Abstract
The spliceosome undergoes dramatic changes in both small nuclear RNA (snRNA) composition and structure during assembly and pre-mRNA splicing. It has been previously proposed that the U2 snRNA adopts two conformations within the stem II region: stem IIa or stem IIc. Dynamic rearrangement of stem IIa into IIc and vice versa is necessary for proper progression of the spliceosome through assembly and catalysis. How this conformational transition is regulated is unclear; although, proteins such as Cus2p and the helicase Prp5p have been implicated in this process. We have used single-molecule Förster resonance energy transfer (smFRET) to study U2 stem II toggling between stem IIa and IIc. Structural interconversion of the RNA was spontaneous and did not require the presence of a helicase; however, both Mg2+ and Cus2p promote formation of stem IIa. Destabilization of stem IIa by a G53A mutation in the RNA promotes stem IIc formation and inhibits conformational switching of the RNA by both Mg2+ and Cus2p. Transitioning to stem IIa can be restored using Cus2p mutations that suppress G53A phenotypes in vivo. We propose that during spliceosome assembly, Cus2p and Mg2+ may work together to promote stem IIa formation. During catalysis the spliceosome could then toggle stem II with the aid of Mg2+ or with the use of functionally equivalent protein interactions. As noted in previous studies, the Mg2+ toggling we observe parallels previous observations of U2/U6 and Prp8p RNase H domain Mg2+-dependent conformational changes. Together these data suggest that multiple components of the spliceosome may have evolved to switch between conformations corresponding to open or closed active sites with the aid of metal and protein cofactors.
Keywords: spliceosome, single-molecule FRET, RNA, snRNP, dynamics
INTRODUCTION
The spliceosome is one of the most compositionally and conformationally dynamic machines inside the cell. With a core of ∼100 proteins and 5 snRNAs, the spliceosome assembles onto precursors to mRNAs (pre-mRNAs) to remove introns while ligating exons together (Wahl et al. 2009). A subset of proteins and the snRNAs preassemble into complexes called small nuclear ribonucleoproteins (snRNPs) that, along with the Prp19p-associated complex, form the main building blocks of the spliceosome. It has long been appreciated that RNA dynamics are linked to splicing. The 5′ exon and lariat-3′ exon intermediate must be repositioned after 5′ splice site cleavage to permit exon ligation (Konarska et al. 2006; Wahl et al. 2009). In addition, base-pairing between the U4 and U6 snRNAs must be disrupted to facilitate a conformational change in U6 to allow its base-pairing with the U2 snRNA and pre-mRNA (Brow 2002). While these dramatic changes in RNA conformation have been studied for a number of years, recent evidence has pointed to a series of additional transitions that occur during splicing.
smFRET and colocalization experiments have provided ample evidence for reversible steps in both spliceosome assembly (Hoskins et al. 2011) and pre-mRNA conformation (Abelson et al. 2010; Crawford et al. 2013; Krishnan et al. 2013). Even during late stages of assembly, the spliceosome-associated pre-mRNA undergoes a number of conformational transitions (Abelson et al. 2010; Crawford et al. 2013; Krishnan et al. 2013). Some of these transitions are likely occurring due to active site remodeling or “toggling” of the active site between different structures. This hypothesis incorporates the two-state model for spliceosomal catalysis in which active site conformations favoring either 5′ splice site cleavage or exon ligation are in competition with one another (Smith et al. 2008). These two catalytic conformations are separated from one another by an “open” state in which the lariat can be repositioned (Smith et al. 2008). Evidence for this hypothesis has come from biochemical genetics studies of the core splicing machinery (Query and Konarska 2004; Konarska et al. 2006; Liu et al. 2007) as well as in vitro studies of partially purified spliceosomes (Tseng and Cheng 2008, 2013). So far, a single active site structure of a spliceosomal complex has emerged (Yan et al. 2015) but conformational dynamics of active site components remain less well-characterized.
