Abstract
Small RNAs regulate gene expression and most genes in the worm Caenorhabditis elegans are subject to their regulation. Here, we analyze small RNA data sets and use reproducible features of RNAs present in multiple data sets to discover a new class of small RNAs and to reveal insights into two known classes of small RNAs—22G RNAs and 26G RNAs. We found that reproducibly detected 22-nt RNAs, although are predominantly RNAs with a G at the 5′ end, also include RNAs with A, C, or U at the 5′ end. These RNAs are synthesized downstream from characteristic sequence motifs on mRNA and have U-tailed derivatives. Analysis of 26G RNAs revealed that they are processed from a blunt end of double-stranded RNAs and that production of one 26G RNA generates a hotspot immediately downstream for production of another. To our surprise, analysis of RNAs shorter than 18 nt revealed a new class of RNAs, which we call NU RNAs (pronounced “new RNAs”) because they have a NU bias at the 5′ end, where N is any nucleotide. NU RNAs are antisense to genes and originate downstream from U bases on mRNA. Although many genes have complementary NU RNAs, their genome-wide distribution is distinct from that of previously known classes of small RNAs. Our results suggest that current approaches underestimate reproducibly detected RNAs that are shorter than 18 nt, and theoretical considerations suggest that such shorter RNAs could be used for sequence-specific gene regulation in organisms like C. elegans that have small genomes.
Keywords: RNA-seq, RNAi, gene silencing
INTRODUCTION
Small RNAs influence gene expression in most eukaryotes. RNAs that are ∼20- to 30-nt long and identify their target genes through base-pairing have been typically characterized as small RNAs (Kim et al. 2009). Some classes of small RNAs can regulate target genes despite imperfect base-pairing (e.g., Bagijn et al. 2012; Montgomery et al. 2012), further expanding their impact on an organism. Although small RNAs are likely to be an integral part of the regulation of all biological processes, their diversity and pervasiveness present considerable challenges for analysis. Thus, even in well-studied organisms (e.g., the simple worm Caenorhabditis elegans), the biogenesis and roles of these RNAs are not well understood.
Four broad classes of regulatory small RNAs have been studied in C. elegans (for review, see Billi et al. 2014). These are miRNAs that are ∼20-nt-long and are processed from individual hairpin transcripts; 21U RNAs or piRNAs that are 21-nt-long, have a 5′ U, and are processed from individual transcripts; 22G RNAs that are ∼22-nt-long, predominantly have a 5′ G and are synthesized by RNA-dependent RNA polymerases (RdRPs) using long transcripts as templates; and 26G RNAs that are ∼26-nt-long, predominantly have a 5′ G and are synthesized by an RdRP and the endonuclease Dicer using long transcripts as templates. Each miRNA (Hammond 2015) and piRNA (Weick and Miska 2014) is transcribed from a well-defined locus and subsequently processed to generate the mature small RNA. In contrast, 22G RNAs and 26G RNAs are made as populations of varying abundance from long RNA transcripts, but the conditions required for the biogenesis and stability of individual RNAs of these classes are not clear.
The identification and analysis of small RNAs relies heavily on next-generation sequencing (small RNA-seq), and approaches to analyze the resultant data are currently not standardized. Despite the approximately 300 data sets describing small RNAs from C. elegans that have been deposited in the Gene Expression Omnibus thus far, the extent of reproducibility between different data sets of RNAs from animals of the same genotype and stage of development obtained from different laboratories is unknown. Therefore, there is a need for analytical approaches that can be used by the community to explicitly compare discoveries from different laboratories. Such comparisons are essential to develop a precise and integrated understanding of the effect of any perturbations (e.g., loss of a protein) on small RNA populations.
Here, we extensively analyze small RNA data sets generated through next-generation sequencing and delineate the reproducible features of RNAs that are present in data sets from different laboratories. These features provide new insights into the biogenesis of 22G RNAs and 26G RNAs. In addition, analysis of RNAs shorter than 18 nt reveals groups of RNAs that differ between data sets from different laboratories and a new class of RNAs that is present in data sets from multiple laboratories.
RESULTS
Strategy for the analysis of small RNAs
The preparation of libraries for small RNA-seq can introduce contaminating RNA fragments that vary from sample to sample and obscure reproducible subsets of small RNAs. To mitigate the effects of such contamination, we removed RNAs that map to loci that generate abundant RNAs such as microRNAs (miRNAs) and ribosomal RNAs (rRNAs), as well as RNAs that were present at very high abundances (0.1% of all RNAs in a data set). We then considered only features that could be detected when RNAs that map to either strand of the genome were examined (see Materials and Methods and Supplemental Fig. S1 for details). For example, when considering 20-nt RNAs from a data set, we divided these RNAs into the two subsets that map to the two different strands of genomic DNA (plus and minus) and only considered features (e.g., a 5′ G bias) that were detectable in both subsets. When comparing small RNA data sets from different laboratories, only data sets of RNA prepared from the same stage of development were considered. For most features presented in this study, observations on RNAs mapping to one genomic strand are presented in figures and similar observations on RNAs mapping to the other genomic strand or in data sets from other laboratories (when applicable) are presented in Supplemental Figures.
