Abstract
Vertebrate DNA is subjected to epigenetic base modifications that have been thought to be limited to methylated and other modified forms of cytidine. A recent study shows that methylation of adenine to form N6-methyladenine is a rare but readily detectable modification that can be mapped to distinct genomic sites in vertebrates.
Background
Epigenetic modifications expand the information content of DNA and have long been known to exist in the genomes of diverse organisms. The most well-studied base modification in vertebrates and higher eukaryotes is methylation of the C-5 position of cytosine residues, which forms the 5-methylcytidine (m5C) nucleotide. m5C is often detected at CpG dinucleotides where it influences transcription by recruiting repressive m5C-binding proteins or preventing the binding of transcription factors [1]. Importantly, cytidine methylation is a reversible modification, and both methylation and demethylation pathways contribute to dynamic regulation of m5C signatures which control both developmentally regulated and activity-dependent gene expression programs [1]. Demethylation of m5C residues has been shown to involve the formation of oxidized intermediates, such as 5-formylcytidine and 5-hydroxymethylcytidine, which also contribute to gene expression control. Thus, the cytidine nucleotide has long been considered to be the major site of DNA base modifications contributing to transcriptional regulation in vertebrates and other eukaryotes.
Unlike higher eukaryotes, many bacteria are known to contain additional DNA base modifications. These include C-4-methylated cytosine (4mC), as well as methylated adenine (N6-methyladenine, or 6 mA). The methylated base, 6 mA, or the corresponding nucleotide N6-methyldeoxyadenosine (m6dA), is an important component of the restriction/modification system used to defend against bacteriophage invasion. This system uses the methylation of host cell DNA to protect it against cleavage by restriction endonucleases, while enabling invading, unmethylated genomic material to be cleaved. In addition to its role in host defense pathways, m6dA is also an important regulator of DNA replication, repair, and transcriptional control in prokaryotes [2].
A new addition to the vertebrate epigenome
Although m6dA is a readily detectable feature of bacterial genomes, it has been more difficult to definitively establish its presence in the genomes of higher organisms. This contrasts with mRNA, where mapping of the ribonucleotide equivalent of m6dA, N6-methyladenosine (m6A), has identified m6A sites in thousands of mammalian mRNAs [3]. Studies aimed at detecting m6dA in the DNA of higher organisms [4] have failed to uncover evidence for the existence of this modification. Although such studies formed the basis for the belief that mammalian DNA is devoid of m6dA, they were done over 40 years ago and were hampered by low sensitivity (limit of detection approximately 0.01 %) In contrast, more recent studies have challenged the notion that eukaryotic DNA does not contain m6dA. In 2006, Wion and colleagues used liquid chromatography coupled with mass spectrometry to interrogate the mouse genome for the presence of m6dA and detected very low levels of m6dA (fewer than 1 m6dA per 106 nucleotides) [5]. Although highly sensitive, such approaches can also lead to artifacts since trace amounts of bacterial contamination in genomic preparations can result in the detection of m6dA due to the high levels of m6dA in their genome. Indeed, bacterial contamination is commonplace in mammalian culture systems, and bacteria are either commensal organisms, or part of the diet of lower organisms such as Caenorhabditis elegans and Drosophila.
A major advance came with the development of m6dA mapping techniques in invertebrate organisms. Three recent studies used global m6dA profiling methods to identify m6dA in the genomes of Chlamydomonas reinhardtii, Drosophila melanogaster, and C. elegans, with levels of m6dA ranging from 0.4 % to 0.001 % of total adenine residues within these genomes [6–8]. Since these methods identified m6dA within a genomic sequence, the m6dA could be definitively assigned to the invertebrate genome rather than caused by bacterial contamination. These studies have been an important advance in our understanding of the repertoire of epigenetic modifications in higher organisms.
