miR-1281 | 1.74 | 0.02 |
| has-miR-4313 | 1.50 | 0.011 |
| has-miR-425 | 1.50 | 0.035 |
| has-miR-1238 | 1.28 | 0.041 |
| Downregulated miRNAs | ||
| has-miR-198 | 0.45 | 0.015 |
| has-miR-623 | 0.25 | 0.014 |
| has-miR-3652 | 0.23 | 0.029 |
| has-miR-658 | 0.20 | 0.016 |
| has-miR-648 | 0.17 | < 0.0001 |
| has-miR-501-3p | 0.07 | 0.04 |
| has-miR-3180-5p | 0.07 | 0.045 |
| has-miR-3605-5p | 0.02 | 0.01 |
| has-miR-1307 | 0.02 | 0.006 |
| has-miR-3202 | 0.01 | 0.02 |
| has-miR-4314 | 0.01 | 0.03 |
| has-miR-3614-5p | 0.01 | < 0.001 |
| has-miR-B4 | 0.01 | < 0.001 |
| has-miR-659 | 0.01 | 0.002 |
| has-miR-3621 | 0.01 | < 0.001 |
| has-miR-3660 | 0.01 | < 0.001 |
| has-miR-936 | 0.00 | < 0.002 |
AAD: acute aortic dissection.
3.2. Baseline characteristics of participators for plasma miRNAs microarray analysis and the differentially expressed miRNAs
The baseline characteristics of participators for plasma miRNAs microarray are displayed in Table 3. The two groups were comparable for age and gender, however, most of AAD patients (70%) have concomitant hypertension and one patient (5%) had atherosclerosis.
Table 3. Baseline characteristics of participators for plasma microarray.
| AAD (n = 20) | Control (n = 20) | |
| Age (years) | 53.3 ± 13.5 | 51.9 ± 11.2* |
| Male | 14 (70%) | 14 (70%)* |
| Hypertension | 14 (70%) | 0 |
| Atherosclerosis | 0 | 0 |
Data are presented as mean ± SD or n (%). *P > 0.05; AAD: acute aortic dissection.
A total of 93 miRNAs were found to be dysregulated in the plasma, of which 33 were up-regulated and 60 were down-regulated in AAD patients compared with that of healthy control subjects.
Differentially expressed miRNAs with fold change > 20 are shown in Table 4. Among these miRNAs, 10 were up-re-gulated and 10 were down-regulated. The most up-re-gulated miRNA was has-miR-4313 with 42.39-fold increase, while the most down-regulated miRNA was has-miR-4454 with 100-fold decrease in AAD patients compared with that of healthy control subjects.
Table 4. Differentially expressed miRNAs with change fold > 20 in AAD patients by plasma microarray analysis.
| Up-regulated miRNAs | Fold change | Down-regulated miRNAs | Fold change |
| has-miR-4313 | 42.39 | has-miR-5703 | 0.03 |
| has-miR-1825 | 39.16 | has-miR-4505 | 0.03 |
| has-miR-4725-5p | 35.32 | has-miR-630 | 0.03 |
| has-miR-4749-3p | 32.02 | has-miR-3663-3p | 0.02 |
| has-miR-425-3p | 31.37 | has-miR-371b-5p | 0.02 |
| has-miR-4652-3p | 30.01 | has-miR-5787 | 0.02 |
| has-miR-933 | 26.75 | has-miR-4530 | 0.02 |
| has-miR-4769-3p | 24.88 | has-miR-6068 | 0.02 |
| has-miR-4323 | 21.43 | has-miR-144-3p | 0.01 |
| has-miR-122-5p | 20.54 | has-miR-4454 | 0.01 |
AAD: acute aortic dissection.
3.3. Differentially expressed miRNAs in both aortic tissue and plasma
Table 5 shows the differentially expressed miRNAs both in aortic tissue and in plasma. Four miRNAs were included, all of which were up-regulated in AAD patients. The relatively higher up-regulated miRNAs were has-miR-4313 and has-miR-933, the former with 1.5- and 42.4-fold increase in aortic tissue and in plasma, and the latter with 10.4- and 26.8-fold increase in AAD patients.
Table 5. Differentially expressed miRNAs in both aortic tissue and plasma in AAD patients.
| miRNAs | miRNA NO. | Fold change |
|
| Aortic tissue miRNA | Plasma miRNA | ||
| has-miR-4313 | MIMAT0016865 | 1.5 | 42.4 |
| has-miR-933 | MIMAT0004976 | 10.4 | 26.8 |
| has-miR-1281 | MIMAT0005939 | 1.7 | 17.8 |
| has-miR-1238 | MIMAT0005593 | 1.3 | 13.8 |
AAD: acute aortic dissection.