22G RNAs and their derivatives are made from characteristic sequence motifs on mRNA
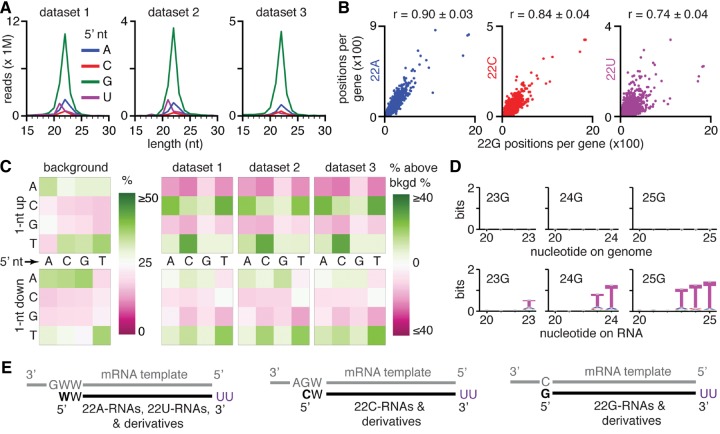
To determine the reproducible features of 22G RNAs, we examined small RNA data sets prepared using the 5′ monophosphate-independent method (Ambros et al. 2003; Pak and Fire 2007) in three different laboratories (Ruvkun lab [SRX193361], Mello lab [SRX154615], Miska lab [SRX892595]) from young adult worms. RNAs from all three data sets showed a comparable distribution with a large peak of 22-nt RNAs with a 5′ G (Fig. 1A). However, smaller peaks of 22-nt RNAs that have a different nucleotide at the 5′ end were also observed in all three data sets (5′ A more than 5′ C or 5′ U; Fig. 1A). A similar distribution was observed when RNAs of each sequence were only considered once (Supplemental Fig. S2A). These observations are consistent with the ability of C. elegans RNA-dependent RNA polymerases (RdRPs) to initiate synthesis at all 4 nt in vitro (Aoki et al. 2007). For each gene in the C. elegans genome, the numbers of positions that generate each of these additional 22-nt RNAs were proportional to the numbers of positions that generate 22-nt RNAs with a 5′ G (Fig. 1B; Supplemental Fig. S2B,C). Finally, the levels of all these RNAs were greatly reduced in animals that lack the RdRPs RRF-1 and EGO-1 (Supplemental Fig. S2D). Together, these observations suggest that the same process generates 22-nt RNAs with each 5′ nucleotide (A, C, G, or U).
FIGURE 1.
22G RNAs with every 5′ nucleotide and longer U-tailed derivatives are reproducibly detected small RNAs in C. elegans. (A) Small RNA reads from three different labs show similar distributions of lengths and 5′ nucleotides. Small RNA reads from populations of adult-staged wild-type C. elegans (N2) captured using 5′ mono-phosphate independent cloning by three different labs (data set 1 to data set 3) were sorted by 5′ terminal nucleotide (A, C, G, U) and length (15 to 30 nt) and plotted. Also see Supplemental Figure S2. (B) The number of positions within each gene where 22G RNAs are made is proportional to the number of positions where 22A RNAs, 22C RNAs, and 22U RNAs are made. For each annotated region (ce6 genes), the number of unique positions with aligned 22G RNAs was plotted against the number of unique positions with aligned 22A RNAs (blue, left), or with aligned 22C RNAs (red, middle), or with aligned 22U RNAs (purple, right). Pearson's correlations (r) from three data sets are indicated as average ± SEM. Also see Supplemental Figure S2. (C) 22G RNAs map to genomic regions with characteristic sequence biases. Nucleotide frequencies (%) 1 nt upstream of and 1 nt downstream from the 5′ end of genomic positions with RNAs that match the plus strand were determined. Changes in frequency above that observed in the background (left, background frequency in data set 1 shown as an example) for three data sets (right) and for each 5′ nucleotide are shown. Also see Supplemental Figure S3. (D) Most 23- to 25-nt-long RNAs are generated by terminal uridylation of 22G RNAs. Nucleotide biases (sequence logos [Schneider and Stephens 1990]) at the 3′ ends of 23- to 25-nt RNAs that align to the genome when only the first 22 nt were required to match the genome and nucleotide biases of the genomic sequences are shown. Also see Supplemental Figures S4–S6. (E) Model for the biogenesis of 22G RNAs and their derivatives. Fragments of mRNA are used as templates to synthesize 22-nt RNAs, from which longer RNAs are generated by untemplated uridylation. (Left) 22A RNAs and 22U RNAs are made at GWW motifs (where W = A or U) on mRNA. (Middle) 22C RNAs are made at AGW motifs on mRNA. (Right) 22G RNAs are made at C nucleotides on mRNA.
To determine whether any sequence motifs are associated with the production of 22G RNAs, we examined genomic sequence biases at the 5′ and 3′ ends of these RNAs (Fig. 1C; Supplemental Fig. S3). We found a characteristic set of biases around the 5′ end of 22G RNAs (Fig. 1C, right) that were detectable above the background sequence bias in the C. elegans genome (Fig. 1C, left; Fire et al. 2006) in all three data sets. The biases detected 1 nt upstream of the 5′ end of the RNA depended on the identity of the 5′ base of the 22-nt RNA (Fig. 1C, right). RNAs with a 5′ A or 5′ U showed an enrichment of C and a depletion of A and G, RNAs with a 5′ C were associated with an enrichment of U and a depletion of A and G. In addition to these upstream biases, a weak depletion of C and G with an accompanying weak enrichment of A and U was observed for the nucleotide following the 5′ nucleotide. These biases could reflect the sequence motifs preferred by RdRPs on mRNA templates for the synthesis of 22G RNAs. Whereas considering all 22-nt RNAs together suggested a modest preference for a C or U immediately upstream of 22-nt RNAs (Gent et al. 2010), examining RNAs with different 5′ nucleotides separately clarifies the observation and reveals that synthesis of RNAs with 5′ nucleotides other than G appear to be promoted by specific upstream bases on mRNA templates.
