Abstract
Previously we showed that in vivo treatment of elderly Fisher 344 rats with acetylcarnitine abolished the age-associated defect in respiratory chain complex III in interfibrillar mitochondria and improved the functional recovery of the ischemic/reperfused heart. Herein we explored mitochondrial protein acetylation as a possible mechanism for acetylcarnitine’s effect. In vivo treatment of elderly rats with acetylcarnitine restored cardiac acetylcarnitine content and increased mitochondrial protein lysine acetylation and increased the number of lysine-acetylated proteins in cardiac subsarcolemmal and interfibrillar mitochondria. Enzymes of the tricarboxylic acid cycle, mitochondrial β-oxidation, and ATP synthase of the respiratory chain showed the greatest acetylation. Acetylation of isocitrate dehydrogenase, long-chain acyl-CoA dehydrogenase, complex V, and aspartate aminotransferase was accompanied by decreased catalytic activity. Several proteins were found to be acetylated only after treatment with acetylcarnitine, suggesting that exogenous acetylcarnitine served as the acetyl-donor. 2-D fluorescence difference gel electrophoresis analysis revealed that acetylcarnitine treatment also induced changes in mitochondrial protein amount; a twofold or greater increase/decrease in abundance was observed for thirty one proteins. Collectively, our data provide evidence for the first time that in the aged rat heart in vivo administration of acetylcarnitine provides acetyl groups for protein acetylation and affects the amount of mitochondrial proteins.
Keywords: Aging, heart, acetylcarnitine, mitochondria, protein acetylation
1. Introduction
Aging decreases cardiac mitochondrial oxidative capacity which, predisposes the heart to greater stress-induced injury. Using the Fisher 344 rat heart as a model of aging, we have shown that aging selectively decreases both mitochondrial content and mitochondrial oxidative metabolism in the interfibrillar mitochondria (IFM) (Fannin et al., 1999; Lesnefsky et al., 2001). The decrease in oxidative phosphorylation is due to decreased CIII and CIV activity in IFM (Lesnefsky et al., 2001). The decreased catalytic activity of CIV is secondary to a defect in the membrane phospholipid environment (Paradies et al., 1994; Fannin et al., 1999). In CIII the aging defect has been localized to heme bL of cytochrome b (Moghaddas et al., 2003) and is not accompanied by decreases either in cytochromes b and c1 or in the iron-sulfur protein, the catalytic subunits of complex III (Lesnefsky et al., 2001).
In contrast to the aging defect that is selective to IFM, ischemia/reperfusion affects both mitochondrial populations. In the IFM the ischemia-induced defect in the iron-sulfur protein of CIII is superimposed on the aging defect, rendering the aged heart more prone to ischemia/reperfusion injury (Lesnefsky et al., 2001) providing a rational explanation why the aged heart sustains greater injury than the young heart after ischemia (Frolkis et al., 1991; Lesnefsky et al., 1994; Lesnefsky et al. 1996).
In addition to the age-associated decreased oxidative phosphorylation, total and free carnitine decreases with aging (Costell et al., 1989; Costell and Grisolia, 1993; Kerner et al., 2001; Tanaka et al., 2004; Noland et al. 2009;) that is corrected by administration of exogenous carnitine and acetylcarnitine (Costell et al., 1989; Costell and Grisolia, 1993; Tanaka et al., 2004). We have shown that a single intraperitoneal injection of acetylcarnitine restored oxidative phosphorylation with glutamate as substrate and CIII activity to values observed in IFM from hearts of adult saline-perfused Fisher 344 rats (Lesnefsky et al., 2006). In addition, acetylcarnitine treatment significantly increased the content of cytochrome b suggesting increased mitochondrial protein synthesis or decreased degradation. This notion is supported by the finding that ablation of SIRT3, the major mitochondrial NAD+-dependent protein deacetylase, is associated with increased mitochondrial translation activity and increasing/decreasing the expression of SIRT3 in C2C12 cells is accompanied by decreased/increased mitochondrial protein synthesis (Yang et al., 2010). More importantly, in vivo acetylcarnitine treatment of elderly rats improved the recovery of cardiac contractile function following ischemia/reperfusion similar to that observed with saline-treated adult hearts (Lesnefsky et al., 2006). Acetylcarnitine treatment had no effect on the above parameters in hearts from six month old adult rats. This proof-of-principle study provides compelling evidence that the age-associated defect in CIII is responsible for the increased ischemia/reperfusion damage in the aged heart.
In the present study we determined the effect of exogenous acetylcarnitine on 1) myocardial acetylcarnitine content as a surrogate for mitochondrial acetylation potential (Pearson and Tubbs, 1967; Oram et al., 1973); 2) acetylation of mitochondrial proteins by immunoblotting and proteomics; 3) the effect of acetylation on enzyme activity; and 4) mitochondrial protein abundance by two-dimensional difference gel electrophoresis (2D-DIGE) in hearts of acetylcarnitine- and saline-treated 24 months old Fischer 344 rats.
2. Materials and methods
2.1. Animals and animal treatment
Male six month old adult and 22 month and 24 month old elderly Fisher 344 rats were obtained from a colony at the National Institute of Aging (Harlan Sprague Dawley, Inc., Indianapolis, IN) and maintained in our animal care facility in a temperature and humidity controlled room and 12 h light/dark cycle. All animal protocols used were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University. The animals had free access to food and water until the experiments. Acetylcarnitine (inner salt, Sigma-Tau, Rome, Italy) was administered intraperitoneally (IP) to 24 month old animals three hours before the experiment as a single dose (1.28 mmoles or 260 mg/kg body weight) (Paradies et al., 1992, Paradies et al., 1994, Lesnefsky et al., 2006) or orally to 22 month old rats in the drinking water (1.5% acetylcarnitine inner salt) over a two-month period (Pesce et al., 2004). Control rats received saline injection or unsupplemented tap water. Six month old adult control rats had free access to food and water and were not treated with acetylcarnitine because acetylcarnitine treatment had no effect on oxidative phosphorylation, electron transport chain enzyme activities, and cytochrome b and cytochrome aa3 content (Lesnefsky et al., 2006). At the time the mitochondria were harvested the mean body weights were 375 ± 18 and 404 ± 25 g in the adult (6 month) and elderly (24 month) groups (n=4), respectively.
2.1. Carnitine and acylcarnitine analysis
At euthanasia approximately 50 mg tissue from the apex of each heart was removed and frozen in liquid nitrogen for later analysis of tissue carnitine and acylcarnitines by HPLC/MS as previously described in detail (Minkler et al., 2008).
2.2. Isolation of heart mitochondria
Following removal of tissue for carnitine analysis the remaining tissue was used for mitochondria isolation. Cardiac SSM and IFM were isolated by differential centrifugation as referenced (Palmer et al., 1977) with the exception that nagarse was replaced by trypsin. Mitochondrial function was assessed with duroquinol, a specific substrate for respiratory chain CIII, as referenced (Hoppel et al., 1987). The remaining mitochondria were stored at minus 60°C until analysis. Mitochondrial protein concentration was determined by the method of Lowry et al. (Lowry et al., 1951).
2.3. SDS-PAGE and immunobloting
Cardiac SSM and IFM from 24 month old saline- and acetylcarnitine-treated (IP and oral) rats were solubilized in SDS sample buffer and 50 μg of protein was loaded per lane. Following separation by SDS-PAGE (4% concentrating and 10% separating gel) the proteins were transferred to nitrocellulose membrane and the membrane incubated with 1000-fold diluted affinity purified rabbit polyclonal acetyl-lysine antibodies (Cell Signaling) followed by horseradish-conjugated goat anti-rabbit antibodies (BioRad, 5000-fold dilution). Acetylated proteins were visualized by enhanced chemiluminescent detection (Amersham Biosciences). For SIRT3 the membrane was incubated with 1000-fold diluted affinity-purified rabbit monoclonal SIRT3 antibodies (Cell Signaling) followed by horseradish-conjugated secondary antibodies.
2.4. Two-dimensional difference gel electrophoresis (2D DIGE)
2D DIGE experiments were carried out as described previously (Yohannes et al., 2008; Yohannes et al., 2010). Briefly, per gel, 150 μg of labeled protein, pooled mixture of Cy2, Cy3 and Cy5 labeled, as well as additional 350 μg of unlabeled protein in the buffer containing 7 M urea, 2 M thiourea, 4% CHAPS w/v, 0.2% DTT w/v, 1 % pharmalyte 3–10 NL were used to passively re-hydrate the 24 cm IPG strips pH 3–10 NL (GE Healthcare, Piscataway, NJ) for 12 hours. The rehydrated strips were then focused under high voltage. Before the second dimension separation, the strips were reduced and alkylated in equilibration buffer (100 mM Tris, pH 8.0, 6 M urea, 30% glycerol) containing 0.5% DTT or 4.5% iodoacetamide, respectively. An IPG strip was then placed onto 12.5% homogeneous Tris-HCl large format SDS-PAGE gel and overlaid with 1% agarose and run in Ettan 12 electrophoresis cell (GE Healthcare, Piscataway, NJ) with Tris/glycine/SDS (25 mM Tris base, 192 mM glycine, and 0.2% SDS) running buffer.
The data analysis was carried out using DeCyder software. Three sets of images from a single gel were loaded into the DIA (Differential in-gel analysis) algorithm within the DeCyder software. Spot detection, spot quantitation, and normalization were performed using the in-gel linked internal standard. Spot resolutions were confirmed from the Gaussian profiles of the spots on the image. Subsequently, log volume ratio for every protein spot of treated sample vs. the control were computed and spots with 2-fold or greater changes were filtered and assigned proteins of interest for the subsequent protein identification.
