Abstract
During microRNA (miRNA) biogenesis, the Microprocessor complex (MC), composed minimally of Drosha, an RNaseIII enzyme, and DGCR8, a double-stranded RNA-binding protein, cleaves the primary-miRNA (pri-miRNA) to release the pre-miRNA stem–loop structure. Size-exclusion chromatography of the MC, isolated from mammalian cells, suggested multiple copies of one or both proteins in the complex. However, the exact stoichiometry was unknown. Initial experiments suggested that DGCR8 bound pri-miRNA substrates specifically, and given that Drosha could not be bound or cross-linked to RNA, a sequential model for binding was established in which DGCR8 bound first and recruited Drosha. Therefore, many laboratories have studied DGCR8 binding to RNA in the absence of Drosha and have shown that deletion constructs of DGCR8 can multimerize in the presence of RNA. More recently, it was demonstrated that Drosha can bind pri-miRNA substrates in the absence of DGCR8, casting doubt on the sequential model of binding. In the same study, using a single-molecule photobleaching assay, fluorescent protein-tagged deletion constructs of DGCR8 and Drosha assembled into a heterotrimeric complex on RNA, comprising two DGCR8 molecules and one Drosha molecule. To determine the stoichiometry of Drosha and DGCR8 within the MC in the absence of added RNA, we also used a single-molecule photobleaching assay and confirmed the heterotrimeric model of the human MC. We demonstrate that a heterotrimeric complex is likely preformed in the absence of RNA and exists even when full-length proteins are expressed and purified from human cells, and when hAGT-derived tags are used rather than fluorescent proteins.
Keywords: miRNA, Microprocessor complex, single-molecule, stoichiometry
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nt-long RNAs that post-transcriptionally regulate their target mRNAs through degradation and translational repression (Guo et al. 2010). They are therefore critical to a diverse array of biological processes ranging from cell growth, survival, and differentiation, to diseased states, such as cancer. MiRNA genes are typically transcribed by RNA polymerase II into long, capped, and polyadenylated primary transcripts (pri-miRNAs), which follow a two-step processing pathway to yield a mature miRNA. The nuclear Microprocessor complex (MC), composed of the ribonuclease (RNase) III enzyme Drosha and its essential cofactor DGCR8, excises a ∼70-nt stem–loop structure (the pre-miRNA) with a 5′ phosphate and a ∼2-nt 3′ overhang (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004; Landthaler et al. 2004). This step is critical for proper miRNA biogenesis because the Drosha cleavage site defines the sequence of the mature miRNA by generating one end of each mature miRNA product. The resulting pre-miRNA is then transported to the cytoplasm by the Exportin5/Ran-GTP complex, where it is further processed by the RNase III enzyme Dicer. Dicer, together with TRBP2—a double-stranded (ds) RNA binding domain (dsRBD)-containing protein—cleaves the upper hairpin stem, generating ∼2-nt 3′ overhangs on the ∼22-nt dsRNA product (Chendrimada et al. 2005; Haase et al. 2005). One strand is then incorporated into an RNA-induced silencing complex (RISC), whose main component is an Argonaute family protein. This complex targets mRNAs via base pairing between the miRNA and mRNA, resulting in the regulation of protein expression.
Important questions concerning the MC are (i) what is the stoichiometry of DGCR8 and Drosha within the MC, and (ii) is the complex formed upon RNA binding or is it preformed in the absence of RNA? Size-exclusion chromatography (SEC) of the MC isolated from mammalian cells yields a large calculated molecular mass, suggesting that there are multiple copies of one or both proteins in the complex (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). Furthermore, V5-tagged DGCR8 constructs expressed in mammalian cells coimmunoprecipitate FLAG-tagged DGCR8, suggesting that DGCR8 is multimeric within the precipitated complex (Han et al. 2004). For Drosha, despite evidence that the processing center is formed intramolecularly by the two RNaseIII domains in a single Drosha monomer, a V5-tagged Drosha construct also coimmunoprecipitates FLAG-tagged Drosha, again suggesting that Drosha is multimeric within the precipitated complex (Han et al. 2004).
DGCR8 purified in the absence of Drosha forms a dimer based on the SEC results (Faller et al. 2007; Senturia et al. 2012). In addition, the dimerization has been crystallized (Senturia et al. 2010). When combined with recombinant Drosha, this isolated DGCR8 dimer showed increased pri-miRNA processing activity compared with monomeric DGCR8 (Faller et al. 2007). Further analysis, based on EM and the cooperativity of RNA binding in filter-binding assays, suggested that this DGCR8 dimer specifically assembles into a trimer of dimers upon binding a single pri-miRNA (Faller et al. 2010). With improved measurement of the extinction coefficient for DGCR8 (Senturia et al. 2012), this same group has, however, modified their model, claiming that DGCR8 binds pri-miRNA hairpin-loop substrates as a dimer of dimers, with each dimer recognizing a single-stranded (ss)–double-stranded (ds) junction (Quick-Cleveland et al. 2014). This conclusion is at odds with a study using gel shifts and NMR to investigate DGCR8 binding to RNA, which found no evidence of higher order cooperativity. Instead, DGCR8's mode of binding appeared to resemble that of other dsRBD-containing proteins, such as protein kinase R (PKR), which interact with RNA through transient and/or noncooperative interactions featuring multiple, potentially overlapping, RNA-binding sites with comparable affinities (Roth et al. 2013). This study, as well as another gel shift-based study, suggests that DGCR8 alone is not able to distinguish pri-miRNA substrates specifically and that substrate specificity is instead created by the concerted action of the Drosha and DGCR8 complex (Roth et al. 2013; Quarles et al. 2015). This interpretation is supported by the fact that Drosha and DGCR8 associate stably even in the absence of added RNA (Han et al. 2004; Roth et al. 2013). However, no laboratories have been able to purify recombinant Drosha and show that it is able to bind to RNA specifically or to cross-link Drosha to pri-miRNA substrates, while DGCR8 can be readily cross-linked to pri-miRNAs (Han et al. 2006). Therefore, the sequential model for binding to pri-miRNAs was created and proposes that DGCR8 binds pri-miRNA substrates specifically and then recruits Drosha to the complex in order to cut.