In addition to 22-nt RNAs, abundant shorter and longer RNAs were also detected in all three data sets (Fig. 1A; Supplemental Fig. S2). For each gene in the C. elegans genome, the numbers of positions that generate 19-, 20-, 21-, 23-, 24-, or 25-nt RNAs were proportional to the numbers of positions that generate 22G RNAs (Supplemental Fig. S4A). This general trend was not affected by 21-nt RNAs with a 5′ U that were abundant in two data sets (data set 1 and data set 2; Fig. 1A), consistent with these RNAs being 21U RNAs or piRNAs produced from a few positions that overlap with genes (Supplemental Fig. S2). Furthermore, removal of all RNAs that share their 5′ end with a 22-nt RNA resulted in a dramatic reduction in RNAs of lengths other than 22 nt (compare Fig. 1A; Supplemental Fig. S4B). These observations suggest that 19- to 25-nt RNAs are largely derivatives of 22-nt RNAs. Abundant RNAs shorter than 22-nt (19- to 21-nt RNAs) could result from the 3′ to 5′ degradation of 22-nt RNAs or from synthesis of shorter RNAs using the mRNA template. On the other hand, abundant RNAs longer than 22-nt (23- to 25-nt RNAs) could result from the untemplated addition of nucleotides to 22-nt RNAs or from synthesis of longer RNAs using the mRNA template. To distinguish between these two possibilities, we required matching of the first 22-nt of 23-, 24-, and 25-nt RNAs to the genome and examined the biases, if any, of nucleotides at the 3′ ends in the RNA sequence and the corresponding genomic sequence (Fig. 1D; Supplemental Fig. S5). In all cases, positions following the 22nd position showed a strong bias for U in the RNA sequence but not in the corresponding genomic position, suggesting that most of the longer RNAs result from the untemplated 3′ uridylation of 22-nt RNAs. These results are consistent with previous studies that detected 3′ uridylated versions of 22G RNAs (van Wolfswinkel et al. 2009; de Albuquerque et al. 2015). Intriguingly, 3′ uridylated 22-nt RNAs (i.e., 23- to 25-nt RNAs with untemplated 3′ U nucleotides) had a different bias at the 5′ end when compared with that of all 22-nt RNAs (Supplemental Fig. S6), raising the possibility that a selected subset of RNAs are subject to untemplated 3′ uridylation in vivo. Thus, an unknown mechanism ensures that 22-nt, and not longer, RNAs are synthesized using stabilized mRNA fragments (Tsai et al. 2015) as templates in vivo. Such precise production of 22-nt RNAs could be enabled by proteins like the Dicer-related helicase DRH-3, which interacts with the RdRP RRF-1 (Aoki et al. 2007), is required for the production of 22G RNAs (Gu et al. 2009), and was recently proposed to bind as a dimer to measure 22 base pairs formed by small RNAs binding mRNA templates (Fitzgerald et al. 2014). Alternatively, cleavage of the mRNA template could generate a precise 5′ end that provides a 22-nt long template for small RNA synthesis.
Taken together, our results suggest that the majority of 22G RNAs are made as RNAs of precise length from mRNA templates at positions with characteristic local sequence biases and are subsequently subject to 3′ to 5′ degradation or untemplated 3′ uridylation to generate the population of RNAs that are reproducibly detected in RNA-seq data sets (Fig. 1E). Additional experiments are necessary to test these hypotheses and to determine how small RNAs of precise lengths are made from long mRNA templates.
26G RNAs are produced upon successive cleavages from a blunt end of dsRNA substrates
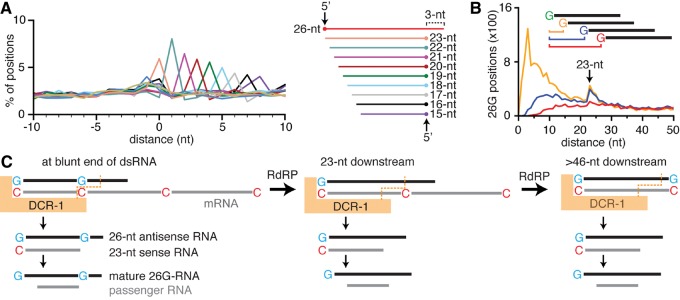
The biogenesis of 26G RNAs is currently unclear (for review, see Billi et al. 2014). The RdRP RRF-3 is required for the synthesis of antisense RNA on mRNA templates, but it is unclear how the resultant dsRNAs are used by Dicer to generate 26G RNAs and their passenger RNAs, which are eliminated by Argonautes to generate the mature 26G RNAs. Purified C. elegans Dicer cuts dsRNA substrates in vitro from a blunt end with an offset such that the 3′ end of 26-nt RNA is 3 nt away from the 5′ end of passenger 23-nt RNA (Welker et al. 2011). A similar 3-nt difference is observed between the 3′ end of 26G RNAs and 5′ end of passenger RNAs in vivo, suggesting that 26G RNAs may be generated from a blunt end of dsRNAs in vivo (Fischer et al. 2011). However, unlike the 23-nt passenger RNAs observed in vitro, the most abundant passenger RNAs observed in vivo are 19-nt long with a variable 3′ end indicative of exonucleolytic processing (Fischer et al. 2011). To gain insight into their biogenesis, we examined 26G RNAs in small RNA data sets with greatly reduced numbers of 22G RNAs and their derivatives. These include data sets obtained using the 5′ monophosphate-dependent capture of small RNAs and those obtained using the 5′ monophosphate-independent capture of small RNAs from animals that lack the RdRPs RRF-1 and EGO-1.