2.5. In-gel digestion and protein identification
The proteins in the gel plugs were digested with trypsin (Promega) and the tryptic peptides analyzed by LC-MS/MS using Fourier Transform LTQ mass spectrometer (Thermo Electron Corp., Bremen, Germany). Separation of peptides via capillary liquid chromatography was performed using a Dionex Ultimate 3000 capillary LC system using 0.1% formic acid in 5% acetonitrile (mobile phase A) and 0.1% formic acid in 85% acetonitrile (mobile phase B). The protein digests were trapped onto a pre-column (C18, PepMap100, 300 μm ×5 mm, 5 μm particle size, 100 Ǻ, Dionex), desalted on-line with mobile phase A at 10 μL/min for 10 min and subsequently loaded onto a Dionex C18 PepMap 75 μm x 15 cm reversed phase column with 5% B. Separation was obtained with a linear gradient of 2% B per min, starting with 100% of A. Subsequently, the peptides were chromatographied at a flow rate of 300 nL/min via PicoTip emitter (New Objective Inc., Woburn, MA) and at a voltage of 1.8 kV. Mass spectrum data were acquired using alternating full MS scan and MS/MS scans. Survey data were acquired from m/z of 400 to 1600 and the precursors are interrogated by MS/MS per switch. The switch into MS/MS was based on precursor intensity and three scans were acquired for every precursor interrogated.
The tandem mass spectra were annotated and peak list files (.MGF files) generated by running Mascot Deamon extract_msn algorithm in Mascot Deamon version 2.2.0 (Matrix Science London, UK). NCBInr database was downloaded in FASTA format via file transfer protocol (FTP) from the website of the NCBI into the local server (http://promas/mascot) and configured for Rodentia to produce faster search times. The resulting peak list files were then used to interrogate sequences (224,004 entries) by running Mascot Deamon Algorithm of Mascot software version 2.2.1 (Matrix Science London, UK). Mascot Deamon searches were performed with maximum peptide and fragment ion mass tolerance of 20 ppm and 0.8 Da, respectively, with partial methionine oxidation, N-termini and lysine acetylation and complete carbamidomethylation of cysteine; three missed cleavage sites were allowed in the search parameters. For each protein identification the criterium was a minimum of two peptides with a significant ion score expectation (P<0.05).
2.5. Isolation and analysis of acetylated peptides by proteomics
The procedure described below has been adopted from methods referenced (Kim et al., 2006; Zhao et al., 2010). Heart SSM and IFM (2.0 mg/100 μl) from adult and elderly control and elderly acetylcarnitine-treated rats were treated with 80% cold acetone, the precipitated proteins resuspended in 200 μl 50 mM NH4CO3, pH 8.5, reduced with dithiothreitol (5 mM DTT, 30 min at room temperature), and alkylated with iodoacetamide (15 mM IAA, 30 min, 50°C). Following quenching of excess IAA with 15 mM DTT, the proteins were precipitated with 80% cold acetone. The precipitated proteins were resuspended in 500 μl 50 mM NH4CO3, pH 8.5, containing 1 mM CaCl2 and digested overnight with trypsin (50:1) at 37°C with gentle mixing using a magnetic stirrer. The tryptic digests were centrifuged (20 min, 16,000xg) and 125 μl of the supernatant containing the tryptic peptides dried (Speed Vac) for isolation of acetylated peptides (see below).
For isolation and enrichment of acetylated peptides six μg affinity-purified polyclonal acetyl-lysine antibodies in 400 μl phosphate-buffered saline, pH 7.5, (PBS) were immobilized to GammaBind Plus (Amersham Biosciences, 40 μl of a 1:1 suspension) previously washed with PBS using Pierce SpinColumns. The conjugated antibodies were washed three times with PBS. The tryptic peptides were reconstituted in 200 μl 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA in 0.5% Tween 20 and the acetylated peptides captured by incubation with the immobilized acetyl-lysine antibodies for five hours at room temperature by end-over-end rotation. The reaction was terminated by centrifugation (1 min, 1000 rpm), the beads washed three times with 400 μl 50 mM Tris, pH 8.0, 100 mM NaCl, 1mM EDTA containing 0.5% Tween 20 and twice with deionized water. The acetylpeptides were eluted with 0.1% trifluoroacetic acid (4×50 μl), brought to dryness (Kim et al., 2006; Zhao et al., 2010) and analyzed by HPLC-MS/MS as described above. Mascot searches were performed using SwissProt_05_2010 database with restriction of the search to the taxon Rattus. Maximum peptide and fragment ion mass tolerance of 15 ppm and 1.0 Da respectively, with methionine oxidation, N-termini and lysine acetylation, carbamidomethylation of cysteine, and two missed cleavage sites were allowed in the search parameters.
2. Enzyme assays
Activities of NAD+- and NADP+-linked isocitrate dehydrogenase (ICDH) (Alp et al., 1976), aspartate aminotransferase (Bergmeyer and Bernt, 1974), citrate synthase (CS) (Srere, 1969), ATPase (Rosca et al., 2009), and acyl-CoA dehydrogenase (LCAD) (Hoppel et al., 1979) were determined as referenced. LCAD activity was measured using palmitoyl-CoA, a substrate for both LCAD and very long-chain acyl-CoA dehydrogenase, as well as with 2,6-dimethylheptanoyl-CoA, a specific substrate of LCAD (Wanders et al., 1998).
2.7. Statistical analysis
Data are presented as mean ± standard error (mean ± SEM). Differences between data sets were tested for statistical significance using Student’s t-test and ANOVA (SigmaStat) with p< 0.05 considered statistically significant.
3. Results
3.1. Myocardial acetylcarnitine content: effect of age and treatment with acetylcarnitine
Because cardiac carnitine and acetylcarnitine content decreases with aging (Costell et al., 1989; Costell and Grisolia, 1993; Kerner et al., 2001; Tanaka et al., 2004; Noland et al. 2009;) we tested whether the beneficial effect of acetylcarnitine treatment on cardiac metabolism is related to restoration of tissue carnitine and acetylcarnitine content. As shown in Table 1 aging significantly decreases the myocardial total carnitine content in Fisher 344 rats that is accounted for by the significant decrease in free carnitine and acetylcarnitine; the latter is quantitatively the most dominant acylcarnitine. Statistically significant age-associated changes also were observed in some other, quantitatively minor acylcarnitines, i.e., isobutiryl-, 3-hydroxyisovaleryl-, ethylmalonylcarnitine, and succinylcarnitine. No significant differences between adult and elderly hearts were observed in medium- and long-chain acylcarnitines. Treatment of elderly Fisher 344 rats with exogenous acetylcarnitine restored the myocardial carnitine and acetylcarnitine content to that in hearts of adult animals. Acetylcarnitine was administered either by intraperitoneal injection three hours before the animals were used (Paradies et al., 1992; Paradies et al., 1994; Lesnefsky et al, 2006), or orally in the drinking water for two months. There was no significant difference in the initial and final body weight after two months of oral acetylcarnitine treatment. The daily acetylcarnitine intake with drinking water in the orally treated animals was 0.483 ± 0.068 g per rat or 1.19 ± 0.18 g per kg body weight. With an estimated 10%–15% bioavailability of acetylcarnitine, this represents a daily acetylcarnitine intake of approximately 119–179 mg/kg body weight.
Table 1.
Myocardial content (nanomoles/g wet weight) of total, free, acetyl-, and other short-, medium-, and long-chain acylcarnitines of 6 months old adult and 24 months old elderly acetylcarnitine-treated (IP and orally) Fisher 344 rats. The animals had free access to regular laboratory chow and water. Data represent the means ± SEM.
| Carnitine species | 6 months (n=4) | 24 months (n=9) | 24 month IP AcCarn (=4) | 24 months Oral AcCarn (n=4) |
|---|---|---|---|---|
| Total | 1078.1 ± 102.3 | 659.2 ± 37.5* | 967.4 ± 90.4# | 879.6 ± 33.0# |
| Free | 520.4 ± 45.7 | 347.3 ± 27.9* | 465.6 ± 51.3# | 406.0 ± 6.6 |
| Acyl | 557.7 ± 45.7 | 311.9 ± 29.8* | 501.8 ± 54.1# | 473.5 ± 33.6# |
| Acyl/free | 1.08 ± 0.07 | 0.94 ± 0.10 | 1.10 ± 0.12 | 1.17 ± 0.09 |
|
| ||||
| Acetyl | 428.6 ± 60.1 | 218.1 ± 28.0* | 374.8 ± 47.3# | 361.8 ± 28.8# |
| Propionyl | 3.9 ± 1.5 | 3.6 ± 1.3 | 7.3 ± 2.3 | 4.8 ± 1.9 |
| Butyryl | 10.7 ± 2.3 | 3.9 ± 1.5* | 3.4 ± 1.3* | 8.6 ± 1.4 |
| Isobutyryl | 1.0 ± 0.4 | nd | 0.2 ± 0.2 | 0.3 ± 0.38 |
| R-3-OH-butyryl | nd | 1.1 ± 0.4 | 0.4 ± 0.3 | 1.6 ± 0.3 |
| S-3-OH-butyryl | nd | 7.7 ± 2.8 | 15.5 ± 1.0 | 20.6 ± 2.6# |
| Isovaleryl | nd | nd | 0.2 ± 0.15 | 0.3 ± 0.32 |
| 3-OH-isovaleryl | 0.9 ± 0.10 | 0.10 ± 0.08* | 0.2 ± 0.15* | 0.2 ± 0.2 |
| 2-Methylbutyryl | 1.1 ± 0.07 | nd | 0.2 ± 0.18* | 0.2 ± 0.28 |
| Hexanoyl | 2.1 ± 0.46 | 1.1 ± 0.32 | 1.1 ± 0.37 | 1.1 ± 0.48 |
| Myristoyl | nd | 0.3 ± 0.18 | nd | 0.9 ± 0.61 |
| Palmitoyl | 2.6 ± 0.68 | 2.6 ± 1.04 | 1.7 ± 0.67 | 6.4 ± 3.47 |
| Stearoyl | 2.0 ± 0.47 | 1.3 ± 0.45 | 0.9 ± 0.30 | 2.7 ± 0.98 |
| Oleoyl | 0.2 ± 0.22 | 0.6 ± 0.43 | 0.2 ± 0.16 | 2.0 ± 1.1 |
| Linoleoyl | nd | 0.1 ± 0.11 | nd | 0.9 ± 0.66 |
| Methylmalonyl | 2.6 ± 2.97 | 2.6 ± 1.39 | 0.8 ± 0.29 | 2.4 ± 0.73 |
| Ethylmalonyl | 1.7 ± 0.71 | 0.3 ± 0.32* | nd | nd |
| Succinyl | 23.4 ± 2.55 | 13.6 ± 3.04 | 3.8 ± 0.87*,# | 7.6 ± 1.42 |
| Adipyl | 0.7 ± 0.58 | 0.5 ± 0.38 | nd | 0.1 ± 0.16 |
| Suberyl | 0.8 ± 0.68 | 1.0 ± 0.81 | nd | 0.4 ± 0.25 |
| Sebacyl | 0.6 ± 0.64 | 0.3 ± 0.27 | nd | nd |
|
| ||||
| Sum of acyls | 485.6 ± 65.6 | 251.8 ± 36.8* | 411.8 ± 57.7# | 423.2 ± 34.4# |
| Sum/Calculated | 0.87 ± 0.04 | 0.82 ± 0.05 | 0.82 ± 0.04 | 0.89±0.02 |
P < 0.05 versus adult;
P < 0.05 versus saline-treated 24 month old;
nd: below detection limit.