3.4. Potential targets of the four differentially expressed miRNAs
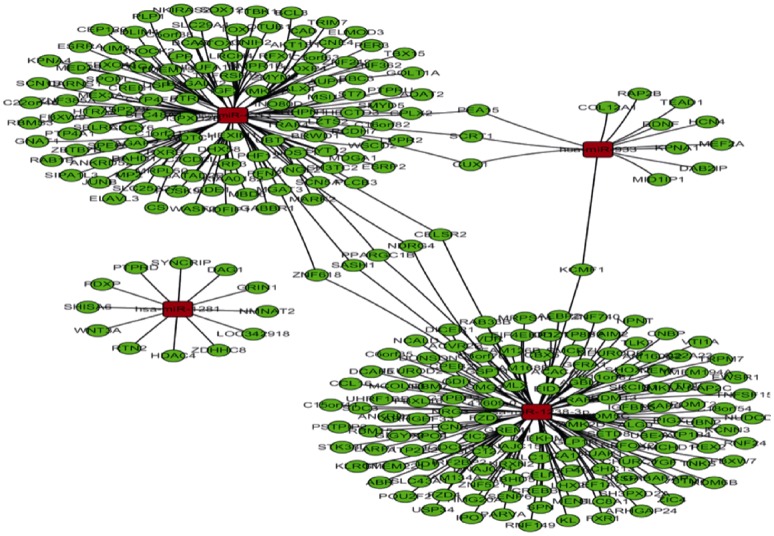
The predicted target genes of the four differentially expressed miRNAs are shown in Figure 2. Computational algorithms show that has-miR-4313 mainly targeted genes related to cell-cell recognition and adhesion, notch signaling, and TGFβ2-ERK signaling pathway that involved in cell proliferation. Has-miR-933 mainly targeted genes related to collagen metabolism, morphogenesis, and cell apoptosis, while the predicted targeted genes of has-miR-1281 and has-miR-933 participate in cytoskeleton organization, inflammation, cell adhesion, and multiple signaling pathways associated with cellular proliferation, differentiation, and apoptosis.
Figure 2. Predicted target genes of the four differentially expressed miRNAs by bioinformatics analysis.
4. Discussion
The present study analyzed the differentially expressed miRNAs by microarray between patients with AAD and healthy control subjects, and further detected four dysregulated miRNAs in both aortic tissue and plasma. Predicted miRNA targets indicated the four dysregulated miRNAs mainly targeted cell-cell adhesion, extracellular matrix metabolism, cytoskeleton organization, inflammation, and multiple signaling pathways related with cellular processes.
In recent years, it was confirmed that circulating miRNA, in contrast to mRNA, is strikingly stable in plasma.[19] Multiple studies have reported circulating miRNAs may be used as biomarkers for clinical diagnosis,[20] novel targets for treatment,[21] drug sensitivity/resistance testing,[22] as well as prognosis judgment.[23] In the cardiovascular field, it also found dysregulation of miRNAs was associated with many clinically relevant cardiovascular conditions including myocardial infarction,[24], heart failure[25], arrhythmia[26], etc.
AAD is the most lethal vascular emergency and there is a lacking of ideal biomarkers for early diagnosis. Although D-dimer is widely used to screen patients with AAD in clinical practice, the pooled specificity for diagnosing AAD by meta-analysis is 0.56 (95%CI: 0.51–0.60).[27] Therefore, a search for novel biomarkers specifically associated with AAD is of great importance for initiation of treatment and improved survival. MiRNAs have been recently found to play an important role in the regulation of vasculature. Both miR-143 and miR-145 are highly expressed in vascular smooth muscle cells (VSMCs) and in vitro experiments indicate the miR-143/145 cluster is required for maintaining VSMC tone.[28] In contrast, miR-145-null mice exhibited blood vessels with a reduced medial layer, abnormalities in actin stress fiber formation and phenotype switch in response to vascular injury.[29] The miR-26a is down -regulated in mouse models of abdominal aortic aneurysm.[30] And over-expression of miR-21, another miRNA that is critical for VSMC phenotype regulation, protects against abdominal aortic aneurysm progression. Conversely, inhibition of miR-21 further augments ongoing abdominal aortic aneurysm formation.[31] These results suggest that some miRNAs perform an important role in the pathologic processes of aorta disease. However, whether miRNAs could be used as biomarkers for diagnosing AAD in clinical practice remains unclear.