To determine the relationships between 26G RNAs and passenger RNAs in vivo, we examined the distance between the 5′ end of 26G RNAs that match one strand of the genome and the 3′ end of RNAs of all other lengths (15–25 nt) that match the other strand of the genome (Fig. 2A; Supplemental Fig. S7). In all cases, if the ends of multiple RNAs matched a single genomic position, that position was considered only once. For all overlapping 23- to 15-nt RNAs, the most common distance between their 5′ end and the 3′ end of 26G RNAs was 3 nt (Fig. 2A; Supplemental Fig. S7). This observation suggests that cleavage from a blunt end of dsRNA by Dicer to generate a 23-nt passenger RNA also occurs in vivo and that the 23-nt RNA is then susceptible to degradation by a 3′ to 5′ exonuclease. If such cleavage occurs in vivo, then subsequent production of 26G RNAs could occur from the cleaved mRNA template 23-nt downstream from the first 26G RNA. Consistent with this expectation, when we examined the distance between the 5′ ends of a 26G RNA and that of the first, second, and third subsequent 26G RNAs, a clear peak at 23 nt was detected (Fig. 2B; Supplemental Fig. S8). This 23-nt phasing of 26G RNAs was also observed in numerous other data sets but not in data sets from animals that lack rrf-3 or eri-1 because these genes are required for the biogenesis of 26G RNAs (Supplemental Fig. S8). In addition to this 23-nt phasing of 26G RNAs, a variable phase of 23- to 29 nt was reported when individual genes were examined (Fischer et al. 2011). Taken together with our observations, cycles of synthesis at an internal C nucleotide on mRNA, 3′ to 5′ degradation of mRNA to generate dsRNA with a blunt end (possibly by the ERI-1 exonuclease, which has such activity in vitro [Kennedy et al. 2004]), cleavage from the blunt end, and synthesis at the next available C nucleotide on mRNA could explain the phasing observed for 26G RNA production in vivo.
FIGURE 2.
Some 26G RNAs are produced with a 23-nt phase from a blunt end of dsRNA. (A) Passenger RNAs of 26G RNAs are subject to 3′–5′ degradation from the blunt end of a dsRNA. (Left) Distance between the genomic position of the 5′ end of each 26G RNA that matches the plus strand of the genome and the 3′ end of overlapping 15- to 25-nt RNAs from rrf-1(-) ego-1(-) mutants was calculated and plotted as a percentage for each length. (Right) Inferred alignments of passenger RNAs and 26G RNAs. Also see Supplemental Figure S7. (B) Some 26G RNAs are produced with a 23-nt phase downstream from other 26G RNAs. Distance between the 5′ end of each 26G position and the first, second, and third subsequent 26G positions were calculated and plotted. Also see Supplemental Figure S8. (C) Model for the phased production of 26G RNAs by successive cleavage of dsRNA from a blunt end. Synthesis by the RdRP RRF-3 generates dsRNA that is cleaved from the blunt end by Dicer to produce a 26G RNA and a 23-nt complementary RNA that is subjected to 3′ to 5′ degradation to generate a 19-nt passenger RNA (left). Subsequent synthesis from a C nucleotide 5′ of the cleaved mRNA and cleavage from the blunt end of the resulting dsRNA can generate another 26G RNA precisely 23 nt downstream from the previous 26G RNA (middle) or more than 23-nt downstream from the previous 26G RNA (right). See Supplemental Figure S9 for the complete model.
These findings reconcile previous in vivo and in vitro observations and suggest the following model for the biogenesis of 26G RNAs (Fig. 2C; Supplemental Fig. S9, and similar to an early model [Ruby et al. 2006]): First, the RdRP RRF-3 synthesizes antisense RNA beginning with a 5′ G; second, a blunt end is generated upon 3′ to 5′ degradation by an exonuclease such as ERI-1; third, the resultant dsRNA is cleaved by Dicer from the blunt end to generate a 26G RNA that is stabilized by an Argonaute protein and a passenger 23-nt RNA that is susceptible to 3′ to 5′ exonucleolytic degradation; and fourth, cleavage of the initial dsRNA by Dicer generates a hotspot 23 nt downstream from the blunt end and thereafter for the phased production of another 26G RNA. Additional experiments are necessary to test this model, to determine how a locus is selected for 26G RNA production, and how 26G RNAs acquire monophosphates at their 5′ ends.