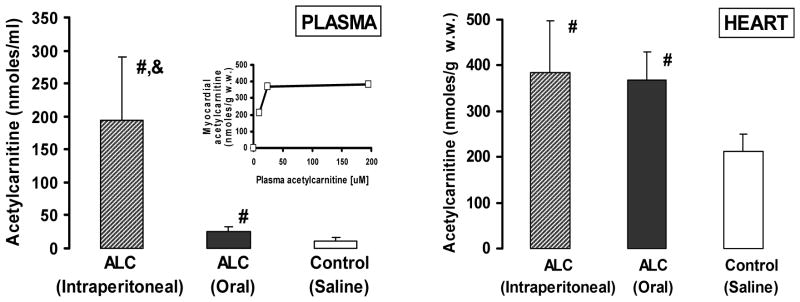
The plasma and heart acetylcarnitine contents of elderly control and acetylcarnitine-treated Fisher 344 rats are shown in Figure 1. Intraperitoneal injection of acetylcarnitine resulted in a dramatic, approximately 20-fold increase in plasma acetylcarnitine content (194.5 ± 96.3 nmoles/ml) and restoration of heart acetylcarnitine to values (383.9 ± 113.6 nmoles/g wet weight) comparable with those in adult heart (Table 1). Oral acetylcarnitine treatment also restored the myocardial acetylcarnitine content (368.5 ± 61.7 nmoles/g. w.w.) despite the much lower plasma acetylcarnitine concentration (25 ± 7.5 nmoles/ml).
Figure 1.
Plasma and myocardial acetylcarnitine content of control and acetylcarnitine-treated (intraperitoneal and oral) elderly rats. Mean ± SEM (n=4). The insert shows the myocardial acetylcarnitine content relative to the plasma acetylcarnitine concentration.
#: significantly different from control
&: significantly different from control and oral acetylcarnitine treatment.
3.2. Acetylation of mitochondrial proteins: effect of acetylcarnitine treatment
The only known reaction of acetylcarnitine is the reversible transacetylation with CoASH to form acetyl-CoA, a reaction catalyzed by carnitine acetyltransferase in the mitochondria. Therefore, increased cardiac acetylcarnitine content results in increased mitochondrial acetyl-CoA. Because the amount of acetylcarnitine given is insufficient to explain the observed beneficial effects of acetylcarnitine on an energetic basis, i.e., production of ATP via oxidation of acetylcarnitine in the tricarboxylic acid cycle and respiratory chain, we focused on mitochondrial protein acetylation as the potential mechanism for acetylcarnitine effect.
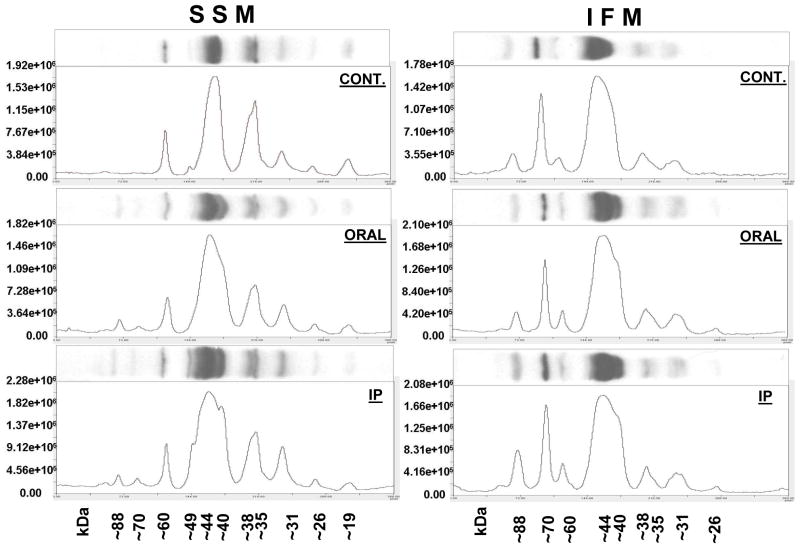
Heart SSM and IFM from elderly saline- and acetylcarnitine-treated Fisher 344 rats were subjected to SDS-PAGE and immunoblotting using affinity-purified polyclonal acetyl-lysine antibodies. As shown in Figure 2, SSM and IFM from hearts of saline-treated elderly rats display qualitative and quantitative differences in their protein acetylation profile. The ~88 kDa and ~70 kDa polypeptides, which are readily recognized by the antibodies in IFM are absent in SSM, whereas the two polypeptides with molecular weights between 29 kDa and 17 kDa observed in SSM are absent in the IFM. Differences in the profile of acetylated proteins between SSM and IFM at baseline further highlights the biochemical differences between these two mitochondrial populations (Palmer et al., 1977). Both oral and intraperitoneal acetylcarnitine treatment results in acetylation of proteins not observed in mitochondria from saline-treated elderly animals, i.e., ~88 kDa, ~76 kDa, and ~49 kDa polypeptides in SSM. These data show that exogenous acetylcarnitine, whether given orally or intraperitoneally, is efficiently taken up by the heart leading to increased mitochondrial protein acetylation.
Figure 2.
Immunoblot analysis of heart SSM and IFM isolated from saline-treated (CONT.) and orally (ORAL) and intraperitoneally (IP) acetylcarnitine-treated F344 elderly rats. SSM and IFM from three separate experiments were pooled and 50 μg aliquots subjected to SDS-PAGE and immunoblotting using affinity-purified acetyllysine antibodies.
The citrate synthase specific activity in the combined mitochondria was as follows: 1890 (cont.), 1550 (oral), and 1718 (IP) nmoles/min/mg mitochondrial protein in SSM and 1891 (control), 1869 (AcCarn-oral) and 2022 (IP-treated) nmoles/min/mg mitochondrial protein in IFM.
To gain insight into the metabolic significance of altered mitochondrial protein acetylation induced by acetylcarnitine treatment, we carried out shotgun proteomics to identify acetylated proteins in the mitochondrial fractions of interest. Acetylcarnitine treatment of elderly rats resulted in an increased number of acetylated peptides and proteins in both mitochondrial populations as compared to SSM and IFM isolated from elderly and adult untreated rats (Table 2 and supplemental Table 1). Furthermore, consistent with the age-associated decreased acetylcarnitine, fewer proteins and peptides were acetylated in both mitochondrial populations isolated from untreated elderly as compared to untreated adult rats. The decreased protein acetylation is observed despite the age-associated decreased SIRT3 expression in SSM and unaltered expression in IFM (Figure 3). These data show that in vivo treatment with acetylcarnitine results in increased acetylation of proteins already acetylated before acetylcarnitine treatment, e.g., ATP synthase α and δ subunits, Mn-superoxide dismutase, mitochondrial aspartate aminotransferase, as well as in acetylation of proteins not acetylated before acetylcarnitine treatment, e.g., long-chain acyl-CoA dehydrogenase, 3-hydroxyacyl-CoA dehydrogenase, NAD-dependent isocitrate dehydrogenase, and phosphate carrier (supplemental Table 1). Congruent with the immunoblotting data, there are differences in protein acetylation specific for SSM or IFM. For example, the triacetylated peptide K.EMF317KCG320KT322K.T unique to S13A1_RAT (solute carrier family 13 member 1) is found only in the IFM in all treatment groups. In contrast, K.QLAAEQE95KDIR.V unique to ETFD_RAT (electron transfer flavoprotein-ubiquinone oxidoreductase) is found only in SSM (supplemental Table 1).
Table 2.
Number of acetylated proteins and peptides in SSM and IFM isolated from 24 month old acetylcarnitine treated (AcCarn) and 24 months and 6 months old saline-treated (Cont.) Fisher 344 rats. Data were obtained on mitochondria pooled from three separate experiments.
|
|
||||||
|---|---|---|---|---|---|---|
| SSM | IFM | |||||
|
| ||||||
| 24m AcCarn | 24m Cont. | 6m Cont. | 24m AcCarn | 24m Cont. | 6m Cont. | |
| Number of acetylated proteins | 38 | 28 | 32 | 44 | 33 | 36 |
| Number of acetylated peptides | 58 | 40 | 42 | 64 | 42 | 47 |
Figure 3.
SIRT3 expression in SSM and IFM from 6month old adult and 24 month old elderly Fisher 344 rats (n=7). Protein load was normalized to citrate synthase activity (100 mU/lane).
&: statistically significant vs. SSM from 6 month old adult;
*: statistically significant vs. SSM from 6 month old adult.