Recently, however, a recombinant deletion construct of Drosha has been purified and shown to bind to the basal ss–ds junctions of pri-miRNA substrates specifically (Nguyen et al. 2015). Using many mutants of both Drosha and DGCR8, Nguyen et al. (2015) were able to reconstitute MC activity and show that DGCR8 plays several roles in the MC. (i) The C-terminal tail of DGCR8 binds and stabilizes Drosha protein, (ii) the dsRBDs of DGCR8 interact with the pri-miRNA stem, and (iii) the RNA-binding heme domain interacts with the apical stem–loop. In the same study, they used a single-molecule photobleaching assay to demonstrate that a fluorescent protein-tagged deletion construct of DGCR8 can bind RNA over a wide range of stoichiometries, with primarily tetramers being formed. In the presence of a fluorescent protein-tagged deletion construct of Drosha, this DGCR8 construct assembled into a heterotrimeric complex on RNA, made up of two DGCR8 molecules and one Drosha molecule. They observed this heterotrimeric complex, containing only one copy of Drosha in the presence of RNA, despite previous evidence from the same laboratory that Drosha is multimeric in immunoprecipitated complexes (Han et al. 2004). No evidence for multimerization, beyond dimers, of DGCR8 on RNA was observed in the presence of Drosha.
Given the various conflicting results, we here extend and confirm the 1 Drosha:2 DGCR8 MC stoichiometry. We also combine single-molecule immunoprecipitation techniques (Jain et al. 2011; Yeom et al. 2011) with subunit counting by photobleaching assays (Ulbrich and Isacoff 2007) to determine the stoichiometry of Drosha and DGCR8 within single MCs. In our study, we use full-length versions of both Drosha and DGCR8 purified from mammalian cells and tagged with human O6-alkylguanine-DNA-alkyltransferase (hAGT)-derived tags rather than fluorescent proteins. We purify our MCs in the presence of micrococcal nuclease to minimize copurifying RNA and immobilize our MCs on microscope coverslips without RNA. By using TIRF microscopy to image single fluorescently labeled MCs and counting the number of photobleaching steps, we confirm that the MC is a heterotrimer composed of two copies of DGCR8 and one copy of Drosha. This stoichiometry is supported by biochemical determination of the MC's molecular mass. Our results show that the heterotrimeric complex assembles when full-length proteins are examined with different tags and is likely preformed, not dependent upon RNA binding.
RESULTS AND DISCUSSION
Single-molecule immunoprecipitation of human MC expressed in human cells
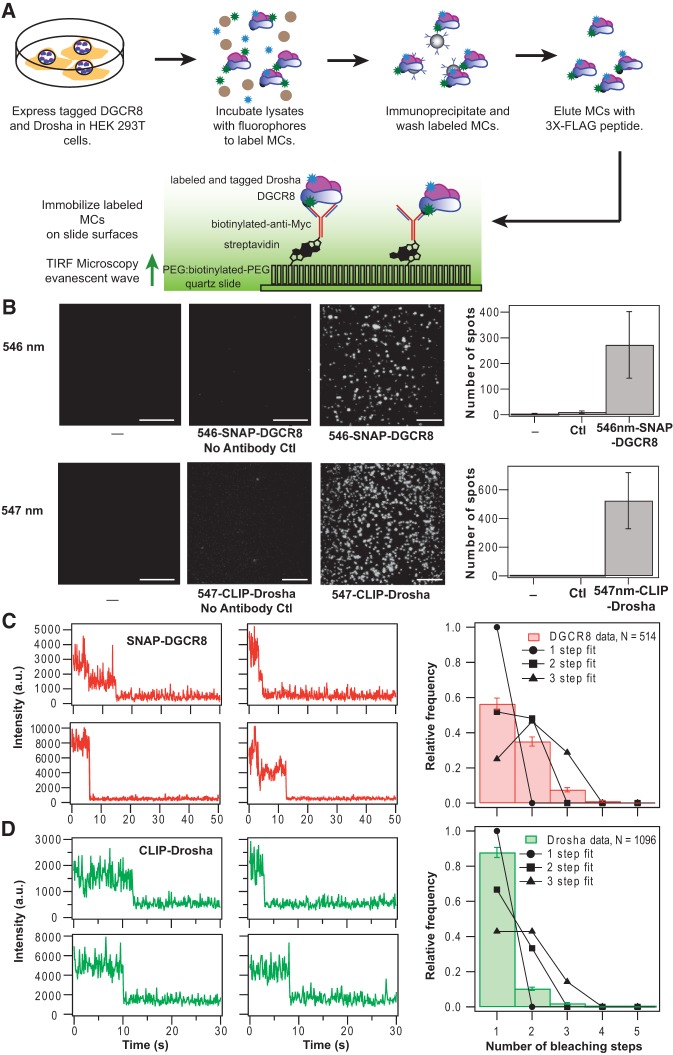
The MC has been suggested to be a multimeric complex containing multiple copies of DGCR8 and/or Drosha because of its large calculated molecular mass based on SEC results (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). To determine the stoichiometry of DGCR8 and Drosha, we used single-molecule subunit counting by photobleaching, a technique that does not depend on the MC's shape or whether there are other proteins within the complex. Full-length human Drosha and DGCR8 proteins were ectopically expressed in human embryonic kidney cells (HEK293T) (Fig. 1A). Drosha proteins were N-terminally tagged with tandem FLAG and Myc (seven copies) in order to affinity-purify MCs on anti-FLAG agarose and allow MC immobilization on slides through anti-Myc antibodies. Additionally, both Drosha and DGCR8 proteins were N-terminally tagged with CLIP and SNAP tags (hAGT derivatives), giving CLIP-6Myc-FLAG-TEV-Myc-Drosha (hereafter referred to as CLIP-Drosha) and SNAP-DGCR8, respectively. The two tags allow incorporation of orthogonal bright, photostable organic benzylcytidine- and benzylguananine-conjugated fluorophores into each protein, avoiding complications due to blinking and low photon output of fluorescent protein tags (Dickson et al. 1997). Labeling was accomplished by incubating lysates with 10 μM CLIP and SNAP fluorophore substrates in the presence of micrococcal nuclease to degrade any copurifying RNA. RNA was extracted from fractions of the sample that were prepared plus or minus micrococcal nuclease treatment and run on an agarose gel to confirm the degradation of RNA in the presence of nuclease. Labeling specificity and efficiency were determined by SDS–PAGE of labeled lysates, where the maximum fluorescence intensity over the 2 h labeling reaction was set to 100% labeling (Supplemental Fig. S1). By this method, the labeling efficiencies were found to be between ∼52% and 60% for fluorophores reacting with CLIP-Drosha and ∼67% and 81% for fluorophores reacting with SNAP-DGCR8. MCs immunoprecipitated with anti-FLAG agarose were washed extensively to reduce excess dye and non-MC protein association. MCs eluted with an excess of 3X-FLAG peptide were then immobilized on anti-Myc antibody-decorated quartz coverslip surfaces. Tagged and fluorescently labeled proteins were active in pri-miR-16 processing either in lysates or after immunoprecipitation (Supplemental Fig. S2).