In addition to toggling of the active site, other snRNA components have been predicted to oscillate between multiple conformations that parallel the active site transitions (Smith et al. 2008). The stem II region of the U2 snRNA (Fig. 1A) can base-pair in two mutually exclusive conformations: stem IIa and IIc (stem IIb is present in both) (Fig. 1A,B; Ares and Igel 1990; Zavanelli and Ares 1991; Zavanelli et al. 1994; Hilliker et al. 2007; Perriman and Ares 2007). Stem IIa facilitates U2 association with the branchsite and prespliceosome formation (Zavanelli and Ares 1991), while stem IIc can inhibit these steps (Zavanelli et al. 1994). Even though stem IIc is not essential for spliceosome assembly, stabilization of stem IIc (or destabilization of stem IIa) increases the splicing of pre-mRNAs with mutant branchsite and 3′ splice site sequences (Hilliker et al. 2007; Perriman and Ares 2007). This suggests that stem IIc may play a role in the catalytic spliceosome during both transesterification reactions. Together with data that stem IIa stabilization (or destabilization of stem IIc) can suppress mutations in a late-stage splicing factor (Prp16p) (Hilliker et al. 2007; Perriman and Ares 2007), a model has emerged in which stem II switches iteratively between stem IIa and IIc as the spliceosome is assembled (IIa), carries out 5′ splice site cleavage (IIc), the active site is rearranged (IIa), and the exons are ligated (IIc) (Hilliker et al. 2007; Perriman and Ares 2007).
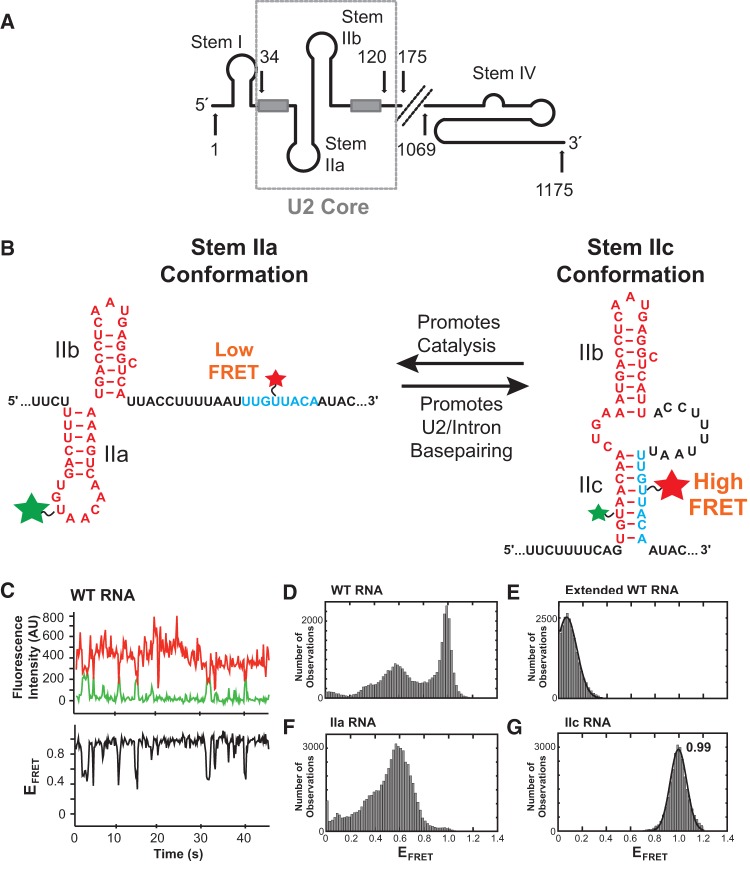
FIGURE 1.
Model RNA design and smFRET data. (A) Model of the yeast U2 snRNA. The U2 core RNA used in smFRET assays (nt 34–120, displayed as stem IIa/b) is boxed. Light gray boxes represent the branch point recognition sequence and Sm protein binding site. (B) Design of a smFRET reporter of stem II conformation. Fluorophores were placed to produce a high FRET signal during stem IIc formation when they come into close proximity. (C, top panel) Example smFRET data from the WT U2 core RNA showing anti-correlated changes in Cy3 donor (green) and Cy5 acceptor (red) fluorescence indicative of FRET. (Bottom panel) Calculated EFRET from the data in the top panel shows fluctuations between mid and high EFRET values. (D) Distribution of EFRET values for the WT U2 core RNA (N = 100 molecules). (E) Distribution of EFRET values after extension by hybridization to a complementary DNA oligo (N = 81). (F) Distribution of EFRET values for a stem IIa RNA (N = 105). (G) Distribution of EFRET values for a stem IIc RNA (N = 72). The data in E and G could be fit by a single Gaussian distribution (black lines) centered at 0.06 and 0.99, respectively.