Different 1-nt staggered clusters of sense RNA fragments are present in data sets from different laboratories
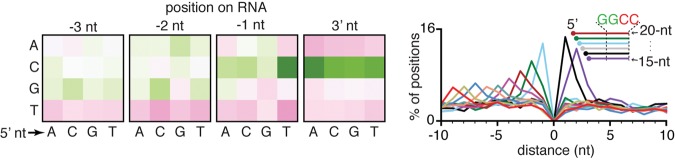
RNAs shorter than ∼18 nt are typically selected against during the preparation of small RNAs for RNA-seq. Nevertheless, we detected substantial numbers of RNAs shorter than 18 nt in some data sets that could not be explained as 3′ to 5′ degradation products of the abundant 22G RNAs. Such RNAs that match the sense orientation of genes had characteristic features that were reproducible within each data set when RNAs that map to either strand of the genome were separately considered but not across data sets from different laboratories. In one data set, we observed RNAs with a sequence bias at their 3′ ends (Fig. 3, left) with 5′ ends arranged in a 1-nt stagger (Fig. 3, right), suggesting that these RNAs are fragments of sense RNA that are subject to 5′ to 3′ exonucleolytic cleavage but protected by factor(s) that bind RNA at the 3′ end. The 1-nt staggered RNAs observed in three additional data sets from the same laboratory had similar sequence features (Supplemental Fig. S10) but these features were not detected in data sets from other laboratories. These observations raise the need for caution when following up on RNAs that appear to be reproducible but were the result of experiments performed in only one laboratory. Further studies are required to determine whether these variable 1-nt staggered sense RNAs result from processes within worms or from experimental procedures required to prepare RNAs for RNA-seq.
FIGURE 3.
One-nucleotide staggered fragments of sense RNAs with 3′ motifs are detected in some data sets. (Left) Changes in nucleotide frequency at the 3′ end (3′ nt, -1 nt, -2 nt, and -3 nt) of 17-nt sense RNAs above that observed in the background for each 5′ nucleotide from one data set (SRX892595) are shown. Background bias and scale are as in Figure 1C. (Right) Distance between the genomic positions of the 5′ terminus of 17-nt sense RNAs and the 5′ terminus of overlapping 15- to 26-nt RNAs that map to the same strand was calculated and plotted as a percentage of each length. Inset indicates inferred arrangement of RNAs. One-nucleotide staggered RNAs could result from the protection of RNAs from 5′ to 3′ degradation by a factor that binds a motif at the 3′ end of RNAs. Also see Supplemental Figure S10.
A new class of antisense RNAs is present in data sets from multiple labs
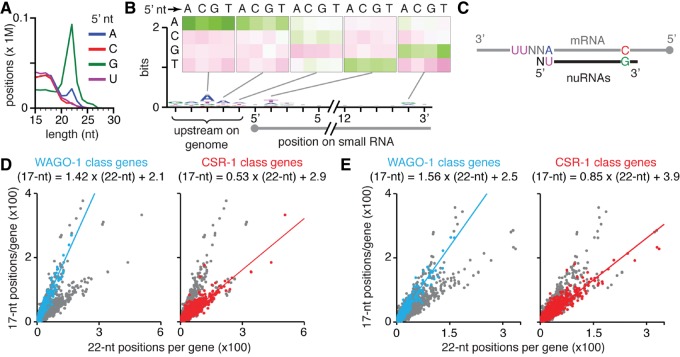
Although 15- to 18-nt sense RNAs could arise from the degradation of mRNAs or pre-mRNAs, 15- to 18-nt antisense RNAs are likely to be either the result of synthesis by RdRPs in vivo or the turnover of known classes of antisense small RNAs. We detected such short antisense RNAs from approximately 0.5 million positions on the genome in some data sets from multiple laboratories (Fig. 4A; Supplemental Fig. S11). These RNAs did not have a strong bias for any base at their 5′ end (Fig. 4A) and were detected when RNAs were captured using 5′ monophosphate-dependent or 5′ monophosphate-independent ligation (Supplemental Fig. S11). RNAs of each length (15–18 nt) originated from genomic positions with similar sequence biases (Fig. 4B; Supplemental Fig. S12). These 15- to 18-nt antisense RNAs appear to be generated downstream from U bases on RNAs and have a preference for a U 1 nt downstream from their 5′ ends and a preference for G 1 nt upstream of their 3′ ends (Fig. 4B; Supplemental Fig. S12). Furthermore, these sequence biases were detected for RNAs with each 5′ nucleotide (Supplemental Fig. S13). Taken together, these results suggest that 15- to 18-nt antisense RNAs are generated downstream from U bases on RNAs and have a 5′ bias of NU, where N is any nucleotide (Fig. 4C). Based on these characteristics, we propose that these RNAs be called NU RNAs (pronounced “new RNAs”).
FIGURE 4.
NU RNAs are a new class of antisense RNAs. (A) Small RNA reads from some data sets show large numbers of RNAs shorter than 18 nt. Small RNA reads in a data set (SRX129660) from populations of adult N2 C. elegans captured using 5′ monophosphate-dependent cloning were sorted by 5′ terminal nucleotide (A, C, G, U) and length (15–30 nt) and plotted. Also see Supplemental Figure S11 for additional data sets with similar distributions. (B) RNAs shorter than 18 nt are associated with characteristic sequence biases in some data sets. Nucleotide biases of 17-nt RNAs that align to the genome from the SRX129660 data set and nucleotide biases at genomic sequences upstream of the aligned RNAs are shown. Inset indicates changes in nucleotide frequency above that observed in the background within and near regions that generate 17-nt RNAs. Background bias and scale are as in Figure 1C. Because these RNAs have a 5′ NU bias, where N is any nucleotide, we refer to them as NU RNAs. Also see Supplemental Figures S11–S15 for additional analysis of NU RNAs. (C) Schematic showing nucleotide biases upstream of NU RNAs on mRNA templates and at the termini within NU RNAs. (D,E) Relative abundance of NU RNAs that match CSR-1 class genes is different from that of NU RNAs that match WAGO-1 class genes in two data sets. For each gene in the C. elegans genome (ce6), in CSR-1 class, and in WAGO-1 class, numbers of unique positions with aligned 17-nt RNAs and the number of unique positions with aligned 22-nt RNAs from two different data sets (D, SRX129660; E, SRX129662) were determined and plotted. WAGO-1 class genes (left in each panel) and CSR-1 class genes (right in each panel) are highlighted separately and regression lines describing the relationship between 17-nt RNAs and 22-nt RNAs of the two classes in each data set are indicated.