FIGURE 1.
Single-molecule photobleaching assay indicates a heterotrimeric model for MC. (A) Scheme for the expression labeling and set-up of the single-molecule assay. The tagged versions of DGCR8 or Drosha being used are CLIP-6Myc-FLAG-TEV-Myc-Drosha and SNAP-DGCR8. See Materials and Methods for details. (B, top left) Three TIRF images of coverslip surfaces in the absence of labeled SNAP-DGCR8 MC samples or with SNAP-surface Alexa fluor 546-nm-labeled SNAP-DGCR8 MC samples plus and minus precoating the surface with anti-Myc antibody. Minus sign indicates no antibody or sample. (Top right) Quantitation of the average number of fluorescent spots detected per imaging area. Error bars denote standard deviation (SD) (n = 5 replicates for no sample, n = 7 for no anti-Myc, n = 15 for anti-Myc surfaces). (Bottom left) Three TIRF images of coverslip surfaces in the absence of labeled CLIP-Drosha MC samples or with CLIP-surface 547-nm-labeled CLIP-Drosha MC samples plus and minus precoating the surface with anti-Myc antibody. Minus sign indicates no antibody or sample. (Bottom right) Quantitation of the average number of fluorescent spots detected per imaging area. Error bars denote standard deviation (SD) (n = 5 replicates for no sample, n = 7 for no anti-Myc, n = 14 for anti-Myc surfaces). Scale bars within the images correspond to 10 μm. (C, left) Four representative SNAP-DGCR8 photobleaching traces showing fluorescence intensity in arbitrary units (a.u.) versus time in sec. (Right) Histogram showing the distribution of bleaching events (N = 514 fluorescence spots analyzed) observed with counting errors compared with normalized probability density fits to the data where n, the number of molecules per complex, is either 1, 2, or 3. The labeling efficiency was set to 65% in all cases. The n = 1 fit yielded a χ2 of 397 and by definition the labeling efficiency is undeterminable. The n = 2 fit yielded a χ2 of 71, while the n = 3 fit yielded a χ2 of 416. For labeling efficiencies <86%, n = 2 yields the best fit. (D, left) Four representative CLIP-Drosha (CLIP-6Myc-FLAG-TEV-Myc-Drosha) photobleaching traces, showing fluorescence intensity in arbitrary units (a.u.) versus time in sec. (Right) Histogram showing the distribution of bleaching events (N = 1096 fluorescence spots analyzed) observed with counting errors compared with normalized probability density fits to the data. As above, in the fits n the number of molecules per complex is 1, 2, or 3, and the labeling efficiency is set to 50%. The n = 1 fit yielded a χ2 of 153 and by definition the labeling efficiency is undeterminable; the n = 2 fit yielded a χ2 of 640 and the n = 3 fit yielded a χ2 of 2253. For labeling efficiencies above 35%, n = 1 yields the best fit. See Supplemental Figure S3 for more details on the fitting.
The immobilization of MCs on surfaces was specific (Fig. 1B) when commercial high-density PEG/biotin-PEG-coated slides were used. When excited with ∼3 mW laser power at 532 nm, the blank slide surface showed ∼2–3 fluorescent spots per 18,500 μm2 imaging area possibly due to surface impurities (Fig. 1B). Adding MCs containing CLIP-Drosha (which contains a Myc tag) and 546-nm-labeled SNAP-DGCR8 to the flow cell without first coating the surface with biotinylated anti-Myc antibodies produced approximately fourfold more spots than background. Adding MCs containing 546-nm-labeled SNAP-DGCR8 to slides coated with biotinylated anti-Myc antibodies yielded >100-fold more spots. When testing surface specificity, as above, but with complexes in which CLIP-Drosha rather than SNAP-DGCR8 had been fluorescently labeled, slightly better specificity was found (Fig. 1B). Lysates of cells in which neither SNAP-DGCR8 nor CLIP-Drosha, but rather YFP, was expressed, were prepared and were incubated with fluorophores and anti-FLAG immunoprecipitated. When this control sample was applied to noncommercial PEG/biotin-PEG-coated slides, which had been made following standard protocols (Bumb et al. 2011; Hardin et al. 2011), little background fluorescence was observed (data not shown). This indicates that on noncommercial PEG/biotin-PEG-coated slides, fluorescent spots were specifically observed only when SNAP and CLIP tags were present.