Ideal biomarkers should be characterized as tissue- or organ-specific in plasma and, indeed, some miRNAs were reported to be heart- and vascular-specific miRNAs, such as miR-499, produced almost exclusively in the heart.[32] Therefore, we screened the differentially expressed miRNAs in both aortic tissue and plasma with the aim of detecting miRNAs specifically associated with AAD. Four miRNAs in AAD patients were found to be up-regulated in both aorta tissue and plasma; moreover, the fold change was striking ranging from over 10- to 40-fold increase in plasma compared with healthy control subjects, making the discrimination between AAD cases and disease-free cases accessible. Furthermore, displays of bioinformatics analyses that the four differentially expressed miRNAs mainly target genes related to collagen metabolic, cytoskeleton organiza-tion, inflammation, cell adhesion, and multiple signaling pathways associated with cellular proliferation, differentiation, and apoptosis, all of which have been considered to be involved in the pathogenesis of AAD. Our study suggests the four miRNAs may not only participate in the pathological processes of AAD, but also be the potential biomarkers for AAD.
Limited studies on the differential expression of miRNAs between aortic dissection patients and healthy control subjects have been reported. In the report of Liao, et al.[17] 18 miRNAs were up-regulated and 56 down-regulated in dissection patients and seven selected miRNAs were verified by quantitative reverse transcription polymerase chain reaction (qRT-PCR) with consistent expression of microarray analysis. Target gene-related pathway analysis showed a significant change in the focal adhesion and the mitogen-activated protein kinase signaling pathways in patients with aortic dissection. Hu, et al.[18] also analyzed the differentially expressed miRNAs in aortic dissection and normal aorta tissue and found five miRNAs were dysregulated, however, only one miRNA was verified by qRT-PCR to be consistent with the results of microarray analysis. This suggests that although microarray technology has an advantage of high throughput screening, it still has relatively high false positive results. Therefore, studies with large sample sizes are needed to reduce the chances of a false positive conclusion. Moreover, the differentially expressed miRNAs were inconsistent, possibly in part due to genetic heterogeneity and the limited sample sizes, while also a reflection of the complexity of pathogenesis in AAD.
In conclusion, our study analyzed the differentially expressed miRNAs between patients with AAD and healthy control subjects, and detected four miRNAs that were up-regulated in both aortic tissue and plasma in AAD patients compared with healthy control subjects. Currently, we are verifying the validity and sensitivity of the four miRNAs in a large sample of AAD patients and controls by qRT-PCR. Meanwhile, we are also testing the specificity of the four miRNAs in AAD by measuring the expression of these miRNAs in a wide spectrum of diseases. It is anticipated some miRNAs will be validated as novel biomarkers for early diagnosis of AAD.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (project 81170286 to FAN XH, project 81300184 to WANG XJ). We thank the patients for their participations in our study.
References
- 1.Spittell PC, Spittell JJ, Joyce JW, et al. Clinical features and differential diagnosis of aortic dissection: experience with 236 cases (1980 through 1990) Mayo Clin Proc. 1993;68:642–651. doi: 10.1016/s0025-6196(12)60599-0. [DOI] [PubMed] [Google Scholar]
- 2.Klompas M. Does this patient have an acute thoracic aortic dissection? JAMA. 2002;287:2262–2272. doi: 10.1001/jama.287.17.2262. [DOI] [PubMed] [Google Scholar]
- 3.Hansen MS, Nogareda GJ, Hutchison SJ. Frequency of and inappropriate treatment of misdiagnosis of acute aortic dissection. Am J Cardiol. 2007;99:852–856. doi: 10.1016/j.amjcard.2006.10.055. [DOI] [PubMed] [Google Scholar]