The levels of NU RNAs detected were different in different data sets. Although NU RNAs with all four bases at their 5′ end could be detected in some data sets (Supplemental Fig. S11), in other data sets, only 15- to 18-nt RNAs with 5′ U could be detected with the characteristics of NU RNAs (Supplemental Fig. S14). One explanation for this observation could be that degraded products of 22G RNAs, of which RNAs with 5′ G, 5′ A, and 5′ C are the most abundant, obscured NU RNAs that have the same 5′ end. Alternatively, the size selection and other preparatory steps used to capture RNAs for sequencing could have prevented the isolation of NU RNAs.
To determine the relationship of NU RNAs to other classes of small RNAs, we looked for them among RNAs that map to three subsets of genes, which have been classified based on the proteins required for the production and/or stability of antisense small RNAs. These included the CSR-1 class genes (Tu et al. 2015) that generate abundant 22G RNAs that are bound by the Argonaute CSR-1, the WAGO-1 class genes (Tu et al. 2015) that generate abundant 22G RNAs that are bound by the Argonaute WAGO-1, and the MUT class genes (Phillips et al. 2014) that require Mutator proteins such as MUT-16 to generate abundant 22G RNAs. In data sets where abundant NU-RNAs could be detected, genes of CSR-1 class, WAGO-1 class, or MUT class all had NU RNAs (Supplemental Fig. S15). Intriguingly, in two data sets, CSR-1 class genes and WAGO-1 class genes could be clearly separated based on the relative numbers of NU RNAs when compared with 22G RNAs (Fig. 4D,E), suggesting that the production or stability of NU RNAs can be independent of the production or stability of 22G RNAs.
In summary, NU RNAs are antisense RNAs that can be shorter than 18 nt and show a genome-wide distribution that is distinct from that of previously known classes of regulatory small RNAs. Additional studies are required to determine the biogenesis and functions, if any, of NU RNAs.
DISCUSSION
We analyzed small RNA-seq data sets and used reproducible features of RNAs present in multiple data sets to gain new insights into known classes of 18- to 26-nt small RNAs and to discover a new class of RNAs shorter than 18 nt.
Analysis of small RNAs is challenging and requires multiple approaches
The millions of sequences that result from typical small RNA-seq experiments present many challenges for analysis. Degradation products of abundant long RNAs can be difficult to distinguish from genuine small RNAs. By only considering features that are detected in multiple data sets from multiple laboratories, our analysis reduces the influence of such RNAs. Derivatives of one class of small RNA can obscure other classes of small RNAs. For example, the abundant tailed and trimmed versions of 22G RNAs obscure the presence of other classes of RNAs (Fig. 1). Different mapping and filtering strategies are needed to even detect some classes of RNAs. For example, U-tailed derivatives of 22G RNAs require mapping of reads while allowing terminal mismatches.
This work presents a starting point for further analyses of small RNAs and likely misses potentially important small RNAs. By requiring features of a class of small RNAs to be present in RNAs that map to either strand of the genome, potentially important RNAs that are made from specific locations on the genome and are different from known classes of small RNAs are ignored. The appropriate method to quantify small RNAs at a locus is not clear. Because different small RNA sequences could make different contributions to function, quantifying the number of different small RNA sequences made at a locus versus the abundance of individual sequences could lead to different conclusions. By making the analysis explicit, the effect of experimental perturbations on small RNAs can be better described and better understood.
RNAs smaller than 18 nt could be used for sequence-specific regulation
The length of RNA necessary for sequence-specific regulation in an organism is expected to be determined by the extent of sequence space that needs to be searched for base-pairing (Supplemental Fig. S16). For example, RNAs as small as 14 nt can be sufficient to identify a unique sequence in organisms with an ∼100 million base-pair genome such as C. elegans. Cellular processes such as transcription, splicing, and cytoplasmic localization can further reduce the sequence space, making 14 nt more than sufficient for specific base-pairing to mRNAs in the cytoplasm. The base-pairing of miRNAs to 3′ UTRs illustrates this effect. Although miRNAs are typically ∼20-nt long, the detection of an mRNA target by a miRNA only requires perfect base-pairing in the ∼7-bp seed region (for review, see Jonas and Izaurralde 2015). The sufficiency of the seed region for sequence-specific gene regulation by miRNAs could be because other mechanisms (e.g., ribosome occupancy on mRNAs) restrict the searchable sequence space to the 3′ untranslated regions (UTRs) of transcripts. As a result, despite such minimal base-pairing, miRNAs have influenced the evolution of sequences in the 3′ UTRs of most expressed genes in mammalian genomes (Farh et al. 2005).
Current approaches typically ignore RNAs smaller than 18 nt such as NU RNAs. Even if NU RNAs are products of small RNA turnover, the clear sequence biases of NU RNAs suggest that they are the result of specific turnover of subsets of antisense small RNAs. Alternatively, the above considerations suggest that it is plausible for the 15- to 18-nt NU RNAs to be used for sequence-specific regulation in C. elegans. Consistent with this possibility, a class of 17-nt RNAs called unusually small RNAs were reported to associate with Argonautes in mammalian cells (Li et al. 2009). Thus, approaches that effectively capture and analyze RNAs shorter than 18 nt are needed to evaluate their potential impact on biology.