Single-molecule photobleaching reveals heterotrimeric MCs
To determine the stoichiometry of DGCR8 and Drosha within the MC, we analyzed photobleaching traces for each protein (Fig. 1C,D). In one case, samples were prepared using both the SNAP-surface 647 Alexa fluorophore and CLIP-surface 547 fluorophore and were then applied to noncommercial PEG/biotin-PEG-coated slides. In another case, samples were prepared using both SNAP-surface 546 Alexa fluorophore and CLIP-surface 488, and were then applied to commercial PEG/biotin-PEG-coated slides. The distributions of DGCR8 and Drosha photobleaching steps from each experiment were fit by normalized frequency distributions for a given stoichiometry and labeling efficiency (Supplemental Fig. S3). For a single bleaching step the labeling efficiency is unknown and immaterial (i.e., no change in the χ2 of the fit is observed upon changing the labeling efficiency in this case) since each complex will contain 1 or 0 labeled subunits, but only the labeled subunits are observed. The labeling efficiencies were held fixed at 50% for Drosha and 65% for DGCR8. These efficiencies are consistent, although slightly lower than we estimate from our SDS–PAGE analysis. There are several reasons for using a lower estimate. (i) Our assumption that the reaction has gone to 100% completion may not be accurate. (ii) Only a fraction of the SNAP tags may be folded properly in order to be labeled. This is similar to previous single-molecule stoichiometry experiments that rely on fluorescent proteins, where generally ∼80% of the fluorescent proteins are folded properly in order to fluoresce (Ulbrich and Isacoff 2007). (iii) Finally, there might be DGCR8 that is not SNAP-tagged present and that gets incorporated into MCs, either from the endogenous pool of DGCR8 or a sample that degrades after gel analysis. Data from both experiments yielded similar results and were therefore pooled (Fig. 1C; Supplemental Fig. S3).
Traces corresponding to fluorophore bleaching of SNAP-DGCR8 showed primarily one or two photobleaching steps (91.4%) (Fig. 1C). The small number of traces showing more than two steps (8.6%) likely results from two MCs localizing within a diffraction-limited spot. The χ2 for the fits was lowest for the binomial distribution corresponding to two DGCR8 molecules per complex (Fig. 1C). Only when the labeling efficiency goes above 86% does the bleaching step distribution corresponding to one DGCR8 molecule per complex yield a better χ2.
Traces corresponding to fluorophore bleaching of CLIP-Drosha showed almost exclusively single photobleaching steps (87.8%) (Fig. 1D), with only a small number of traces showing more than a single step (12.2%). The distribution of photobleaching steps is best fit by a binomial distribution with one Drosha protein (Fig. 1D). Only when the labeling efficiency falls below 35% does a binomial distribution with two Drosha proteins yield a better χ2. Taken together, these results support a heterotrimeric model for the MC, composed of two copies of DGCR8 protein and a single Drosha protein.
A heterotrimeric complex is consistent with the biochemically determined molecular mass for human MCs
The molecular mass of a heterotrimeric MC composed of two DGCR8s and one Drosha (∼331 kDa) protein did not agree with the experimentally determined molecular mass of the MC from SEC (∼670 kDa) (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). However, SEC separates proteins based on their frictional coefficient or Stokes radius and therefore provides accurate molecular mass estimates only for globular proteins (Erickson 2009). SEC was already reported to systematically overestimate the molecular mass of DGCR8 proteins, likely due to their elongated, nonglobular shape (Faller et al. 2007).
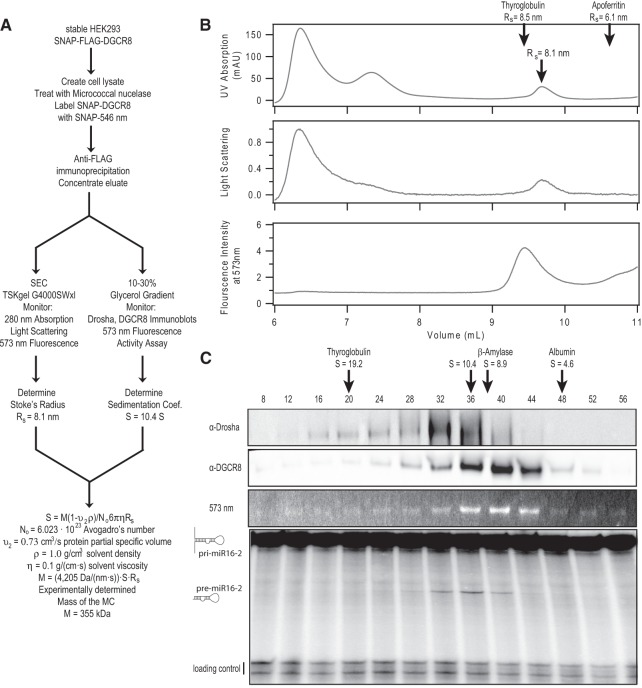
To biochemically determine the molecular mass of the MC, the Svedberg formula can be solved for the molecular mass of a protein complex in terms of the Stokes radius and the sedimentation coefficient; these can be experimentally determined using SEC and glycerol gradient centrifugation, respectively (Fig. 2A; Siegel and Monty 1966; Erickson 2009). We isolated the MC from HEK293 cells that had been made to stably express FLAG-SNAP-DGCR8. Cell lysates were treated with micrococcal nuclease to minimize the contribution of RNA bound to the MCs, and a portion of the lysates were labeled with SNAP-surface 546 Alexa, providing an additional means by which to track the protein. MCs were then isolated by anti-FLAG immunoprecipitation. Complexes were eluted with a 3X-FLAG peptide and subjected to either SEC or centrifugation in a 10%–30% glycerol gradient.