3.3. Effect of acetylcarnitine treatment on mitochondrial enzyme activities and oxidative phosphorylation
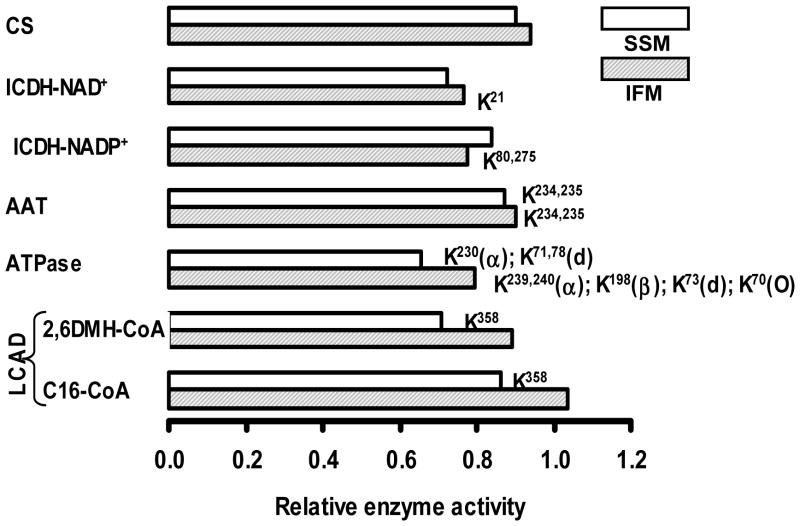
It has been shown that acetylation of enzymes impacts their catalytic activity resulting either in inhibition or stimulation (North and Sinclair, 2007; Lombard et al., 2011; Fritz et al., 2012; Pereira et al., 2012). In order to determine the effect of protein acetylation on enzyme activity we measured the activity of enzymes whose acetylation status was affected by acetylcarnitine treatment (Figure 4 and supplemental Table 1). Acetylcarnitine administration led to acetylation of LCAD on K358 in SSM with decreased activity, but not in IFM. Complex V was acetylated in both SSM and IFM but at different lysine residues. In IFM, subunits alpha and delta of ATP synthase were acetylated on K239, 240 and K70, respectively, and in SSM on residues K230 and K78. Acetylation of complex V was associated with decreased catalytic activity in both mitochondrial populations. Following acetylcarnitine treatment, aspartate aminotransferase was acetylated on residues K234, 235 in both mitochondrial populations that was accompanied by decreased activity. NADP+-dependent ICDH was acetylated on multiple lysine residues in the two mitochondrial populations; acetylcarnitine treatment lead to the acetylation of two additional sites, i.e., K80,275 only in IFM. The activity of NADP+-dependent ICDH was decreased in both, SSM and IFM. NAD+-dependent ICDH was only acetylated following acetylcarnitine treatment and only in the IFM. Citrate synthase activity was determined as control as acetylcarnitine treatment had no effect on the enzyme’s acetylation status. Thus, acetylation of the examined enzymes revealed a tendency for decreasing catalytic activity. This trend in decrease was more prevalent in SSM than in IFM.
Figure 4.
Comparison of the effect of acetylation on the activity of enzymes in SSM (open bars) and IFM (shaded bars) isolated from hearts of 24 month-old acetylcarnitine- (n=4) and saline-treated (n=2) Fisher 344 rats. Acyl-CoA dehydrogenase was assayed using palmitoyl- (C16), and 2,6-dimethylheptanoyl-CoA (2,6DMH-CoA) as substrates. Acetylated lysine residues are shown next to the respective columns.
Abbreviations used: CS - citrate synthase; ICDH-NAD+ - NAD+-dependent isocitrate dehydrogenase; ICDH-NADP+ - NADP+-dependent isocitrate dehydrogenase; AAT - aspartate aminotransferase; ATPase - complex V; LCAD - long-chain acyl-CoA dehydrogenase.
The activities are normalized to values obtained with mitochondria from saline-treated rats and are listed
Specific activities (nmoles/min/mg mitochondrial protein) in control SSM: CS=2100; ICDH-NAD+=12.6; ICDH-NADP+=1616; AAT=3923; ATPase=2064; LCAD-2,6DMH-CoA=187; LCAD-C16-CoA=91.
Specific activities (nmoles/min/mg mitochondrial protein) in control IFM: CS=2508; ICDH-NAD+=9.7; ICDH-NADP+=1941; AAT=4671; ATPase=2673; LCAD-2,6DMH-CoA=139; LCAD-C16-CoA=127.
In order to assess if changes in mitochondrial enzyme activities, specifically complex V activity, affected mitochondrial respiration we determined oxidative phosphorylation in SSM and IFM from 6 month and 24 month old control animals and from 24 month old acetylcarnitine treated rats. Mitochondrial oxidative phosphorylation assessed with glutamate and duroquinol as substrates, is significantly and selectively decreased in cardiac IFM with aging, but without affecting the ADP/O ratio and maximum respiratory capacity and is restored by in vivo acetylcarnitine treatment to values observed with mitochondria from hearts of adult animals (Table 3). Since the ADP/O ratios are unaffected ATP production is returned along with the oxidative rates. This finding is in agreement with previous data from our laboratory (Lesnefsky et al 2006). The effect of acetylcarnitine on restoration of CI- and CIII-dependent mitochondrial respiration was not reproduced by in vivo treatment of elderly Fisher 344 rats with propionylcarnitine despite restitution of free and total carnitine content to that of adult levels (manuscript in preparation).
Table 3.
Respiratory properties of subsarcolemmal and interfibrillar mitochondria determined with complex I (glutamate) and complex III (duroquinol) substrates. Oxygen consumption rates are expressed as nano Atom oxygen/minute/mg protein. The numbers represent the mean±SE of five (6m and 24m control) and three (24m AC) separate experiments.
| Subsarcolemmal mitochondria (SSM) | |||||||
|---|---|---|---|---|---|---|---|
| State 3 | State 4 | RCR | ADP/O | 2mM ADP | Uncoupled | ||
| Glutamate | 6m | 163.0±14.9 | 11.0±2.2 | 17.0±2.0 | 2.81±0.13 | 178.0±19.6 | 170.0±14.8 |
| 24m | 164.0±9.5 | 12.0±1.4 | 14.0±1.7 | 2.84±0.07 | 170.0±13.6 | 167.0±14.7 | |
| 24m AC | 181.0±29.5 | 16.0±3.5 | 12.0±0.7 | 2.58±0.06 | 195.0±36.2 | 191.0±29.7 | |
| DHQ | 6m | 376.0±24.4 | 85.0±4.1 | 4.0±0.2 | 1.58±0.02 | 346.0±17.6 | 418.0±26.5 |
| 24m | 391.0±16.4 | 91.0±3.2 | 4.0±0.2 | 1.54±0.03 | 392.0±18.9 | 455.0±20.2 | |
| 24m AC | 416.0±12.2 | 108.0±3.2 | 4.0±0.01 | 1.53±0.01 | 423.0±1.6 | 455.0±67.8 | |
| Interfibrillar mitochondria (IFM) | |||||||
| State 3 | State 4 | RCR | ADP/O | 2mM ADP | Uncoupled | ||
| Glutamate | 6m | 249.0±22.3 | 19.0±3.2 | 14.0±2.7 | 2.91±0.09 | 263.0±31.0 | 263.0±36.0 |
| 24m | 202.0±8.9* | 19.0±2.5 | 11.0±1.2 | 2.88±0.07 | 192.0±18.6* | 218.0±13.4 | |
| 24m AC | 264.0±4.3** | 20.0±3.5 | 13.0±2.1 | 2.94±0.37 | 268.0±10.8** | 264.0±17.1 | |
| DHQ | 6m | 554.0±28.6 | 98.0±6.8 | 6.0±0.32 | 1.62±0.06 | 535.0±36.9 | 620.0±28.0 |
| 24m | 437.0±22.6* | 105.0±5.9 | 4.0±0.1 | 1.59±0.04 | 452.0±18.8* | 553.0±40.6 | |
| 24m AC | 574.0±72.9** | 169.0±62.3 | 4.0±0.8 | 2.08±0.5 | 538.0±14.2** | 732.0±46.3 | |
2.0mM ADP was used to measure maximal oxidative phosphorylation.
and ** significantly different (P<0.05) from 6 month old control and 24 month old acetylcarnitine (AC) treated and 24 month old acetylcarnitine (AC) treated animals.
3.4. Effect of acetylcarnitine treatment on mitochondrial protein abundance
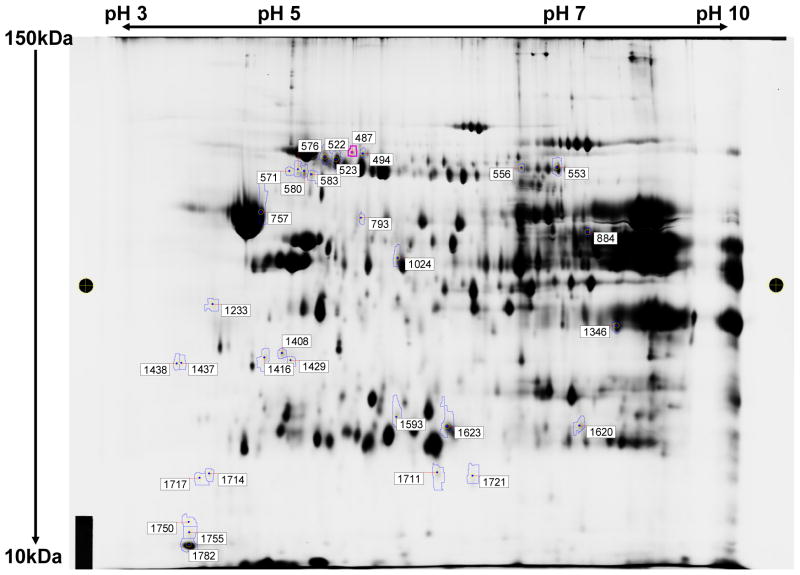
To appraise if in vivo acetylcarnitine treatment affected the amount of mitochondrial proteins we carried out 2D-DIGE analysis. In this experiment, heart IFM harboring the aging defect, isolated from elderly saline-treated and acetylcarnitine-treated (IP) Fisher 344 rats were subjected to 2D-DIGE for quantitative evaluation of the effect of acetylcarnitine treatment on mitochondrial protein abundance. This technique is useful for separation and determination of the expression of soluble and peripheral proteins but not for the hydrophobic integral membrane proteins such as the mitochondrial-encoded subunits of the respiratory chain complexes. As shown in Figure 5 acetylcarnitine treatment altered the abundance of 31 protein spots two-fold or greater (boxed numbers). Among the proteins with altered amounts, 23 were decreased and eight increased in abundance. Interestingly, in a recent study on rat liver mitochondrial acetylome, it has been shown that in vivo treatment of elderly rats with acetylcarnitine altered the abundance of 26 proteins with 22 being decreased (Musicco et al., 2011; Pesce et al., 2012). In this latter study several proteins with decreased amount were identical to those found in the present study (marked by asterisks in Table 4). All proteins that showed altered abundance were nuclear encoded and, with the exception of voltage-dependent anion channel 2, were soluble or peripheral membrane proteins. These data show that exogenous acetylcarnitine affects the abundance of mitochondrial proteins in the heart.