MATERIALS AND METHODS
Data sets and analysis overview
RNA-seq data sets (Supplemental Table S1) were obtained from the European Nucleotide Archive (Leinonen et al. 2011) using the Galaxy platform (Giardine et al. 2005; Blankenberg et al. 2010, 2014; Goecks et al. 2010) for biomedical research. The analysis pipelines were created using the Galaxy platform (v. 14.10) and were run using the BioBlend library (v. 0.5.3) (Sloggett et al. 2013) and custom Python scripts (Jones et al. 2001; Oliphant 2007). Sixteen workflows were created and linked together as shown in Supplemental Figure S1. Plots were generated using the Matplotlib library (v. 1.4.3) (Hunter 2007) and custom python scripts. All scripts and workflows are available upon request.
Data analysis
Each FASTQ file was processed with FASTQ Groomer. The 3′ adapter was removed using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) Trimmer, discarding any read that was untrimmed or contained only adapter sequences, and keeping only reads 15 nt or longer. Bowtie (v.1.1.1) was used to align the reads to the Ce6 C. elegans genome. The reads were aligned three distinct times to the genome using three different settings: The first allowed up to two mismatches; the second allowed no mismatches in the first 22 nt of the read but allowed unlimited mismatches in the remaining nucleotides; and the third allowed no mismatches anywhere. For each alignment, every read was allowed to align to up to four locations on the genome and unaligned reads were discarded. The two-mismatch alignment was filtered to keep only the alignment(s) for each read with the fewest number of mismatches. For each set of alignments, all reads that align to a region known to produce miRNA (miRbase 21) (Griffiths-Jones 2004; Kozomara and Griffiths-Jones 2014) and rRNA or snRNA (ChrI:15046730-15070972 and ChrV:17113353-17134036) were removed. Finally, any sequence that makes up >0.1% of the total number of alignments from each set was removed. These processes resulted in three sets of alignments in SAM format. These alignments were processed in multiple ways (Supplemental Fig. S1) as described below for one of the alignment sets.
Remove other lengths with shared 5′ ends
The set of alignments was split into reads that were 15 nt long and reads that were not 15 nt long. Any alignment that was not 15 nt long but shared its 5′ position with a 15-nt-long alignment was removed. The process was repeated for 16- to 26-nt length reads to generate 12 alignments in SAM format.
Identify reads sense and antisense to genes
A set of alignments and a set of gene intervals from the ce6 genome that included exons, introns, and UTRs for each gene were filtered by strand (plus strand vs. minus strand). Plus-strand reads were joined to plus- and minus-strand intervals to obtain alignments sense and antisense to these intervals, respectively. Minus-strand reads were joined to plus- and minus-strand intervals to obtain alignments antisense and sense to these intervals, respectively. Sets of alignments from each strand that align sense to genes were concatenated to form one set. Sets of alignments from each strand that align antisense to genes were concatenated to form another set.
Assign 5′ end position to longest RNA
A set of alignments was split by length. All 26-nt alignments were kept. Starting with 25-nt alignments, any 25-nt-length alignment that shares the 5′ position with a 26-nt alignment was discarded. The remaining 25-nt alignments were added to the 26-nt alignments. Any 24-nt alignment that shares the 5′ position with the 25- or 26-nt alignment was discarded, and the remaining 24-nt alignments were added to the 25- and 26-nt alignments. This process of subtraction followed by concatenation was repeated with the 23- to 15-nt alignments.
5′ Nucleotide and length plots
Sets of alignments that were sense and antisense to genes were identified as in the “Identify reads sense and antisense to genes” section above. The sense sets and the antisense sets were split by length and 5′ nucleotide. The number of alignments with each 5′ nucleotide for each length was plotted (e.g., Fig. 1A). Plots for unique genomic positions were also similarly created (e.g., Fig. 4A).
22 Versus non-22 and 22G versus 22 non-G plots
Sets of alignments that were antisense to genes were identified as in the “Identify reads sense and antisense to genes” section above. These alignments were collapsed to identify the genomic positions of the 5′ ends of the aligning reads. All reads and genomic positions were split by length. The 22-nt reads and positions were in addition also split by the identity of the 5′ nucleotide. For each length, the numbers of reads and genomic positions that align to each gene was counted and plotted against those of 22-nt reads and genomic positions (e.g., Supplemental Fig. S3). Additionally, the number of 22-nt reads and genomic positions with a 5′ G were compared with the number of 22-nt reads and positions without a 5′ G that align to each gene, respectively, and plotted (e.g., Fig. 1B).
26G Overlap plots
A set of alignments was filtered to obtain the genomic positions of the 5′ ends of reads that were 26 nt long and had a 5′ G nucleotide. These positions were filtered to obtain plus-strand and minus-strand aligned positions using the process described in the “Identify reads sense and antisense to genes” section above. For each strand, the positions were sorted by chromosome and genomic position. For each 26G, the distances in nucleotides to the first, second, and third subsequent 26G position were calculated and plotted (e.g., Fig. 2B).
Weblogos
For a set of alignments, the sequences of aligned genomic positions for each read along with that of 5-nt upstream and 5-nt downstream regions were obtained. The aligned reads and the genomic positions were filtered by length, 5′ nucleotide, and strand. A sequence logo using a bit-score to calculate the frequency of each nucleotide (Weblogo [v 3.4] [Schneider and Stephens 1990; Crooks et al. 2004]) was generated for the filtered reads and genomic positions (e.g., Fig. 1D).