FIGURE 2.
Biochemical confirmation of the heterotrimeric MC. (A) Scheme for the sample preparation and analysis to determine the molecular mass of the MC. See Materials and Methods for details. (B) SEC analysis of the MC. UV absorption, light scattering, and 573-nm fluorescence were recorded. The fluorimeter was on a separate instrument upstream of the UV spectrometer, causing the slight shift in time between peaks on these two channels. Arrows indicate the position where standards with known Stokes radii (Rs) eluted and where the MC eluted on the UV spectrometer with an extrapolated Stokes radius of 8.1 ± 1.7 nm. (C) Glycerol gradient analysis of the MC. Fractions collected from the gradient were analyzed for the presence of Drosha and DGCR8 by Western blot, for 573 nm fluorescence and for in vitro processing of a 32P-labeled pri-miR16 substrate. Arrows indicate the positions where standards with known Svedberg coefficients (S) migrated and where the MC migrated with an extrapolated Svedberg coefficient of 10.4 ± 4.1 S.
The 280-nm absorption SEC profile of the FLAG-affinity eluate shows two major peaks besides the void volume, one of high-molecular mass and one migrating similarly to the thyroglobulin standard (∼670 kDa) (Fig. 2B). These two complexes were previously observed by SEC analysis of the MC isolated from cells stably expressing FLAG-Drosha, where only the low-molecular mass complex showed efficient pri-miRNA processing activity (Gregory et al. 2004). The 573-nm fluorescence profile, on the other hand, showed only a single peak corresponding to the lower-molecular mass complex, indicating that the larger complex either does not contain significant amounts of SNAP-DGCR8 or that SNAP-DGCR8 in this complex is misfolded or inaccessible to fluorophores in the lysate. The low-molecular mass complex observed on the UV channel containing fluorescently labeled SNAP-DGCR8 corresponds to a Stokes radius of 8.1 ± 1.7 nm (Fig. 2B; Supplemental Fig. S4).
Fractions from the FLAG-affinity eluate run over a 10%–30% glycerol gradient were probed by immunoblotting for Drosha and SNAP-DGCR8 (Fig. 2C). Drosha and DGCR8 displayed broad and overlapping elution profiles. The fraction displaying peak pri-miRNA processing activity corresponded to a sedimentation coefficient of 10.4 ± 4.1 S (Fig. 2C; Supplemental Fig. S4).
Given the experimentally determined Stokes radius and sedimentation coefficient, we calculate that the SNAP-tagged MC has a molecular mass of ∼355 kDa. While the errors in this measurement are large, previous studies have shown that this technique generally leads to an experimentally determined molecular mass that is within 10% of the expected molecular mass of a complex (Erickson 2009). This is within 10% of the calculated molecular mass of a 2 SNAP-DGCR8:1 endogenous Drosha heterotrimeric complex (∼383 kDa). It is more than 20% greater than the predicted molecular mass of a heterodimer (∼271 kDa) and even less compatible with any complex that contains more than two copies of Drosha (2 Drosha:1 SNAP-DGCR8 ∼430 kDa). Therefore, our biochemical characterization of the MC further supports the heterotrimeric MC model that we derived from our single-molecule results.
Concluding remarks
Using a single-molecule photobleaching assay, we confirmed and extended recent results (Nguyen et al. 2015) showing that the MC expressed in human cells is a heterotrimer composed of two DGCR8 proteins and one Drosha protein. Our results demonstrate that the MC is likely a preformed heterotrimer when full-length proteins are examined, with hAGT-derived instead of fluorescent protein tags. The stoichiometry of the complex had been an outstanding question in the field, as it was previously suggested that the MC is not a heterodimer of Drosha and DGCR8 (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). The heterotrimeric model is consistent not only with that conclusion, but also with the multimerization of DGCR8 seen in co-IP experiments (Han et al. 2004), reported dimerization of purified recombinant DGCR8 (Faller et al. 2007), and the Kim laboratory's model of the MC that positions a DGCR8 dimer binding to the apical ss-loop junction of the pri-miRNA hairpin (Nguyen et al. 2015).
The fact that the MC contains a single Drosha protein is also congruent with data from the Kim laboratory indicating that the MC processing center is formed intramolecularly by the two RNaseIII domains of the single Drosha polypeptide (Han et al. 2004), as well as their current model of the MC, which has a single Drosha molecule binding the basal ss–ds junction of the pri-miRNA hairpin. A single Drosha protein is, however, inconsistent with the Kim laboratory's observation, which we have also repeated (data not shown), that Drosha can multimerize in co-IP experiments independent of added RNA (Han et al. 2004). It is possible that Drosha exists in complexes other than the MC that contain more than a single copy of Drosha. Supporting this hypothesis is the fact that the original SEC, performed using immunopurified FLAG-Drosha stably expressed from HEK293 cells, revealed a large-molecular mass complex and a lower-molecular mass complex that migrates similarly to thyroglobulin at ∼670 kDa, while DGCR8 was present only in the smaller-molecular mass complex (Gregory et al. 2004). Whether the large-molecular mass complex and the Drosha multimerization in co-IP experiments are functionally important or the result of protein aggregation remains to be determined.