Figure 5.
2-DIGE of rat heart IFM from saline- and acetylcarnitine-treated 24month old Fischer 344 rats. Equal amounts of IFM from three separate experiments were combined and processed for analysis as described under the Methods section. The protein molecular mass scale in kDa is depicted on the y-axis while the x-axis shows the range of isoelectric points. Thirty-one protein spots (numbers in boxes) revealed a two-fold or greater change in protein expression.
Table 4.
Identification of IFM proteins with two-fold or greater altered abundance induced by intraperitoneal injection of 24 month old Fisher 344 rats with acetylcarnitine. Data were obtained from IFM pooled from three separate experiments.
| Spot number | Protein name | Volume ratio |
|---|---|---|
| 523 | *Glucose regulated protein (mortalin/heat shock protein 70) | −3.07 |
| 553 | *Aconitase 2 (mitochondrial) | −2.04 |
| 556 | *Aconitase 2 (mitochondrial) (identified with three peptides | −2.03 |
| 576 | *Inner membrane protein (mitofilin) | −2.73 |
| 580 | *Inner membrane protein (mitofilin) Inner membrane protein, isoform CRA_a |
−3.41 |
| 757 | ATP synthase beta subunit | −3.85 |
| 793 | Aldehyde dehydrogenase 2 (mitochondrial) | −2.79 |
| 884 | NADH dehydrogenase ubiquinone flavoprotein 1 Isocitrate dehydrogenase 2 (NADP+) *Long-chain acyl-CoA dehydrogenase, very long chain |
2.3 |
| 1233 | Alfa-tropomyosin (sequence not found in rat) | −3.62 |
| 1346 | L-3-hydroxyacyl-coenzyme A dehydrogenase Voltage-dependent anion channel 2 Electron transfer flavoprotein, alfa |
2.03 |
| 1408 | Hydroxysteroid dehydrogenase like 2 | 2.05 |
| 1416 | ATP synthase beta subunit | −2.55 |
| 1429 | Hydroxysteroid dehydrogenase like 2 | 2.2 |
| 1437 | Complement component1, q subcomponent binding protein | −6.39 |
| 1438 | Complement component1, q subcomponent binding protein | −6.09 |
| 1593 | Keratin | 2.04 |
| 1620 | Glioblastoma amplified sequence Malate dehydrogenase, mitochondrial Electron transfer flavoprotein, alfa |
3.46 |
| 1623 | *Peroxiredoxin 3 Lysophospholipase 1 |
−2.43 |
| 1711 | 3-oxoacid CoA transferase | 3.74 |
| 1721 | Isocitrate dehydrogenase 2 (NADP+) | 2.52 |
Asterisk: proteins with similar changes in expression level found in rat liver mitochondria after in vivo acetylcarnitine treatment (Musicco et al., 2011).
4. Discussion
Using the Fisher 344 rat as an aging model in previous studies we have shown that the age-associated defect in CIII activity is selective to IFM and that the defect is abolished by in vivo treatment of elderly rats with acetylcarnitine. Following treatment with acetylcarnitine oxidative phosphorylation and CIII activity were restored to values observed in IFM from adult saline-treated rats. Furthermore, compared to hearts from saline-treated animals, acetylcarnitine treatment prevented the additional ischemia/reperfusion-induced decrease in contractile function as assessed by developed pressure, +dP/dt, and dP/dt. Herein we show that acetylcarnitine treatment restores cardiac acetylcarnitine to levels of adult hearts, increases mitochondrial protein acetylation, and affects enzyme activities and mitochondrial protein abundance.
In the present study we demostrate that with age there is a decrease in cardiac acetylcarnitine (as well as free and total carnitine) that is restored to adult values by either intraperitoneal or oral administration of exogenous acetylcarnitine (Table 1 and Figure 1). Exogenous acetylcarnitine administered IP or orally enters tissues by means of the Na+-dependent organic cation transporter (OCTN2) present in the plasma membrane (reviewed in Tein, 2003). The markedly different plasma acetylcarnitine concentration after intraperitoneal and oral acetylcarnitine administration, i.e., 194 μM vs. 25 μM, reflects the low bioavailability of oral acetylcarnitine (approx. 10–15%). Despite this large difference in plasma acetylcarnitine between IP and oral administration, the myocardial acetylcarnitine content is the same. Because the Km for OCTN2 is around 2–3μM, the transporter already is operating at Vmax at 25 μM plasma acetylcarnitine concentration (see insert in Figure 1). The increase in myocardial free carnitine content is most likely due to intracellular metabolism of the administered exogenous acetylcarnitine.
The rapid restoration of cardiac acetylcarnitine (three hours after IP injection of acetylcarnitine) is expected considering the published transport rates of carnitine in isolated perfused working hearts and the substrate specificity of OCTN2. It has been shown that the unidirectional (Na+-dependent) transport rates of carnitine range from 111 nmol/g dry weight/hr to 210 nmol/g dry weight/hr, depending on carnitine concentration (Vary, T.C. and Neely, J.R. 1982a; Vary, T.C. and Neely, J.R. 1982b; Vary, et al. 1983). Furthermore, OCTN2 catalyzes the uptake of acetylcarnitine with an affinity comparable to that of free carnitine (Wu et al. 1999). Thus, IP administered acetylcarnitine is readily taken up by the aged heart and affects mitochondrial protein acetylation and enzyme activity.
Cytosolic acetylcarnitine enters the mitochondria through the carnitine:acylcarnitine translocase (CACTL) localized in the mitochondrial inner membrane (reviewed in Zammit et al., 2009; Indiveri et al., 2011). In the mitochondrial matrix, acetylcarnitine rapidly equilibrates with the acetyl-CoA pool via reversible transesterification catalyzed by carnitine acetyltransferase (CAT) (Pearson and Tubbs, 1967; Oram et al., 1973; Lysiak et al., 1988). Because all three of these proteins, i.e., OCTN2, CACTL, and CAT, are highly expressed in heart and skeletal muscle as compared to other tissues, acetylcarnitine can be considered as a specific intramitochondrial acetyl-donor in muscle. The acetyl group of acetyl-CoA then is available for several reactions, including protein acetylation. In fact, acetylcarnitine treatment leads to acetylation of mitochondrial proteins that differ both quantitatively and qualitatively from the acetylation pattern observed in mitochondria from untreated animals (Figure 2 and supplemental Table 1). Following in vivo acetylcarnitine treatment of elderly rats, the number of acetylated proteins as well as the number of acetylated peptides was increased in both mitochondrial populations as compared to mitochondria isolated from saline-treated elderly and adult rats (Table 2 and supplemental Table 1). Furthermore, the number of acetylated proteins and peptides was lower in both SSM and IFM from elderly control as compared to untreated adult rats despite the decreased expression of the major mitochondrial NAD+-dependent deacetylase, SIRT3, in SSM (Figure 3). SIRT3 expression was unaltered by age in the IFM. These findings are in apparent conflict with the expected inverse change in Sirt3 expression and mitochondrial protein acetylation. The likely reason or explanation for this conundrum is the age-induced decrease in mitochondrial acetylation potential. In heart, acetylcarnitine is in equilibrium with acetyl-CoA (Kerner et al. 2014), the content of acetylcarnitine reflects that of acetyl-CoA (Pearson and Tubbs 1967), and over 90% of acetyl-CoA is confined to the mitochondrial matrix (Oram et al. 1973). Thus, decreased cardiac acetylcarnitine mirrored by decreased mitochondrial acetyl-CoA and consequently decreased mitochondrial acetylation potential leads to decreased mitochondrial protein acetylation via mitochondrial protein acetyltransferase(s), i.e., GCN5L1 (vide infra). This also could explain the decreased protein acetylation in IFM despite that Sirt3 expression was not being affected by aging.
Mitochondrial protein acetyltransferase activity has been recently identified and characterized (Scott et al., 2012). The presence of GCN5L1 with protein-lysine acetyltransferase activity in mitochondria provides the basis for the finding that mitochondrial-encoded proteins, such as the hydrophobic respiratory chain subunits of CI, as well as ATP synthase Fo subunit 8, are acetylated (Kim et al., 2006). Because the Km values (0.64–8.5μM) for acetyl-CoA on several select protein acetyltransferases (Albaugh et al., 2011) fall into the range of cardiac acetyl-CoA concentrations, i.e., 2–10μM, depending on the experimental conditions (Latipaa et al., 1985; Latipaa et al., 1989; Kudo et al., 1995; Gilibili et al., 2011) it is reasonable to propose that following acetylcarnitine treatment the increased intramitochondrial acetyl-CoA concentration increases the velocity of protein acetyltransferase(s). The fact that during fasting the hepatic acetyl-CoA content is increased, combined with the finding that fasting leads to the acetylation of a greater number of mitochondrial proteins in liver (Kim et al., 2006; Pougovkina et al., 2014), is consistent with the notion that mitochondrial acetyl-CoA content affects the acetylation of mitochondrial proteins. Alternatively, increased acetylation of mitochondrial proteins after acetylcarnitine treatment could arise from inhibition of protein deacetylation or a combination of increased acetylation and decreased deacetylation. Nonetheless, if the acetylcarnitine-induced increase in mitochondrial protein acetylation was due to inhibition of deacetylation, no new, previously non-acetylated proteins should have been observed after acetylcarnitine treatment. However, acetylcarnitine treatment led to acetylation of proteins not observed in mitochondria from untreated animals as well as to acetylation on additional lysine residues of already acetylated proteins (Figure 2 and in supplemental Table 1). These data indicate that exogenous acetylcarnitine was the acetyl-donor for protein acetylation.