- 4.Asouhidou I, Asteri T. Acute aortic dissection: be aware of misdiagnosis. BMC Res Notes. 2009;2:25. doi: 10.1186/1756-0500-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggebrecht H, Naber CK, Bruch C, et al. Value of plasma fibrin D-dimers for detection of acute aortic dissection. J Am Coll Cardiol. 2004;44:804–809. doi: 10.1016/j.jacc.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 6.Katoh H, Suzuki T, Hiroi Y, et al. Diagnosis of aortic dissection by immunoassay for circulating smooth muscle myosin. Lancet. 1995;345:191–192. doi: 10.1016/s0140-6736(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T, Katoh H, Watanabe M, et al. Novel biochemical diagnostic method for aortic dissection. Results of a prospective study using an immunoassay of smooth muscle myosin heavy chain. Circulation. 1996;93:1244–1249. doi: 10.1161/01.cir.93.6.1244. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T, Katoh H, Kurabayashi M, et al. Biochemical diagnosis of aortic dissection by raised concentrations of creatine kinase BB-isozyme. Lancet. 1997;350:784–785. doi: 10.1016/S0140-6736(05)62569-X. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, Distante A, Zizza A, et al. Preliminary experience with the smooth muscle troponin-like protein, calponin, as a novel biomarker for diagnosing acute aortic dissection. Eur Heart J. 2008;29:1439–1445. doi: 10.1093/eurheartj/ehn162. [DOI] [PubMed] [Google Scholar]
- 10.Shinohara T, Suzuki K, Okada M, et al. Soluble elastin fragments in serum are elevated in acute aortic dissection. Arterioscler Thromb Vasc Biol. 2003;23:1839–1844. doi: 10.1161/01.ATV.0000085016.02363.80. [DOI] [PubMed] [Google Scholar]
- 11.Erbel R, Aboyans V, Boileau C, et al. 2014 ESC Guidelines on the diagnosis and treatment of aortic diseases: Document covering acute and chronic aortic diseases of the thoracic and abdominal aorta of the adult. The task force for the diagnosis and treatment of aortic diseases of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2873–2926. doi: 10.1093/eurheartj/ehu281. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Distante A, Eagle K. Biomarker-assisted diagnosis of acute aortic dissection: how far we have come and what to expect. Curr Opin Cardiol. 2010;25:541–545. doi: 10.1097/HCO.0b013e32833e6e13. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Hata A. Functions of microRNAs in cardiovascular biology and disease. Annu Rev Physiol. 2013;75:69–93. doi: 10.1146/annurev-physiol-030212-183737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Yu Q, Wang Q, et al. Roles of miRNA in cardiovascular development and dysfunction. Curr Med Chem. 2013;20:3613–3622. doi: 10.2174/0929867311320290007. [DOI] [PubMed] [Google Scholar]
- 17.Liao M, Zou S, Weng J, et al. A microRNA profile comparison between thoracic aortic dissection and normal thoracic aorta indicates the potential role of microRNAs in contributing to thoracic aortic dissection pathogenesis. J Vasc Surg. 2011;53:1341–1349. doi: 10.1016/j.jvs.2010.11.113. [DOI] [PubMed] [Google Scholar]
- 18.Hu ZY, Luo JF, Zhong SL, et al. MicroRNAs expression in normal and dissected aortic tissue. Zhonghua Xin Xue Guan Bing Za Zhi. 2012;40:406–410. [PubMed] [Google Scholar]
- 19.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Lv M, Wang H, et al. Identification of circulating microRNAs as novel potential biomarkers for gastric cancer detection: a systematic review and meta-analysis. Dig Dis Sci. 2014;59:911–919. doi: 10.1007/s10620-013-2970-9. [DOI] [PubMed] [Google Scholar]
- 21.To KK. MicroRNA: a prognostic biomarker and a possible druggable target for circumventing multidrug resistance in cancer chemotherapy. J Biomed Sci. 2013;20:99. doi: 10.1186/1423-0127-20-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Zhang K, Bi M, et al. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs. 2014;25:346–352. doi: 10.1097/CAD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 23.Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18:268–274. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Pei F, Zhu X, et al. Circulating microRNAs as novel and sensitive biomarkers of acute myocardial Infarction. Clin Biochem. 2012;45:727–732. doi: 10.1016/j.clinbiochem.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira-Carvalho V, Da SM, Guimaraes GV, et al. MicroRNAs: new players in heart failure. Mol Biol Rep. 2013;40:2663–2670. doi: 10.1007/s11033-012-2352-y. [DOI] [PubMed] [Google Scholar]
- 26.Kim GH. MicroRNA regulation of cardiac conduction and arrhythmias. Transl Res. 2013;161:381–392. doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimony A, Filion KB, Mottillo S, et al. Meta-analysis of usefulness of d-dimer to diagnose acute aortic dissection. Am J Cardiol. 2011;107:1227–1234. doi: 10.1016/j.amjcard.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 28.Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leeper NJ, Raiesdana A, Kojima Y, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maegdefessel L, Azuma J, Toh R, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-aug-mented expansion. Sci Transl Med. 2012;4:122r. doi: 10.1126/scitranslmed.3003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adachi T, Nakanishi M, Otsuka Y, et al. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183–1185. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]