Because the predominant theory for MC binding to pri-miRNA substrates involved the sequential binding of DGCR8 followed by the recruitment of Drosha for cleavage (Han et al. 2004), many investigations have been done to study how DGCR8 alone binds RNA (Faller et al. 2007; Sohn et al. 2007; Senturia et al. 2010, 2012; Quick-Cleveland et al. 2014; Quarles et al. 2015). Several recent results have questioned this sequential model for substrate binding. Both Roth et al. (2013) and Quarles et al. (2015) suggest that DGCR8 alone is not able to distinguish pri-miRNA substrates specifically, but that substrate specificity is instead created by the concerted action of the Drosha and DGCR8 complex. This idea is supported by the fact that Drosha and DGCR8 stably associate with each other even in the absence of RNA (Han et al. 2004; Roth et al. 2013), which makes a sequential model for pri-miRNA recognition by DGCR8 and subsequent recruitment of Drosha unlikely (Han et al. 2006). Additionally, the work from the Kim laboratory has now shown that Drosha can indeed bind RNA in the absence of DGCR8. While DGCR8 stabilizes Drosha protein and increases substrate specificity, it is not strictly required (Nguyen et al. 2015), again making a sequential model for RNA binding unlikely. In this same study, the Kim laboratory showed that the MCs bound to pri-miRNAs are heterotrimeric. However, their analyses were limited to traces in which both Drosha and DGCR8 bound coincidently to pri-miRNA molecules within the time resolution of their assay (Nguyen et al. 2015). While a stable complex exists before RNA binding, it was still formally possible that the complex in the absence of RNA exists in a different stoichiometry than it adopts upon binding to RNA. In our study, we have shown that a heterotrimeric complex is very likely preformed in the absence of RNA. Not only do we investigate the stoichiometry of the complex in the absence of added RNA, but we also treat the lysates from which we purify MCs with micrococcal nuclease to degrade any RNA present. We consider it unlikely that the MC will copurify with protected RNA, since in the Nguyen et al. (2015) single-molecule study, they observed dissociation of the MC from RNA on the 1–2 min timescale of a photobleaching trace, while we treat our lysates with nuclease for 30 min. Taken together with the Kim laboratory result, this implies that the preformed heterotrimeric complex binds pri-miRNAs and that no further multimerization on RNA occurs. Future study of intact MC binding to RNA rather than DGCR8 alone is necessary.
We also attempted to study the binding of MCs to single pri-miRNA molecules in our photobleaching assays, but interestingly were not able to observe significant colocalization with Cy5-labeled pri-miRNAs at the low nM concentrations of protein that single-molecule experiments necessitate. This was true even on our commercial high-density PEG/biotin-PEG-coated slides, which showed low nonspecific binding of proteins to the surface. The constructs used in those experiments were, however, full-length proteins, unlike the deletion constructs used in the Kim laboratory paper. This may imply that the terminal regions of Drosha and/or DGCR8, which were present in our analyses or the hAGT-derived tags, could alter the binding affinity of the MC to pri-miRNA substrates.
Establishing the stoichiometry of the MC is a first step toward building a structural model of the complex with and without bound RNA. Structural information is available for pieces of DGCR8—the dsRBDs (Sohn et al. 2007) and the dimerization domain (Senturia et al. 2010)—while the structure of Drosha can be inferred only by comparison with other prokaryotic RNaseIII family enzymes or the unicellular eukaryotic Giardia Dicer (Jinek and Doudna 2009). The orientation of the dsRBDs relative to these RNaseIII domains remains unknown. A heterotrimeric model of the MC predicts that there are five dsRBDs, four contributed by the two DGCR8 proteins (two each), and one from Drosha, but whether all RBDs engage in RNA contacts or some might function as protein–protein interacting domains is yet to be determined. In the prokaryotic RNaseIII enzymes, a large-scale rearrangement of the dsRBDs upon RNA binding is observed. Whether a similar structural rearrangement occurs in the context of the entire MC warrants further study. It also will be interesting to learn whether the proposed bending of the pri-miRNA structure occurs when bound by the intact MC.
In addition, previous studies concluded that heme binds DGCR8 across its dimerization interface and that heme availability affects pri-miRNA processing both in vivo and in vitro (Faller et al. 2007; Senturia et al. 2010; Weitz et al. 2014). Here we developed an assay that allowed us to establish MC stoichiometry and have shown that under normal cellular growth conditions, the MC is a heterotrimer composed of two DGCR8 proteins and one Drosha protein. This is an essential first step toward determining whether the cell might regulate MC stoichiometry in response to heme availability or cellular signaling events.