While it is clear that stem II rearrangement is important for splicing, how this rearrangement occurs and is regulated is not known. One possibility is that protein cofactors could be influencing stem II conformation. Two potential candidates for this role are the RNA recognition motif-containing protein Cus2p and the DEAD-box ATPase Prp5p. Cus2p was originally isolated as cold-sensitive U2 snRNA suppressor 2 of the stem IIa-destabilizing G53A mutation (Yan et al. 1998). Although Cus2p is nonessential under normal growth conditions, it becomes essential when stem IIa is destabilized by mutation (Yan et al. 1998). Prp5p also shows a genetic interaction with stem IIa mutants. In this case, the ATP hydrolysis activity of Prp5p can be bypassed by either deletion of Cus2p or by stabilization of stem IIa (Perriman and Ares 2000; Perriman et al. 2003). Competing models have been proposed for how Cus2p and Prp5p regulate stem II structure. It was first proposed that Cus2p may bind to and stabilize a stem IIa-folded form of the U2 snRNA (Yan et al. 1998) based on isolation of Cus2p suppressors of stem IIa mutants. An alternate model proposed that Cus2p may bind more stably to stem IIc RNAs. This was based on coimmunoprecipitation (co-IP) assays in which Cus2p coimmunoprecipitated U2 snRNAs with destabilized stem IIa more efficiently than those in which stem IIc formation was prevented (Perriman and Ares 2007). In both models, Prp5p could then displace Cus2p and stabilize the stem IIa conformation prior to branchsite duplex formation (Liang and Cheng 2015). It is not clear if Prp5p may also actively destabilize stem IIc, facilitate annealing of stem IIa, or both. How stem IIa and IIc interconvert during the catalytic stages of splicing is also unclear. It has been speculated that Prp5p could be involved in this transition (Hilliker et al. 2007; Perriman and Ares 2007; Kosowski et al. 2009); however, recent work suggests that there is no obligate role for Prp5p after prespliceosome formation (Liang and Cheng 2015).
We have used smFRET to study the conformational dynamics of the stem II-containing core region of the U2 snRNA and how these dynamics are influenced by Cus2p and Mg2+. In the absence of Mg2+ and protein, we found that the most common FRET efficiency (EFRET) is consistent with stem IIc formation. Most frequently, stem IIc spontaneously transitions to conformations with similar EFRET values to those found in stem IIa-stabilized RNAs. Mg2+ or Cus2p alter this equilibrium to prevent stem IIc accumulation and induce stem IIa-specific dynamics. A stem IIa-destabilizing mutation, G53A, results in accumulation of stem IIc and suppression of both Cus2p- and Mg2+-dependent transitioning. Addition of mutant Cus2p proteins that suppress G53A in cells restores IIc to IIa conformational switching. Our data suggest that Mg2+ and Cus2p may work together to regulate stem II dynamics and IIa formation during prespliceosome formation. The spliceosome may subsequently use Mg2+ or functionally equivalent interactions with proteins to aid in stem IIa/IIc conformational transitions that occur during the catalytic stages of splicing.
RESULTS
The U2 core RNA is conformationally heterogeneous
To study conformational dynamics of U2 stem II by smFRET, we first designed a model RNA of the U2 core region (nucleotides 31–126; Fig. 1A,B). FRET donor (Cy3) and acceptor (Cy5) fluorophores were incorporated at positions U56 and U101 by derivatization of amino modifier C6 deoxythymidine nucleotides (Fig. 1B) in two separate RNA fragments. Additionally, the 3′ RNA fragment contained a terminal biotin for surface immobilization. The final U2 core model RNA was prepared by splinted RNA ligation of these two fragments. In our design, we predicted that stem IIc could be identified by a high smFRET signal since the two labeled nucleotides approach one another within the stem IIc duplex (Fig. 1B, U56-Cy3 base-pairs with A103 while A58 base-pairs with U101-Cy5). Stem IIa formation was predicted to give rise to lower smFRET. The model RNA was immobilized with streptavidin onto a PEG and PEG-biotin-derivatized quartz slide. Single-molecule fluorescence was observed by illumination on a prism-based total internal reflection fluorescence (TIRF) microscope with laser excitation at 532 and 640 nm.
We first collected data in the absence of Mg2+ and protein. Under these conditions, the immobilized RNAs showed many anti-correlated changes in FRET donor and acceptor intensities (Fig. 1C; Supplemental Fig. S1). These data were then used to calculate apparent EFRET (Fig. 1C). Analysis of trajectories from individual RNA molecules showed many transitions (Fig. 2A), most frequently oscillating between mid EFRET values of 0.4–0.6 and high EFRET values of ∼1.0. In order to interpret the observed transitions, we collected EFRET trajectories from several hundred molecules and plotted the resulting EFRET values as a histogram. The histogram showed several features: a sharp peak at ∼0.96 and less defined peaks centered near 0.35 and 0.54 (Fig. 1D).
FIGURE 2.