Most of the acetylated proteins found in both SSM and IFM are enzymes of fatty acid β-oxidation, the tricarboxylic acid cycle (TCA cycle), and different subunits of ATP-synthase/complex V. Several of these proteins are acetylated at multiple sites (supplemental Table 1). The few studies where protein acetylation was directly linked to function were carried out in cells in culture or on recombinant proteins. Zhao et al. (Zhao et al., 2010) found that in human liver the peroxisomal enoyl-CoA/3-hydroxyacyl-CoA dehydrogenase (EHHADH) was acetylated on multiple lysine residues. Treatment of cultured Chang human liver cells and HEK293T cells ectopically expressing EHHADH with histone deacetylase (HDAC) and sirtuin inhibitors doubled the enzymatic activity. Similarly, adding fatty acid to the culture medium increased EHHADH activity in HEK293T cells, suggesting a physiological role of acetylation in regulation of hepatic peroxisomal fatty acid oxidation. In contrast to activation of EHHDAC by acetylation, ablation of SIRT3 increased the acetylation of LCAD and decreased mitochondrial fatty acid oxidation in mouse liver (Hirshey et al., 2010). Deletion of SIRT3 resulted in hyperacetylation of LCAD with K42 showing the greatest acetylation among the eight acetylated lysine residues (K42,156,189,240,254,318,322,358). Other studies showed that deletion of SIRT3 caused a significant increase in the acetylation of K81, K322 and K358 (Hebert et al., 2013; Rardin et al., 2013), whereas the acetylation state of K42 was unaltered (Rardin et al., 2013). By expressing mouse liver LCADs in which the putative lysine residues were mutated to arginine one study implicated K42 (Hirshey et al., 2010) and another study K318/322 (Bharati et al., 2013 Rardin et al., 2013) as crucial for regulation of LCAD activity. However, liver LCAD activity was not reported in either of these studies. In contrast to the above referenced studies on the inhibitory effect of protein acetylation on mitochondrial fatty acid oxidation, recent studies have shown that increased protein acetylation induced by obesity and SIRT3 knockout increased mitochondrial fatty acid oxidation (Alrob et al. 2014). These data highlight the complexity of the effect of protein acetylation on protein function.
In the present study we found that seven enzymes involved in mitochondrial fatty acid oxidation, including LCAD, are acetylated on one or two lysine residues (supplemental Table 1). Since acetylation of LCAD has been reported to affect its catalytic activity in mouse liver we examined the activity of this enzyme in SSM and IFM isolated from hearts of 24 month saline and acetylcarnitine treated Fisher 344 rats. After acetylcarnitine treatment LCAD is acetylated at K358 in SSM, the same site found to be acetylated in rat liver, but is not acetylated in IFM. Acetylation of K358 is accompanied by ~30% decreased activity with the LCAD-specific substrate 2,6-dimethylhepatnoyl-CoA and by ~15% with palmitoyl-CoA as the substrate for LCAD as well as for very long-chain acyl-CoA dehydrogenase. In IFM, where K358 was not acetylated, no difference in LCAD activity was found between IFM isolated from hearts of saline and acetylcarnitine treated rats.
In addition to fatty acid oxidation enzymes, we found that six out of the eight TCA cycle enzymes are acetylated in rat heart. In human liver, all eight TCA cycle enzymes are acetylated (Zhao et al., 2010). Again, the effect of acetylation on any of the TCA cycle enzymes in heart mitochondria is unknown. In liver ICDH is acetylated on multiple sites (Kim et al.,2006; Choudhary et al., 2009; Zhao et al., 2010). In a recent study the regulated site was mapped to K143 by showing that acetylation inhibits and deacetylation stimulates ICDH activity (Yu et al., 2012). In the present study we found that NADP+-dependent ICDH is acetylated on multiple sites in both mitochondrial populations (supplemental Table 1). Additional acetylation at K275 in IFM has no significant effect on activity. Similarly, no effect of acetylation on activity of NAD+-dependent ICDH was found.
Among all the acetylated proteins identified in the present study, including those of oxidative phosphorylation, ATP synthase (complex V) was acetylated to the greatest extent. This finding is in line with the few published data on complex V acetylation in heart (Nguyen et al. 2013, Foster et al. 2013). In hearts from saline-treated rats in our studies five subunits of complex V were acetylated either on a single or on multiple lysine residues and acetylcarnitine treatment led to acetylation of additional lysine side chains (supplemental Table 1). We found that acetylation of complex V was accompanied by a slight decrease in activity in both mitochondrial populations. However, this had no impact on oxidative phosphorylation, i.e., state 3 respiration at low and high ADP concentration, respiratory coupling ratios, and ADP/O ratios as shown by data presented in Table 3.
In contrast, skeletal muscle of SIRT3 knockout mice have increased complex V protein acetylation and decreased ATP production. Site-directed mutagenesis has shown that acetylation of K139 of oligomycin sensitivity conferring protein (OSCP) is at least in part responsible for decreased ATP production (Vassilopoulos et al. 2014). Acetylation of OSCP in regulation of complex V activity was implicated also in earlier studies; however, no specific lysine residue has been linked to inhibition (Wu et al. 2013). Similarly, deacetylation of human ATP synthase β by SIRT3 and its Drosophila homologue, dSIRT2, resulted in decreased acetylation and increased complex V activity. Deletion of dSIRT2 resulted in decreased respiration and ATP production and mutagenesis studies implicated residue K259 and K480 of the ATP synthase β subunit (Rahman et al. 2014). Again, as with LCAD, these data show the complexity of protein acetylation on catalytic activity.
Analysis of the acetylation status of aspartate aminotransferase revealed the presence of two acetylated lysine residues (K234, 235) in both mitochondrial populations detected only after acetylcarnitine treatment; however acetylation at these two sites had no significant effect on catalytic activity. Neither the acetylation status nor the catalytic activity of CS was affected by acetylcarnitine treatment and consequently was used as a control.
Besides mitochondrial proteins, several nuclear, and cytoplasmic proteins which co-purified with mitochondria during isolation also were acetylated. Many of these proteins are involved in cell signaling and transcriptional regulation and were acetylated only after acetylcarnitine treatment (supplemental Table 1) suggesting that the acetyl group of acetylcarnitine affects protein acetylation in the cytoplasm and nucleus in addition to the mitochondria. This would require the synthesis of acetyl-CoA from acetylcarnitine via CAT or from citrate through ATP-citrate lyase. In heart, CAT is most abundant in mitochondria but also is present in microperoxisomes (Connock and Perry, 1983; Usuda et al. 2006) and in nuclei (Madiraju et al., 2009), but is absent from the cytosol (Abbas et al., 1998). The contribution of acetylcarnitine via CAT to histone acetylation in cultured fibroblasts has been recently reported (Madiraju et al., 2009). The existence of an alternative route for extramitochondrial protein acetylation via ATP-citrate lyase also has been documented (Wellen et al., 2009). This pathway would require the conversion of acetylcarnitine via acetyl-CoA to citrate, export of citrate from the mitochondria into the cytosol and nucleus and regeneration of acetyl-CoA by the ATP-citrate lyase present in both compartments, at least in lipogenic tissues. In support of the latter pathway, silencing of ATP-citrate lyase (Wellen et al., 2009) or the mitochondrial citrate transporter [solute carrier 25 A1 (SLC25A1)] dramatically decreases histone acetylation (Morciano et al., 2009).
The acetylation of nuclear proteins functioning as transcriptional activators and repressors could possibly explain the finding that acetylcarnitine administration affected the abundance of 31 nuclear-encoded mitochondrial proteins in rat heart (present study) and 26 proteins in rat liver (Mussico et al., 2011). Most of the proteins whose expression was affected by acetylcarnitine administration were enzymes of mitochondrial fatty acid oxidation, TCA cycle, and subunits of ATP synthase. In this respect, it is interesting that many proteins with altered expression level also showed increased acetylation following acetylcarnitine treatment (Table 4 and supplemental Table 1). Alternatively, acetylation of proteins also could affect their turnover by increasing/decreasing their stability (Caron et al., 2005, Simonsson et al., 2005, Webster et al., 2014).
5. Conclusion
In the present study we have demonstrated that in vivo treatment of elderly rats with acetylcarnitine restored the myocardial acetylcarnitine content to adult values. Because acetylcarnitine and acetyl-CoA are in equilibrium, increased myocardial acetylcarnitine leads to increased mitochondrial acetylation potential and increased mitochondrial protein acetylation. Moreover, acetylcarnitine administration altered the expression of 31 proteins in mitochondria, possibly by altered transcription/translation or altered protein turnover. Although intraperitoneal administration of acetylcarnitine ameliorated the CIII defect in IFM (Lesnefsky et al., 2006), no acetylated peptide assigned to cytochrome b or any other CIII subunit was found by mass spectrometric analysis of the tryptic digests in the present study. Nevertheless, the fact that cardiac metabolism of propionylcarnitine involves the same enzymatic steps as acetylcarnitine and restored cardiac carnitine content to adult levels suggests that it is the acetyl moiety of acetylcarnitine that is involved in restoration of CI- and CIII-dependent respiration and in IFM.
Further studies are needed to define in more detail the mechanism of acetylcarnitine action at the molecular level and to explore the translational aspects of acetylcarnitine treatment. Acetylcarnitine diminishes the aging defect in different tissues (Paradies et al., 1992; Paradies et al., 1994; Paradies et al., 1995; Hagen et al., 1998; Iossa et al., 2002; Virmani and Binienda, 2004; Virmani et al., 2004; Lesnefsky et al., 2006; Ames, 2010) and has been tested in Clinical Phase I, II, and III studies on humans with no side effects. Since orally administered acetylcarnitine was as effective in increasing the mitochondrial acetylation potential as was intraperitoneal injection, it provides a convenient, non-invasive, and safe way to treat and to prevent the adverse cardiac effects associated with aging.
Supplementary Material
Table 1S: Acetylated proteins and peptides in SSM and IFM isolated from 6 and 24 month old saline- and 24 month acetylcarnitine-treated Fisher 344 rats.
Highlights.