MATERIALS AND METHODS
Plasmids
pSNAP-tag(m) and pCLIP-tag(m) vectors were purchased from NEB. pSNAP-DGCR8: A thrombin site was cloned into the BamHI site of pSNAP-tag(m) to give pSNAP-thrombin. Then DGCR8 was PCRed from pFLAG-HA-DGCR8 (Herbert et al. 2013) with primers that attached an XhoI and a NotI site onto the 5′ and 3′ ends, respectively. This product was inserted into the XhoI and NotI sites of the pSNAP-thrombin vector to give pSNAP-DGCR8. pCS3–6Myc-FLAG-TEV-Myc-Drosha: Oligonucleotides with the FLAG and TEV sequences were purchased in order to have HindIII and KpnI compatible overhangs after annealing. This dsDNA product was treated with PNK and then inserted into the HindIII and KpnI sites of pcDNA3 to create pcDNA3-FLAG-TEV. Myc-Drosha was PCR amplified from pcDNA4-TO-cmycDrosha (Landthaler et al. 2004) with primers that append SfoI and XbaI sites to either end, and this PCR product was inserted between the SfoI and XbaI sites of pcDNA3-FLAG-TEV to give pcDNA3-FLAG-TEV-Myc-Drosha. The pCS3-MT(6Myc) plasmid was digested with EcoRI while FLAG-TEV-Myc-Drosha was excised from pcDNA3-FLAG-TEV-Myc-Drosha using XbaI, HindIII, and FspI. Both digests were subsequently treated with T4 DNA polymerase to fill in the 3′ overhangs. The FLAG-TEV-Myc-Drosha fragment was ligated into the pCS3-MT(6Myc) plasmid to give pCS3-6Myc-FLAG-TEV-Myc-Drosha. The CLIP sequence was excised from CLIP-tag(m) vector using XhoI/NheI, and then PCR amplified with primers that add ClaI sites onto either end. The CLIP sequence was inserted into the ClaI site of the pCS3-6Myc-FLAG-TEV-Myc-Drosha plasmid to give pCS3-CLIP-6Myc-FLAG-TEV-Myc-Drosha.
Expression, purification, and labeling of MCs
HEK293T cells were cultured as in Herbert et al. (2013). Four 10-cm plates of cells were transfected with 10.5-μg Drosha plasmid DNA and 1.3-μg DGCR8 plasmid DNA using TransIT-293 reagent (Mirus Bio) per manufacturer's instructions. Cells were harvested 30–48 h later and frozen on dry ice for at least 30 min until further processing. Cells were lysed in ∼650-μL Lysis buffer (2% NP-40, 10% glycerol, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA) supplemented with 1 mM DTT, EDTA-free protease inhibitor tablets (Roche), and Phosphatase Inhibitor Cocktails 2 and 3 (Sigma) for 45 min on ice. Lysates were cleared by spinning for 15 min at 16k RCF. SNAP- or CLIP-tag fluorophore substrates (NEB) were added to lysates to a final concentration of 10 μM. 5 U/μL micrococcal nuclease (Thermo Scientific) and 3 mM CaCl2 were added to lysates. The final volume of the lysate was adjusted with buffer and DTT up to 700 μL. Labeling was allowed to precede at 20°C for 2 h, removing aliquots periodically for assessment of labeling efficiency. At 30 min, the majority of the lysate, ∼500 μL, was removed for immunopurification of the labeled MCs as in Herbert et al. (2013) except that 1 mM DTT was added to the wash buffer.
The extent of SNAP and CLIP dye labeling was assessed as in Hoskins et al. (2011). Aliquots (5 μL) from the 2-h labeling reaction were frozen on dry ice. Later, the samples were diluted in 4X NuPAGE LDS sample buffer, heated to 95°C for 5 min and analyzed by SDS–PAGE on a 4%–12% Bis-Tris polyacrylamide gel (Invitrogen) followed by fluorescence imaging in a Syngene G:Box Chemi XT4 System. Intensities of the fluorescent bands corresponding to the SNAP- and CLIP-labeled proteins were determined using the GeneSys software and normalized to maximum observed intensity. For the determination of complex stoichiometry, dual labeled complexes were used. The various fluorophore combinations used were SNAP-surface Alexa fluor 647 and CLIP-surface 547 or SNAP-surface Alexa fluor 546 and CLIP-surface 488. The complexes used to determine the specificity of immobilization in Figure 1B were singly labeled on either DGCR8 or Drosha with either CLIP-surface 547 or SNAP-surface Alexa fluor 546.
In vitro processing assays
32P-labeled pri-miR-16 was synthesized as previously described and used for in vitro processing assays with purified MCs or fractions from glycerol gradient fractions as in Herbert et al. (2013).
Photobleaching experiments
Single-molecule measurements were performed using a home-built prism-type TIRF microscope based on an Olympus IX71 (Bumb et al. 2011; Hardin et al. 2011). Flow cells with a PEG/biotin-PEG-coated surface were created either exactly as in Bumb et al. (2011) or in some cases commercial high-density PEG/biotin-PEG-coated slides from MicroSurfaces, Inc. were used. Wash buffer (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 125 mM NaCl, 0.5 mM DTT, 5% glycerol, 13.3 mM MgCl2, and 5 mM EGTA) was used to wash slides and dilute proteins for coating slides. Streptavidin (Invitrogen) was bound to the biotin-PEG slide surface by flowing a 0.2 mg/mL stock of streptavidin through the cell and incubating for ∼20 min. Surfaces were further protected by coating with 2 mg/mL BSA (Bio-rad) for ∼30 min. Then an 8 μg/mL solution of monoclonal anti-c-Myc-biotin, clone 9E10 (Sigma), was allowed to bind to the surface for 30 min. Immunopurified fluorescently labeled MCs were diluted 5000–10,000-fold, flowed onto the surface, and allowed to bind. After allowing complexes to bind the slide surface, slides were washed with a 250 mM NaCl version of the wash buffer. MCs were excited and fluorescence collected as in Bumb et al. (2011). The fluorescent particles were selected from the first two frames of each movie using the MOSAIC particle tracker plugin for ImageJ (Sbalzarini and Koumoutsakos 2005), so that in order to be a valid particle it must be selected in both 100 msec frames. Then the fluorescence intensity versus time traces were extracted from each region of interest using custom written LabView software. Bleaching step-finding and statistical analyses were performed using custom IGOR programs.
Size exclusion chromatography and glycerol gradient centrifugation
Stable HEK293 Flp-In cell lines were created using a Flipase (Flp)/Flp recognition target site-directed recombination system (Invitrogen). Flp-In 293 cells were cotransfected with pcDNA5/FRT-SNAP-FLAG-DGCR8 and pOG44. Stable clones were selected using 200 μg/mL hygromycin (EMD-Millipore).