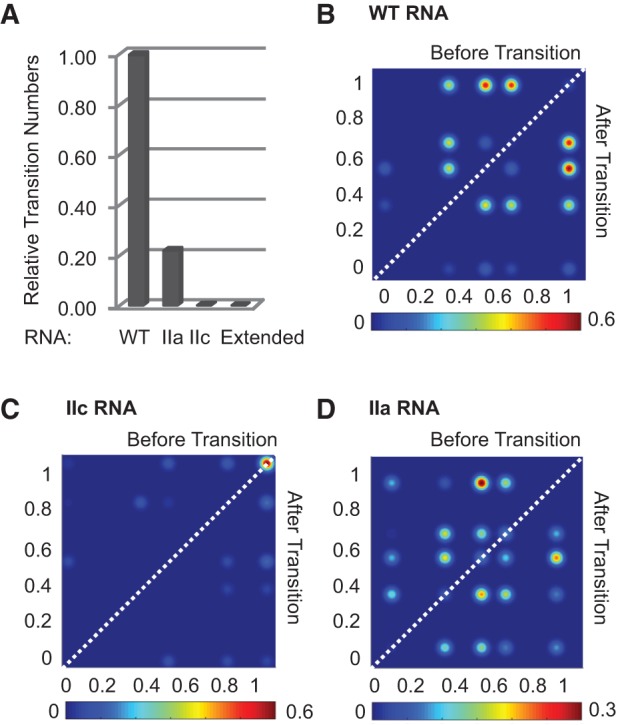
Analysis of smFRET dynamics from different stem II RNAs. (A) Relative number of changes in EFRET value (transitions) observed with WT and mutant U2 core RNAs. The WT RNA undergoes many more transitions than stabilized RNAs. (B) TODP of the WT U2 core RNA. Most frequently, this RNA transitions between EFRET of 0.54 or 0.67 and 0.96. (C) TODP of stem IIc RNA shows a single high EFRET of 0.99. (D) TODP of stem IIa RNA shows many transitions. In B–D, transitions are colored based on their observed frequency with red and blue colors representing more and less common transitions, respectively.
To assign EFRET values to specific U2 conformers we constructed a series of U2 core RNA variants. Few dynamics were observed when the RNA was extended by annealing to a complementary DNA oligo. This duplexed RNA produced EFRET signals centered at 0.06 (Fig. 1E) and suggests that the higher EFRET values observed in Figure 1D correspond to more compact RNA structures. We then prepared two different mutant U2 core RNAs: one stabilized into the stem IIa conformer and the other into stem IIc. Stem IIa was stabilized by introducing U50G, A52G, U63C, A65C mutations that have been used in vivo and substitute A:U for G:C base pairs (Perriman and Ares 2007). While molecules of the stem IIa RNA were less dynamic than wild type (WT) (Fig. 2A; Supplemental Fig. S1), they nonetheless showed interconversion between multiple EFRET states. Unlike WT, the stem IIa RNA rarely sampled the highest EFRET state (∼0.96). Instead, a broad EFRET distribution was observed that peaked at ∼0.54 (Fig. 1F). The FRET data alone do not allow us to predict potential structures that may be present in the region downstream from IIb, but it is possible that flexibility of this region may contribute to the observed IIa EFRET distribution—potentially through pseudoknot formation or other tertiary interactions. These experiments indicate that even though the RNA was “stabilized” into favoring stem IIa, it was still conformationally heterogeneous.
The stem IIc conformer was also stabilized using mutations previously used in vivo albeit not simultaneously: G53A, A95C, U97G (Perriman and Ares 2007). In this case, G53A is proposed to destabilize stem IIa by removing a G:C base pair, while A95C and U97G increase base-pairing of stem IIc. In contrast to the stem IIa RNAs, stem IIc-stabilized RNAs showed few dynamics (Fig. 2A; Supplemental Fig. S1). A histogram of stem IIc EFRET values showed a sharp distribution centered at EFRET of 0.99 (Fig. 1G). Together, these data support a model for the WT RNA at equilibrium in which the predominant structure is the stem IIc state (EFRET∼1) along with a mixture of conformers with similar EFRET values to those observed in a stabilized stem IIa RNA (EFRET 0.4–0.8).
U2 can spontaneously interconvert between stem IIc and stem IIa conformers
We next determined the routes by which the WT U2 core RNA was sampling the conformers observed in Figure 1D. Hidden Markov modeling (Mckinney et al. 2006) was used to define the distribution of EFRET states observed on individual U2 core RNA molecules. Models from multiple RNAs were then combined in transition occupancy density plots (TODPs [Abelson et al. 2010; Blanco and Walter 2010]) that depict the transitions occurring within the entire data set, each scaled based on its frequency of observation. While the most frequent transitions occurred between mid EFRET states of 0.67 or 0.54 and a high EFRET state of 0.96 (Fig. 2B), the conformational landscape sampled by the WT U2 RNA core was complex (Fig. 2B; Supplemental Table S1). As with our histogram analysis, we used mutant U2 core RNAs stabilized into stem IIa or IIc to guide interpretation of the TODP.