Acetylcarnitine administration reverses the age-induced mitochondrial defect
In vivo acetylcarnitine treatment increases mitochondrial protein acetylation
Acetylcarnitine affects mitochondrial protein expression and enzyme activity
Acknowledgments
We thank Maria Stoll and Paul Minkler for the carnitine and acylcarnitine analysis, Dr. Mariana Rosca for carrying out the mitochondrial respiratory studies and Dr. Bernard Tandler for editorial comments. The work presented was supported in part by NIH Grant PO1 AG15885 and Sigma-Tau Pharmaceuticals and RostaQuo (Rome, Italy).
Footnotes
Disclosures
Ashraf Virmani, Aleardo Koverech, and Claudio Cavazza are employees of Sigma Tau Pharmaceuticals (Rome, Italy).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas AS, Wu G, Schulz H. Carnitine acetyltransferase is not a cytosolic enzyme in rat heart and therefore cannot function in the energy-linked regulation of cardiac fatty acid oxidation. J Mol Cell Cardiol. 1998;30:1305–1309. doi: 10.1006/jmcc.1998.0693. 1998. [DOI] [PubMed] [Google Scholar]
- Alp PR, Newsholme EA, Zammit VA. Activities of citrate synthase, and NAD+-linked and NADP+-linked isocitrate dehydrogenase in muscles from vertebrates and invertebrates. Biochem J. 1976;154:689–700. doi: 10.1042/bj1540689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrob OA, Sankaralingam S, Ma C, Wagg CS, Fillmore N, Jaswal JS, Sack MN, Lehner R, Gupta MP, Michelakis ED, Padwal RS, Johnstone DE, Sharma AM, Lopaschuk GD. Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signaling. Cardiovasc Res. 2014;103:485–497. doi: 10.1093/cvr/cvu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN. Optimal micronutrients delay mitochondrial decay and age-associated diseases. Mech Ageing Dev. 2010;131:473–479. doi: 10.1016/j.mad.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU, Bernt E. Glutamate-oxaloacetate transaminase in Methods of enzymatic analysis. 2. Vol. 2. Verlag Chemie/Academic Press; 1974. pp. 727–733. [Google Scholar]
- Bharati SS, Zhang Y, Mohsen A-W, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, Verdin E, Gibson BW, Hirshey MD, Goetzman E. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J Biol Chem. 2013;288:33837–3384. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C, Boyault C, Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. BioEssays. 2005;27:408–415. doi: 10.1002/bies.20210. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Connock MJ, Perry SR. Detection of acyl-CoA beta-oxidation enzymes in peroxisomes (microperoxisomes) of mouse heart. Biochem Int. 1983;6:545–551. [PubMed] [Google Scholar]
- Costell M, O’Connor JE, Grisolia S. Age-dependent decrease of carnitine in muscle of mice and humans. Biochem Biophys Res Commun. 1989;161:1135–1143. doi: 10.1016/0006-291x(89)91360-0. [DOI] [PubMed] [Google Scholar]
- Costell M, Grisolia S. Effect of carnitine feeding on the levels of heart and skeletal muscle carnitine of elderly mice. FEBS Lett. 1993;315:43–46. doi: 10.1016/0014-5793(93)81129-n. [DOI] [PubMed] [Google Scholar]
- Fannin SW, Lesnefsky EJ, Slabe TJ, Hassan MO, Hoppel CL. Aging selectively decreases oxidative capacity in rat heart interfibrillar mitochondria. Arch Biochem Biophys. 1999;37:399–407. doi: 10.1006/abbi.1999.1508. [DOI] [PubMed] [Google Scholar]
- Foster DB, Liu T, Rucker J, O’Meally RN, Devine LR, Cole RN, O’Rourke B. The cardiac acetyl-lysine proteome. PLOS ONE. 2013;8(7):e67513. doi: 10.1371/journal.pone.0067513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolkis VV, Frolkis RA, Mkhitarian LS, Fraifeld VE. Age-dependent effects of ischemia and reperfusion on cardiac function and Ca2+ transport in myocardium. Gerontology. 1991;37:233–239. doi: 10.1159/000213266. [DOI] [PubMed] [Google Scholar]
- Gilibili RR, Kandaswamy M, Sharma K, Giri S, Rajagopal S, Mullangi R. Development and validation of a highly sensitive LC-MS/MS method for simultaneous quantitation of acetyl-CoA and malonyl-CoA in animal tissues. Biomed Chromat. 2011;25:1352–1359. doi: 10.1002/bmc.1608. [DOI] [PubMed] [Google Scholar]
- Hagen TM, Ingersoll RT, Wehr CM, Lykkesfeldt J, Vinarsky V, Bartholomew JC, Song MH, Ames BN. Acetyl-L-carnitine fed to old rats partially restores mitochondrial function and ambulatory activity. Proc Natl Acad Sci USA. 1998;95:9562–9566. doi: 10.1073/pnas.95.16.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphal MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshey MD, Shimazu T, Goetzman E, Jiang E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Alt F, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–126. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppel C, DiMarco JP, Tandler B. Riboflavin and rat hepatic cell structure and function. J Biol Chem. 1979;254:4164–4170. [PubMed] [Google Scholar]
- Hoppel CL, Kerr DS, Dahms B, Roessmann U. Deficiency of the reduced nicotinamide adenine nucleotide dehydrogenase component of complex I of mitochondrial electron transport. J Cin Invest. 1987;80:71–77. doi: 10.1172/JCI113066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiveri C, Iacobazzi V, Tomazzi A, Giangregorio N, Infantino V, Convertini P, Console L, Palmieri F. The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol Aspects Med. 2011;32:223–233. doi: 10.1016/j.mam.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Iossa S, Mollica MP, Lionetti L, Crescenzo R, Botta M, Barletta A, Liverini G. Acetyl-L-carnitine supplementation differently influences nutrient partitioning, serum leptin concentration and skeletal muscle mitochondrial respiration in young and old rats. J Nutr. 2002;132:636–642. doi: 10.1093/jn/132.4.636. [DOI] [PubMed] [Google Scholar]
- Kerner J, Turkaly PJ, Minkler PE, Hoppel CL. Aging skeletal muscle mitochondria in the rat: decreased uncoupling protein-3 content. Am J Physiol Endocrinol Metab. 2001;281:E1054–E1062. doi: 10.1152/ajpendo.2001.281.5.E1054. [DOI] [PubMed] [Google Scholar]
- Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL. Fatty acid chain elongation in palmitate-perfused working rat heart. J Biol Chem. 2014;289:10223–10234. doi: 10.1074/jbc.M113.524314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molec Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kudo N, Barr AJ, Barr RL, Desai S, Lopaschuk GD. High rates of fatty acid oxidation during reperfusion of ischemic hearts are associated with a decrease in malonyl-CoA levels due to increase in 5′-AMP-activated protein kinase inhibition of acetyl-CoA carboxylase. J Biol Chem. 1995;270:17513–17520. doi: 10.1074/jbc.270.29.17513. [DOI] [PubMed] [Google Scholar]
- Latipaa PM, Peuhkurinen KJ, Hiltunen JK, Hassinen IE. Regulation of pyruvate dehydrogenase during infusion of fatty acids of varying chain lengths in the perfused rat heart. J Mol Cell Cardiol. 1985;17:1161–1171. doi: 10.1016/s0022-2828(85)80112-7. [DOI] [PubMed] [Google Scholar]
- Latipaa PM. Energy –linked regulation of mitochondrial fatty acid oxidation in isolated perfused rat heart. J Mol Cell Cardiol. 1989;21:765–771. doi: 10.1016/0022-2828(89)90715-3. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Gallo DS, Ye J, Whittingham TS, Lust WD. Aging increases ischemia-reperfusion injury in the isolated, buffer-perfused heart. J Lab Clin Med. 1994;124:843–851. [PubMed] [Google Scholar]
- Lesnefsky EJ, Lundergan CF, Hodgson JM, Nair R, Reiner JS, Greenhouse SE, Califf RM, Ross AM. Increased left ventricular dysfunction in elderly patients despite successful thrombolysis: the GUSTO-I angiographic experience. J Am Coll Cardiol. 1996;28:331–337. doi: 10.1016/0735-1097(96)00148-9. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Gudz TI, Moghaddas S, Migita CT, Ikeda-Saito M, Turkaly PJ, Hoppel CL. Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome c binding site. J Mol Cell Cardiol. 2001;33:37–47. doi: 10.1006/jmcc.2000.1273. 2001. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. Mitochondrial dysfunction in cardiac disease: Ischemia-reperfusion, aging, and heart failure. J Mol Cell Cardiol. 2001;33:1065–1089. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Hoppel CL. Oxidative phosphorylation and aging. Ageing Res Rev. 2006;5:402–433. doi: 10.1016/j.arr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, He DC, Moghaddas S, Hoppel CL. Reversal of mitochondrial defects before ischemia protects the aged heart. FASEB J. 2006;20:E840–E845. doi: 10.1096/fj.05-4535fje. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Tishkoff DX, Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb Exp Pharmacol. 2011;206:163–188. doi: 10.1007/978-3-642-21631-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AC, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lysiak W, Lilli K, DiLisa F, Toth PP, Bieber LL. Quantitation of the effect of L-carnitine on the levels of acid-soluble short-chain acyl-CoA and CoASH in rat heart and liver mitochondria. J Biol Chem. 1988;263:1151–1156. [PubMed] [Google Scholar]
- Madiraju P, Pande SV, Prentki M, Madiraju SRM. Mitochondrial acetylcarnitine provides acetyl groups for nuclear histone acetylation. Epigenetics. 2009;4:399–403. doi: 10.4161/epi.4.6.9767. [DOI] [PubMed] [Google Scholar]
- Minkler PE, Stoll MS, Ingalls ST, Yang S, Kerner J, Hoppel CL. Quantification of carnitine and acylcarnitines in biological matrices by HPLC electrospray ionization-mass spectrometry. Clin Chem. 54:1451–1462. doi: 10.1373/clinchem.2007.099226. [DOI] [PubMed] [Google Scholar]