Approximately fifty 15-cm plates of stable HEK293 SNAP-FLAG-DGCR8 cells were lysed in 15 mL Lysis buffer. The lysate was cleared by spinning as above and micrococcal nuclease (Thermo Scientific) and 3 mM CaCl2 were added to lysates. To 500 μL of lysate SNAP-532nm and DMSO were added. The entire 15 mL of lysate (with and without SNAP-532nm) were incubated at 20°C to allow micrococcal nuclease activity and for labeling in the small fraction to occur. After 30 min, the lysates were mixed again and incubated with 5 mL of anti-FLAG MS2 agarose for immunoaffinity purification as above. After eluting proteins with five times 5 mL of wash buffer plus 3X FLAG peptide, the eluate was concentrated in an Amicon Ultra with a 50 kDa molecular weight cut off down to ∼250 μL. Half of this concentrated eluate was loaded onto a TSK gel G4000SWxl SEC column. 280 nm absorption, light scattering, and 573 nm fluorescence were monitored. To the other half, MW standards were added: 10 mg/mL albumin, 8 mg/mL thyroglobulin, and 4 mg/mL β-amylase. The combined sample was layered on top of a 10%–30% glycerol gradient and spun at 40 K for 20 h in a SW41 rotor in a Beckman L8-M ultracentrifuge. A syringe needle was used to puncture the bottom of the centrifuge tube and six drops (∼250 μL) were collected per fraction. Fractions were run on SDS-PAGE and Coommassie stained to identify the elution peaks for each of the standards. Another SDS-PAGE gel was imaged on the Syngene G:Box Chemi XT4 System for 573 nm fluorescence. Immunoblots using anti-DGCR8 (Protein Tech) and anti-Drosha (Cell Signaling) monitored the elution peaks of both DGCR8 and Drosha. Finally, fractions were used in in vitro processing assays as described above.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to E. Abbondanzieri for initial discussions of labeling strategies, G. Piszczek of the NHLBI Biophysics core for help running SEC, A. Ditmore and Y. Seol of the Neuman laboratory for advice and software for fluorescent spot detection, N. Darricarrère of C. Crews’ laboratory for the kind gift of the HEK293 Flp-In cells, and D. Liu and A. Miccinello of the Steitz laboratory for editorial help. J.A.S. is an investigator of the Howard Hughes Medical Institute. K.M.H. was an HHMI Fellow of the Damon Runyon Cancer Research Foundation. A Burroughs Wellcome Fund 2011 Collaborative Research Travel Grant was awarded to K.M.H. This work was supported by grant GM026154 from the National Institute of General Medical Sciences (NIGMS, NIH) to J.A.S. This research was supported in part by the Intramural Research Programs of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054684.115.
REFERENCES
- Bumb A, Sarkar SK, Wu XS, Brechbiel MW, Neuman KC. 2011. Quantitative characterization of fluorophores in multi-component nanoprobes by single-molecule fluorescence. Biomed Opt Express 2: 2761–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Dickson RM, Cubitt AB, Tsien RY, Moerner WE. 1997. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 388: 355–358. [DOI] [PubMed] [Google Scholar]
- Erickson HP. 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online 11: 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. 2007. Heme is involved in microRNA processing. Nat Struct Mol Biol 14: 23–29. [DOI] [PubMed] [Google Scholar]
- Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F. 2010. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. RNA 16: 1570–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901. [DOI] [PubMed] [Google Scholar]
- Hardin AH, Sarkar SK, Seol Y, Liou GF, Osheroff N, Neuman KC. 2011. Direct measurement of DNA bending by type IIA topoisomerases: implications for non-equilibrium topology simplification. Nucleic Acids Res 39: 5729–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Pimienta G, DeGregorio SJ, Alexandrov A, Steitz JA. 2013. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep 5: 1070–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. 2011. Ordered and dynamic assembly of single spliceosomes. Science 331: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. 2011. Probing cellular protein complexes using single-molecule pull-down. Nature 473: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167. [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. 2015. Functional anatomy of the human microprocessor. Cell 161: 1374–1387. [DOI] [PubMed] [Google Scholar]
- Quarles KA, Chadalavada D, Showalter SA. 2015. Deformability in the cleavage site of primary microRNA is not sensed by the double-stranded RNA binding domains in the microprocessor component DGCR8. Proteins 83: 1165–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick-Cleveland J, Jacob JP, Weitz SH, Shoffner G, Senturia R, Guo F. 2014. The DGCR8 RNA-binding heme domain recognizes primary microRNAs by clamping the hairpin. Cell Rep 7: 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BM, Ishimaru D, Hennig M. 2013. The core Microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein. J Biol Chem 288: 26785–26799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol 151: 182–195. [DOI] [PubMed] [Google Scholar]
- Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. 2010. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci 19: 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturia R, Laganowsky A, Barr I, Scheidemantle BD, Guo F. 2012. Dimerization and heme binding are conserved in amphibian and starfish homologues of the microRNA processing protein DGCR8. PLoS One 7: e39688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta 112: 346–362. [DOI] [PubMed] [Google Scholar]
- Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. 2007. Crystal structure of human DGCR8 core. Nat Struct Mol Biol 14: 847–853. [DOI] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. 2007. Subunit counting in membrane-bound proteins. Nat Methods 4: 319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz SH, Gong M, Barr I, Weiss S, Guo F. 2014. Processing of microRNA primary transcripts requires heme in mammalian cells. Proc Natl Acad Sci 111: 1861–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C. 2011. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep 12: 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.