A TODP of stem IIc-stabilized RNA confirmed that the IIc RNA was rarely transitioning and adopted a high EFRET state of 0.99 (Fig. 2A,C). This suggests that the high EFRET state observed in the TODP of the WT RNA corresponds to stem IIc formation, in agreement with the histogram analysis. TODP analysis of the stem IIa RNA revealed a conformationally complex landscape with the RNA transitioning between a number of EFRET states (Fig. 2D; Supplemental Table S1). Visual inspection of the TODP plots revealed that the transitions occurring in the WT and stem IIa RNAs were similar. The major differences centered on transitions to the highest EFRET state, which was 0.96 in the WT RNA and 0.91 in the stem IIa-stabilized mutant. It is possible that in both cases this EFRET value represents stem IIc base-pairing, since the stem IIa-stabilizing mutations would not prevent stem IIc from forming even though they prevent its accumulation (Fig. 1F). These results are consistent with the WT RNA transitioning from stem IIc to a subset of conformations also observed upon stabilization of stem IIa. Stem IIa stabilization, in turn, changes the transitions occurring out of the mid- EFRET states.
Subtle changes in stem IIb impact IIa/IIc dynamics
Stem IIb is predicted to remain unchanged as stem II interconverts between IIa and IIc (Fig. 1B). Serendipitously, we were able to study smFRET transitions occurring in stem II when IIb contains a single deoxyuridine nucleotide at position U73. Removal of this single hydroxyl group appears to destabilize stem IIa and increase the abundance of stem IIc (Supplemental Fig. S2). In addition, this substitution appears to alter the dynamic landscape of the RNA (Supplemental Fig. S2C,E). These results indicate that while stem IIb is not predicted to change its base-pairing during stem IIa/IIc transitions, it is nonetheless participating in the reaction in more than just a bystander role. This supports previous data demonstrating that mutations that destabilize stem IIb (G69A and C84U) can also suppress the cold-sensitive and stem IIa-destabilizing G53A mutation (Zavanelli et al. 1994).
Surrounding RNA structures influence stem II conformational dynamics
It is possible that other U2 RNA structures surrounding stem II can influence stem II conformation and dynamics. To test this hypothesis, we constructed a smFRET reporter RNA corresponding to U2 snRNA nt 1–126. This reporter RNA is capable of forming stem I and the branchsite stem loop (BSL; Perriman and Ares 2010) in addition to stem II. As with the smaller RNA construct, smFRET analysis revealed that this longer RNA was conformationally dynamic. However, we observed a loss in the highest EFRET signals corresponding to stem IIc conformation (Supplemental Fig. S3). While these results could be consistent with coexistence of the BSL and stem IIa, further dissection of the RNA dynamics and structure will be needed to determine whether any conformational rearrangements in U2 stem loop regions are coupled to one another.
Mg2+ decreases stem IIc abundance and induces stem IIa dynamics
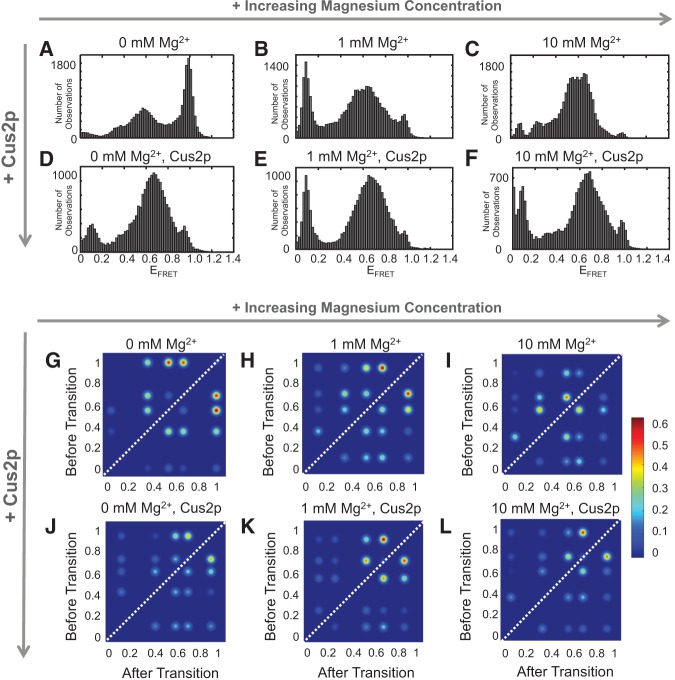
We next wondered what role cofactors could play in the conformational dynamics observed in the WT U2 core RNA. Near physiological levels of Mg2+ (1 mM) we observed a dramatic change in EFRET values (Fig. 3A versus 3B) and an overall decrease in RNA dynamics (Supplemental Fig. S4). Instead of the stem IIc conformation with a high EFRET predominating the WT histogram, this conformation was greatly reduced and mid EFRET states were most abundant. Notably, the TODP of the WT RNA in the presence of 1 mM Mg2+ more strongly resembles that of the stem IIa-stabilized RNA in the absence of Mg2+ (Fig. 2D versus 3H; Supplemental Table S1). Low EFRET signals also begin to appear in the presence of Mg2+, suggesting that Mg2+ could assist in transitions to more extended conformations from the mid-EFRET states, although thorough analysis of these transitions is complicated since EFRET < 0.1 approaches our lower limit of detection. When the Mg2+ concentration was further increased to 10 mM, transitions to the highest EFRET states were suppressed further (Fig. 3I; Supplemental Table S1; Supplemental Fig. S5). Mg2+ impacts the structure of the WT U2 core in two ways: It prevents accumulation of the stem IIc conformation and induces a set of conformations and dynamics that also occur when stem IIa is stabilized by mutation.