- Moghaddas S, Hoppel CL, Lesnefsky EJ. Aging defect at the QO site of complex III augments oxyradical production in rat heart interfibrillar mitochondria. Arch Biochem Biophys. 2003;414:59–66. doi: 10.1016/s0003-9861(03)00166-8. [DOI] [PubMed] [Google Scholar]
- Morciano P, Carrisi C, Capobianco L, Mannini L, Burgio G, Cestra G, DeBenedetto GE, Corona DF, Musio A, Cenci G. A conserved role for the mitochondrial citrate transporter Sea/SLC25A1 in the maintenance of chromosomal integrity. Hum Mol Genet. 2009;18:4180–4188. doi: 10.1093/hmg/ddp370. [DOI] [PubMed] [Google Scholar]
- Musicco C, Capelli V, Pesce V, Timperio AM, Calvani M, Mosconi L, Cantatore P, Gadaleta MN. Rat liver mitochondrial proteome: Changes associated with aging and acetyl-L-carnitine treatment. J Proteomics. 2011;74:2536–2547. doi: 10.1016/j.jprot.2011.05.041. [DOI] [PubMed] [Google Scholar]
- Nguyen TTM, Wong R, Menazza S, Sun J, Chen Y, Wang G, Gucek M, Steenbergen C, Sack MN, Murphy E. Cyclophilin D modulates mitochondrial proteome. Circ Res. 113:1308–1319. doi: 10.1161/CIRCRESAHA.113.301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland RC, Koves TR, Seiler SE, Lum H, Lust RM, Ilkayeva O, Stevens RD, Hegardt F, Muoio DM. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J Biol Chem. 2009;284:22840–22852. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B, Sinclair DA. Sirtuins: a conserved key unlocking AceCS activity. TIBS. 2007;32:1–4. doi: 10.1016/j.tibs.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram JF, Bennetch SL, Neely JR. Regulation of fatty acid utilization in isolated perfused heart. J Biol Chem. 1973;248:5299–5309. [PubMed] [Google Scholar]
- Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- Paradies G, Ruggioero FM, Gadaleta MN, Quagliariello E. The effect of aging and acetyl-L-carnitine on the activity of the phosphate carrier and on phospholipid composition in rat heart mitochondria. Biochim Biophys Acta. 1992;1103:324–326. doi: 10.1016/0005-2736(92)90103-s. [DOI] [PubMed] [Google Scholar]
- Paradies G, Ruggiero FM, Petrosillo G, Gadaleta MN, Quagliariello E. The effect of aging and acetyl-L-carnitine on the activity of cytochrome oxidase and adenine nucleotide translocase in rat heart mitochondria. FEBS Lett. 1994;350:213–215. doi: 10.1016/0014-5793(94)00763-2. [DOI] [PubMed] [Google Scholar]
- Paradies G, Ruggiero FM, Petrosillo G, Gadaleta MN, Quagliariello E. Carnitine-acylcarnitine translocase activity in cardiac mitochondria from aged rats: the effect of acetyl-L-carnitine. Mech Aging Dev. 1995;84:103–112. doi: 10.1016/0047-6374(95)01636-8. [DOI] [PubMed] [Google Scholar]
- Pearson DJ, Tubbs PK. Carnitine and derivatives in rat tissues. Biochem J. 1967;105:953–963. doi: 10.1042/bj1050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CV, Lebiedzinska M, Wieckowski MR, Oliviera PJ. Regulation and protection of mitochondrial physiology by sirtuins. Mitochondrion. 2012;12:66–76. doi: 10.1016/j.mito.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Pesce V, Fracasso F, Musicco C, Lezza AMS, Cantatore P, Gadaleta MN. Acetyl-L-carnitine dietary supplementation to old rats increases mitochondrial transcription factor A content in rat hindlimb skeletal muscle. Ann NY Acad Sci. 2004;1019:430–433. doi: 10.1196/annals.1297.077. [DOI] [PubMed] [Google Scholar]
- Pesce V, Nicassio L, Fracasso F, Musicco C, Cantatore P, Gadaleta MN. Acetyl-L-carnitine activates the peroxisome proliferator-activated receptor-γ coactivators PGC-1α/PGC-1β-dependent signaling cascade of mitochondrial biogenesis and decreases the oxidized peroxiredoxins content in old rat liver. Rejuvenation Res. 2012;15:136–139. doi: 10.1089/rej.2011.1255. [DOI] [PubMed] [Google Scholar]
- Pougovkina O, te Brinke H, Ofman R, van Cruchten AG, Kulik W, Wanders RJA, Houten SM, de Boer VCJ. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Human Mol Genetics. 2014:1–10. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- Rahman M, Niraka NK, Singh A, Zhu LL, Taguchi K, Bamba T, Fukusaki E, Shaw LM, Lambright DG, Acharya JK, Acharya UR. Drosophila Sirt2/mammalian SIRT3 deacetylates ATP synthase β and regulates complex V activity. J Cell Biol. 2014;206:289–305. doi: 10.1083/jcb.201404118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci USA. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosca MG, Okere IA, Sharma N, Stanley WC, Recchia FA, Hoppel CL. Altered expression of the adenine nucleotide translocase isoforms and decreased ATP synthase activity in skeletal muscle mitochondria in heart failure. J Mol Cell Cardiol. 2009;46:927–935. doi: 10.1016/j.yjmcc.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 2005;280:21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- Srere PA. In: Citrate synthase, in Methods in Enzymology. Lowenstein JM, editor. Vol. 13. Academic Press; 1969. pp. 7–11. [Google Scholar]
- Tanaka Y, Sasaki R, Fukui F, Waki H, Kawabata T, Okazaki M, Hasegawa K, Ando S. Acetyl-L-carnitine supplementation restores decreased tissue carnitine levels and impaired lipid metabolism in aged rats. J Lipid Res. 2004;45:729–735. doi: 10.1194/jlr.M300425-JLR200. [DOI] [PubMed] [Google Scholar]
- Tein I. Carnitine transport: pathophysiology and metabolism of known molecular defect. J Inherit Metab Dis. 2003;26:147–169. doi: 10.1023/a:1024481016187. [DOI] [PubMed] [Google Scholar]
- Usuda N, Nakazawa A, Terasawa M, Reddy JK, Nagata T. Immunocytochemical study of the ultrastructure of peroxisomes and the effects of peroxisome proliferators. Ann NY Acad Sci. 2006;804:297–309. doi: 10.1111/j.1749-6632.1996.tb18624.x. [DOI] [PubMed] [Google Scholar]
- Vary TC, Neely JR. Characterization of carnitine transport in isolated perfused adult rat hearts. Am J Physiol Heart Circ Physiol. 1982;242:H585–H592. doi: 10.1152/ajpheart.1982.242.4.H585. [DOI] [PubMed] [Google Scholar]
- Vary TC, Neely J. A mechanism for reduced myocardial carnitine levels in diabetic animals. Am J Physiol Heart Circ Physiol. 1982;243:H154–H158. doi: 10.1152/ajpheart.1982.243.2.H154. [DOI] [PubMed] [Google Scholar]
- Vary TC, Neely JR. Sodium dependence of carnitine transport in isolated perfused adult rat heart. Am J Physiol Heart Circ Physiol. 1983;244:H247–H252. doi: 10.1152/ajpheart.1983.244.2.H247. [DOI] [PubMed] [Google Scholar]
- Vassiloloulos A, Pennington JD, Andersson T, Rees DM, Bosley AD, Feamley IA, Harn A, Flynn CR, Hill S, Rose KL, Kim S-S, Deng C-X, Walker JE, Gius D. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21:551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virmani A, Binienda Z. Role of carnitine esters in brain pathology. Mol Aspects Med. 2004;25:533–59. doi: 10.1016/j.mam.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Virmani A, Gaetani F, Binienda Z, Xu A, Duhart H, Ali SF. Role of mitochondrial dysfunction in neurotoxicity of MPP+: partial protection of P12 cells by acetyl-L-carnitine. Ann NY Acad Sci. 2004;1025:267–273. doi: 10.1196/annals.1316.033. [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Denis S, Ruiter JPN, Ijlst L, Dacremont G. 2,6-Dimethylheptanoyl-CoA is a specific substrate for long-chain acyl-CoA dehydrogenase (LCAD): evidence for a major role of LCAD in branched chain fatty acid oxidation. Biochim Biophys Acta. 1998;1393:35–45. doi: 10.1016/s0005-2760(98)00053-8. [DOI] [PubMed] [Google Scholar]
- Webster BR, Scott I, Traba J, Han K, Sack MN. Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim Biophys Acta. 2014;1841:525–534. doi: 10.1016/j.bbalip.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Huang W, Prasad PD, Seth P, Rajan DP, Leibach FH, Chen J, Conway SJ, Ganapathy V. Functional characteristcs and tissue distribution pattern of organic cation transporter 2 (OCTN2), an organic cation/carnitine transporter. J Pharmacol Exptl Therap. 1999;290:1482–1492. [PubMed] [Google Scholar]
- Wu W-T, Lee H-C, Liao C-C, Wei Y-H. Regulation of mitochondrial F0F1ATPase activity by Sirt3-catalyzed deacetylation and its deficiency in human cells harboring 4977 bp deletion of mitochondrial DNA. Biochim Biophys Acta. 2013;1832:216–227. doi: 10.1016/j.bbadis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Yang Y, Cimen H, Han M-J, Shi T, Deng J-H, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, Koc EC. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010;285:7417–7429. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohannes E, Chang EJ, Christ GJ, Davies KP, Chance MR. Proteomics analysis identifies molecular targets related to diabetes mellitus-associated bladder dysfunction. Mol Cell Proteomics. 2008;7:1270–1285. doi: 10.1074/mcp.M700563-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohannes E, Chang J, Tar MT, Davies KP, Chance MR. Molecular targets for diabetes mellitus-associated erectile dysfunction. Mol Cell Proteomics. 2010;9:565–578. doi: 10.1074/mcp.M900286-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit VA, Ramsey RR, Bonomini M, Arduini A. Carnitine, mitochondrial function and therapy. Adv Drug Delivery Rev. 2009;61:1353–1362. doi: 10.1016/j.addr.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1S: Acetylated proteins and peptides in SSM and IFM isolated from 6 and 24 month old saline- and 24 month acetylcarnitine-treated Fisher 344 rats.