The outstanding question was whether m6dA is found in higher organisms, including humans. A recent study from Gurdon and colleagues [9] provides the first analysis of vertebrate m6dA residues genome-wide. The study, published in Nature Structural & Molecular Biology, examined the genomes of frogs, mice, and human cells using ultra-high-performance liquid chromatography with tandem mass spectrometry and revealed low levels of m6dA in all three genomes (approximately 1 m6dA for every 1.2 × 106 deoxyadenosine residues, or 0.00009 % of deoxyadenosine residues).
The authors then went on to globally profile m6dA distribution within Xenopus and mouse genomes using an m6dA antibody-based DNA immunoprecipitation (DIP) method. In brief, this method involves immunoprecipitating DNA fragments that contain m6dA using a m6dA-specific antibody. Their analysis revealed a large number of reads that cluster to form m6dA peaks in these genomes (approximately 20,000–50,000 depending on the tissue). Notably, only a small number of m6dA peaks were located within genes (approximately 7–21 %). Peaks within genes were largely excluded from exons, but a higher number were located within intronic regions. In addition, the authors observed a relative paucity of m6dA sites immediately after transcription start sites (TSSs), which contrasts with the marked enrichment of m6dA within this region in C. elegans [8].
The authors validated their results by repeating the global mapping studies in Xenopus using two additional m6dA antibodies. There was a high degree of overlap of individual m6dA peaks as well as overall m6dA distribution among all three antibodies tested, indicating that the DIP-seq mapping technique is likely detecting valid m6dA sites. When comparing m6dA sites across different Xenopus tissues, there were subsets of m6dA sites that were common to two or more tissues, but others that were unique. This may indicate tissue-specific m6dA patterns, but warrants further investigation to rule out sample-to-sample variability. Similarly, comparison of m6dA peaks in frog and mouse genomes revealed some peaks that overlapped and others that were distinct, which may be due to different tissues examined in each species (testes, fat, and oviduct in frog and kidney in mouse) or to species-specific methylation patterns. Additional detailed analyses of m6dA distribution across various tissues and in diverse species will likely lend further insights into the tissue-/cell type-specific distribution of m6dA as well as the consistency of m6dA sites across species.
The overall pattern of m6dA localization observed in the Gurdon study, as well as in the other recent m6dA profiling studies, provides the first insights into the potential function of m6dA. Although Gurdon and colleagues observe a slight enrichment of m6dA peaks just upstream of TSSs, the most defining feature of m6dA observed in this region is an absence of m6dA just after the TSS. In contrast, Greer et al. report no clear distribution of m6dA near genes in C. elegans [8], whereas m6dA is enriched at and just after TSSs in Ch. reinhardtii [6]. These distinct distribution patterns might indicate different roles for m6dA in vertebrates than in lower organisms. The fact that m6dA exhibits variable localization around TSSs further suggests potential functions for this modification in transcriptional regulation. Indeed, m6dA in Ch. reinhardtii is associated with actively transcribed genes [6]. However, further studies will be necessary to determine whether m6dA is repressive or permissive for transcription in vertebrates. Such analyses will also be useful for understanding how relevant functional studies in lower organisms are likely to be for understanding m6dA in vertebrates.
Moving forward: m6dA regulatory pathways and functional insights
A major priority right now for m6dA research is to uncover the function of m6dA, a task which will be facilitated by a more detailed understanding of the readers and writers of this mark. A putative m6dA methyltransferase, DAMT-1, has been identified in C. elegans [8], although the closest vertebrate homolog of this protein (METTL4) has not yet been explored for m6dA-forming potential. Notably, Gurdon and colleagues identify an AG-rich motif which was enriched in m6dA peaks in Xenopus. Two AG-containing motifs were also detected at m6dA sites in C. elegans [8], suggesting that the m6dA methyltransferase machinery in these organisms might share a similar recognition sequence. However, efforts to identify m6dA motifs in other higher eukaryotes, such as mice and flies, have failed to identify consistent consensus sequences [7, 9], suggesting the existence of other mechanisms for m6dA formation in addition to simple sequence recognition elements.