FIGURE 3.
Influence of cofactors on formation of U2 stem IIa and IIc. (A–F) Histograms of EFRET states observed for the WT RNA in the presence or absence of Mg2+ and/or Cus2p. Increasing concentrations of Mg2+ diminish the abundance of the high EFRET values near 1.0 and increase the abundance of mid-EFRET values. Addition of Cus2p causes the same effect and further reduces the number of high EFRET values observed at 1 mM Mg2+. These data are consistent with Mg2+ and Cus2p suppressing stem IIc formation and increasing the abundance of stem IIa. (G–L) TODPs for data shown in A–F. Both Mg2+ and Cus2p induce patterns in the TODPs also observed when stem IIa was stabilized by mutation. Transitions to EFRET states of ∼0.3 were suppressed in the presence of Cus2p. In G–L, data are colored as described in Figure 2. Each histogram and TODP represents data collected from N = 100–175 molecules.
Cus2p binds to U2 stem II core RNAs
Given the dramatic change in dynamics elicited by Mg2+, we wondered what impact a protein cofactor could have on RNA behavior. We cloned and purified Saccharomyces cerevisiae Cus2p containing an amino-terminal 6× polyhistidine affinity tag (Supplemental Fig. S6A) and tested its RNA binding ability. Previous work demonstrated that Cus2p changes the mobility of a U2 RNA transcript in an electrophoretic mobility shift assay (EMSA; Yan et al. 1998). To determine whether Cus2p could bind the stem II-containing core region, we carried out EMSAs using this smaller RNA domain of U2. Radiolabeled RNAs containing nucleotides 34–120 of the U2 snRNA were prepared by in vitro transcription. Stem IIa was stabilized by including the same mutations used in the smFRET assays and deletion of nucleotides 97–105, which form the 3′ half of stem IIc (the ΔCC mutation [Perriman and Ares 2007]). Stem IIc was stabilized using the same mutations used in the single-molecule assays. RNase T1 digests of these RNAs showed a high cleavage sensitivity at G100 in the WT RNA but not in the stem IIc-stabilized RNA, confirming that this nucleotide was base-paired in the latter case (Supplemental Fig. S7). No additional bands were detected in the stem IIc-stabilized RNA from U97G or G64, indicating that these nucleotides were also engaged in base-pairing interactions to extend and stabilize stem IIc. Similarly, no bands were detected from U50G, A52G, or G53 in the stem IIa-stabilized construct, indicating that stem IIa was intact and these nucleotides were also base-paired (Supplemental Fig. S7).
Upon addition of Cus2p, we observed a change in mobility with each of the RNAs that were tested (Fig. 4). Most often the shifted RNA traveled just above the free RNA as a diffuse band—likely due to weak interaction with Cus2p as well as conformational heterogeneity of the RNA (Fig. 4, Shift-1). Addition of cold WT stem II RNA was able to abolish the gel shift (Supplemental Fig. S8) as did use of a previously described Cus2p Y48D mutant in RRM1, as expected from previous results (Supplemental Fig. S6A,B; Yan et al. 1998). While the diffuse gel shift we observed makes quantification of Cus2p/RNA interaction difficult, no significant differences in the fraction of WT or stem IIa RNAs appearing in Shift-1 relative to the unshifted RNA were apparent (Supplemental Fig. S9). Together these results suggest that Cus2p is interacting with these radiolabeled RNAs and these interactions resulted in shifted RNA bands.