Distance to overlapping positions or reads on same strand
The set of alignments was filtered by length. One length of alignments was aligned to all other length alignments on the same strand to identify those that overlap. The distance in nucleotides between the 5′ end of the first length and the 5′ end of each overlapping alignment was calculated. The number of overlapping alignments at each 5′ to 5′ distance was counted for each length. The percentage of each overlapping length that overlapped with each 5′ to 5′ distance was calculated and plotted. This process was repeated for unique genomic positions of the alignments of each length (e.g., Fig. 3, right).
Distance to overlapping positions/read on opposite strand
The set of alignments was filtered by length. One length of alignments was aligned to all other length alignments on the opposite strand to identify those that overlap. The distance in nucleotides between the 5′ end of the first length and the 3′ end of each overlapping alignment was calculated. The number of overlapping alignments at each 5′ to 3′ distance was counted for each length. The percentage of each overlapping length that overlapped with each 5′ to 3′ distance was calculated and plotted. This process was repeated for unique genomic positions of the alignments of each length (e.g., Fig. 2A).
Upstream and downstream bias
Genomic positions sense and antisense to genes were identified as in the “Identify reads sense and antisense to genes” section above, and split into plus- and minus-strand positions. For each alignment position, a 3-nt sequence upstream of and downstream from each terminus (5′ and 3′) was obtained from the genomic DNA sequence. For each nucleotide of the 5′ flanking sequences (3 nt upstream and 3 nt downstream), the frequency of each nucleotide base was calculated given each 5′ terminus nucleotide base. For each nucleotide of the 3′ flanking sequences (3 nt upstream and 3 nt downstream), the frequency of each nucleotide base was calculated given each 3′ terminus nucleotide base. Finally, the 3′ terminus nucleotide base frequency was calculated given the identity of the 5′ base. Background frequencies were determined by performing each of the above three sets of calculations on aligned genomic positions for 22-nt reads after the positions were randomized within exons using BEDTools shuffle (v. 2.22.1) (Quinlan and Hall 2010). These calculated background frequencies were subtracted from the nucleotide frequencies calculated for each length using the three sets of calculations above. These final frequencies above background frequencies were plotted as heat maps for each position and length (e.g., 22 nt shown in Fig. 1C).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Mount and members of the Jose laboratory for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (R01GM111457) to A.M.J.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054551.115.
REFERENCES
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818. [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26: 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SE, Kim JK. 2014. Endogenous RNAi pathways in C. elegans. WormBook 7: 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Prot Mol Biol 19: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Bouvier E, Baker D, Afgan E, Stoler N, Galaxy Team, Taylor J, Nekrutenko A. 2014. Dissemination of scientific software with Galaxy ToolShed. Genome Biol 15: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BF, Placentino M, Ketting RF. 2015. Maternal piRNAs are essential for germline development following de novo establishment of endo-siRNAs in Caenorhabditis elegans. Dev Cell 34: 448–456. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. 2005. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310: 1817–1821. [DOI] [PubMed] [Google Scholar]
- Fire A, Alcazar R, Tan F. 2006. Unusual DNA structures associated with germline genetic activity in Caenorhabditis elegans. Genetics 173: 1259–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G. 2011. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet 7: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald ME, Vela A, Pyle AM. 2014. Dicer-related helicase 3 forms an obligate dimer for recognizing 22G-RNA. Nucleic Acids Res 42: 3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. 2004. The microRNA Registry. Nucleic Acids Res 32: D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. 2015. An overview of microRNAs. Adv Drug Deliv Rev 87: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. 2007. Matplotlib: a 2D Graphics Environment. Comp Sci Eng 9: 90–95. [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant E, Peterson P, et al. 2001. SciPy: open source scientific tools for Python. http://www.scipy.org/ [Online; accessed November 24, 2014].
- Kennedy S, Wang D, Ruvkun G. 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tárraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, et al. 2011. The European nucleotide archive. Nucleic Acids Res 39: D28–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B. 2009. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J Virol 83: 12751–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G. 2012. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 8: e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. 2007. Python for scientific computing. Comp Sci Engin 9: 10–20. [Google Scholar]
- Pak J, Fire A. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Montgomery BE, Breen PC, Roovers EF, Rim YS, Ohsumi TK, Newman MA, van Wolfswinkel JC, Ketting RF, Ruvkun G, et al. 2014. MUT-14 and SMUT-1 DEAD box RNA helicases have overlapping roles in germline RNAi and endogenous siRNA formation. Curr Biol 24: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloggett C, Goonasekera N, Afgan E. 2013. BioBlend: automating pipeline analyses within Galaxy and CloudMan. BMC Bioinformatics 29: 1685–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Chen CC, Conte D Jr, Moresco JJ, Chaves DA, Mitani S, Yates JR III, Tsai MD, Mello CC. 2015. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Wu MZ, Wang J, Cutter AD, Weng Z, Claycomb JM. 2015. Comparative functional characterization of the CSR-1 22G-RNA pathway in Caenorhabditis nematodes. Nucleic Acids Res 43: 208–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148. [DOI] [PubMed] [Google Scholar]
- Weick EM, Miska EA. 2014. piRNAs: from biogenesis to function. Development 141: 3458–3471. [DOI] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. 2011. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell 41: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.