How does m6dA regulate DNA function? One possibility is that m6dA mediates the recruitment of transcription factors and/or m6dA binding proteins (Fig. 1). Although readers of m6A in RNA have been identified, proteins that specifically recognize methylated adenosine residues in DNA remain to be uncovered. The major epigenetic mark in vertebrates, m5C, is known to act in part by recruiting m5C-binding proteins such as MECP2 [10], and it is possible that m6dA functions in an analogous manner. Thus, a better understanding of the proteins that read and write the m6dA mark will be necessary to perform knockout studies that are likely to reveal m6dA function.
Fig. 1.
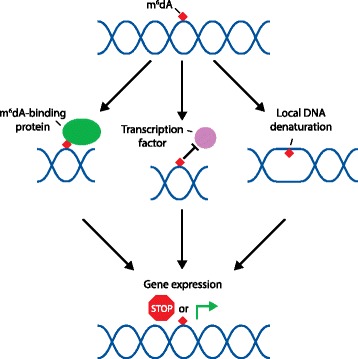
Methylated deoxyadenosine (m 6 dA) residues in DNA have the potential for multiple functions within the eukaryotic genome. m6dA residues might recruit m6dA-binding proteins (left), or m6dA might prevent the binding of transcription factors (middle). In addition, because N 6 methylation of adenosines can influence Watson:Crick base pairing, m6dA might act by inducing local DNA denaturation (right). Any or all of these possible functional roles for m6dA have the potential to influence transcription or other modes of gene expression. Whether m6dA acts primarily by repressing or promoting gene expression remains to be determined
Another possibility is that m6dA functions to destabilize DNA duplexes. Although m6dA forms standard Watson:Crick base pairs with thymidine, the base pairs between m6dA and thymidine are less stable than canonical adenosine:thymidine base pairs. Thus, m6dA may facilitate DNA unwinding or the open state of DNA needed for transcription initiation and other processes (Fig. 1).
An intriguing feature of m6dA across diverse organisms is that its abundance is markedly decreased in more evolved organisms. Could this be due to the evolution of active m6dA demethylation pathways? In other words, m6dA could occur frequently, but its rapid removal may account for its low overall abundance in the genome. NMAD-1, a homolog of the m6A RNA demethylase ALKBH5, has shown evidence of m6dA demethylation in C. elegans [8]. Interestingly, the Drosophila homolog of Tet (called Dmad), which normally functions as a m5C demethylase in vertebrates [11], was identified as a m6dA demethylase in flies [7]. Its potential as a m6dA demethylase in vertebrates, however, remains to be tested.
In addition to uncovering m6dA demethylases in vertebrates, it will be important to determine whether these enzymes produce demethylated intermediates, as is seen with the Tet-catalyzed production of 5-hydroxymethylcytidine (hm5C) from m5C residues in eukaryotic DNA [11]. Thus, there remains the possibility that the repertoire of modified nucleotides expands beyond m6dA. Such intermediate forms of modified adenine, if present, might also perform unique functions in regulating gene expression and thus represent important areas of future exploration.
Acknowledgements
This work was supported by NIH grants K99MH104712 (KDM) and R01CA186702 (SRJ).
Abbreviations
- 6 mA
Methylated adenine
- DIP
DNA immunoprecipitation
- m5C
5-Methylcytidine
- m6A
N6-methyladenosine
- m6dA
N6-methyldeoxyadenosine
- TSS
Transcription start site
Footnotes
Competing interests
The authors declare that they have no comepteing interests.
Authors’ contributions
KDM and SRJ wrote and edited the manuscript. Both authors read and approved the final version of the manuscript.
References
- 1.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 2.Løbner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8:154–60. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunthert U, Schweiger M, Stupp M, Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci U S A. 1976;73:3923–7. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratel D, Ravanat JL, Charles MP, Platet N, Breuillaud L, Lunardi J, et al. Undetectable levels of N6-methyl adenine in mouse DNA: cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006;580:3179–84. doi: 10.1016/j.febslet.2006.04.074. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, et al. N6-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell. 2015;161:879–92. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, et al. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161:868–78. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–14. doi: 10.1016/0092-8674(92)90610-O. [DOI] [PubMed] [Google Scholar]
- 11.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]