FIGURE 4.
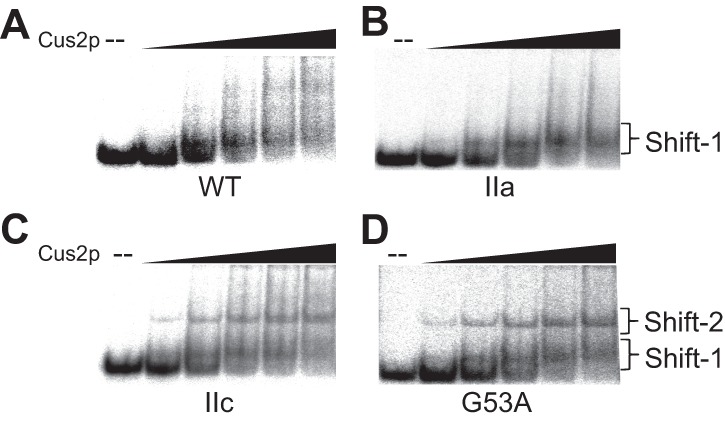
(A–D) EMSA analysis of Cus2p interactions with various U2 stem II core model RNAs. Each RNA was incubated with a variable amount of Cus2p (0 to 2 µM). A single prominent shift (Shift-1) was observed with the WT and stem IIa-stabilized RNAs (A,B), while the stem IIc and G53A RNAs (C,D) also produced a higher mobility shift (Shift-2).
In the case of the stem IIc-stabilized RNA, a second, higher gel shift could also be observed (Fig. 4C, Shift-2). This indicates that while Cus2p can interact with WT, stem IIa-, and stem IIc-stabilized RNAs an additional interaction could be observed when stem IIc was stabilized. This observation could indicate specific binding of stem IIc by Cus2p; however, from this EMSA analysis alone it is unclear which of the shifts observed may be the most relevant to Cus2p function during prespliceosome formation.
Cus2p decreases stem IIc abundance and induces a subset of stem IIa dynamics
When Cus2p was added to the WT U2 model RNA in a smFRET assay, we also observed a dramatic change in EFRET signals (Fig. 3D). Like Mg2+, Cus2p prevented accumulation of the high EFRET state (0.96) corresponding to stem IIc formation. This effect is not generally due to inclusion of protein in the assay since addition of a control protein (BSA) did not prevent our observation of stem IIc accumulation (Supplemental Fig. S10A,C). Loss of the high EFRET state was observed both in the presence of the polyhistidine tag and after its removal by protease cleavage (data not shown). TODP analysis showed that Cus2p also induced a distribution of EFRET states similar to that observed in the stem IIa-stabilized mutant and in the WT RNA in the presence of Mg2+ (Fig. 3J; Supplemental Fig. S5; Supplemental Table S1). All EFRET states observed in the stem IIa mutant were also present in the WT RNA in the presence of Cus2p.
Despite the appearance of similar EFRET states in the presence of Cus2p, Mg2+, or stem IIa-stabilizing mutations, TODPs revealed certain transitions varied in abundance under each condition. In particular, Cus2p strongly suppressed observation of transitions to the 0.30 EFRET state (Fig. 3J). In contrast, transitions between these EFRET states were more abundant in the presence of Mg2+ and absence of Cus2p (Fig. 3I). When Cus2p and Mg2+ were combined, transitions to the 0.30 EFRET state continued to be less frequent (Fig. 3K,L). These results indicate that Cus2p can decrease the prevalence of some Mg2+-dependent transitions and conformations under physiological conditions.
We then tested whether stem IIa or IIc mutations could counteract the action of Cus2p. Addition of Cus2p to the stem IIc-stabilized RNA had no impact on the observed transitions and the high EFRET state of 0.99 remained. Cus2p was unable to induce formation of EFRET states consistent with stem IIa, and TODPs in the presence or absence of Cus2p were nearly identical (Supplemental Fig. S11A). These stem IIc-stabilizing mutations antagonized Cus2p's ability to suppress stem IIc formation. Furthermore, these results indicate that if Cus2p is forming a specific interaction with the stem IIc-stabilized RNA (as evidenced by Shift-2 in Fig. 4C), it does so in a manner that does not result in a change in EFRET.
Addition of Cus2p to the stem IIa-stabilized RNA slightly changed the complex pattern of transitions, but transitions to the 0.33 state continued to be less frequently observed by TODP analysis (Supplemental Fig. S11B). Therefore, stem IIa-stabilizing mutations do not antagonize the ability of Cus2p to suppress observation of certain conformations or transitions.
Unexpectedly, while the Y48D mutant of Cus2p did not induce a gel shift of the WT U2 RNA (Supplemental Fig. S6), this protein did suppress formation of stem IIc in smFRET experiments (Supplemental Fig. S10A,B). The Y48D mutant was also able to suppress observation of the 0.33 EFRET state in TODPs. Based on these observations, we wondered if Y48D Cus2p was still interacting with stem II to induce changes in smFRET but perhaps this interaction was too weak to observe by EMSA. Since we observed an additional interaction only upon stabilization of stem IIc (Shift-2, Fig. 4), we tested whether or not Y48D Cus2p could produce this particular gel shift. We incubated Y48D Cus2p with the stem IIc RNA and observed a single complex being formed with mobility in between Shifts 1 and 2 (Supplemental Fig S10D,E). These results suggest that while the Y48D mutation weakens Cus2p's interaction with non-stem IIc RNAs, it does not impact the protein's ability to associate with stem IIc-stabilized RNAs. Thus, changes in smFRET observed with Y48D Cus2p could arise from binding of the protein to stem IIc and facilitating conversion of the RNA to other conformations to which it binds less well. Our combined EMSA and smFRET results are consistent with a model in which Cus2p can bind to both stem IIc and stem IIa and in which Cus2p binding to stem IIc facilitates formation of stem IIa.
Destabilization of stem IIa can alter stem II response to cofactors
As previously described, Cus2p was originally isolated as a suppressor of the cold-sensitive G53A stem IIa mutation (Yan et al. 1998). This mutation was proposed to destabilize stem IIa by introduction of an A/C mismatch in stem IIa and can be suppressed by mutations found close to the carboxy-terminus of Cus2p (the cus2–9 and cus2–25 alleles, corresponding to L284F and D282N mutations, respectively). We wondered how the G53A mutation would impact stem II dynamics as well as interactions with both Mg2+ and Cus2p.
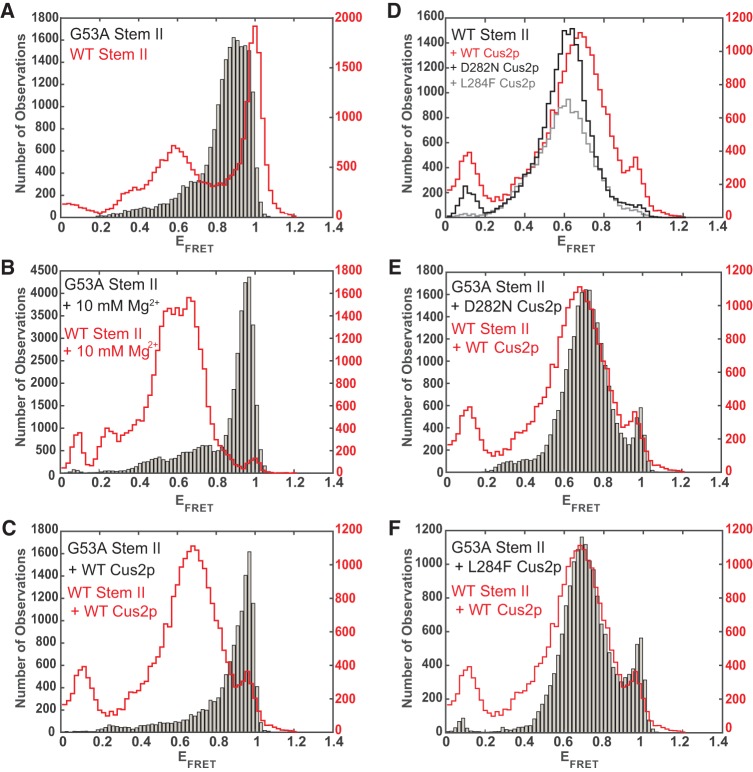
As expected for a stem IIa destabilizing mutation, histogram analysis of the G53A RNA showed a decrease in the mid-FRET signals that correspond to stem IIa (Fig. 5A). The distribution of the highest EFRET signals were not as closely clustered around an EFRET of ∼1 as in the WT or stem IIc stabilized RNAs. This suggests that while G53A destabilizes stem IIa, the distribution of RNA conformations that result may not be identical to those formed with other RNAs.
FIGURE 5.
Results from smFRET analysis of G53A RNAs show suppression of Mg2+ and Cus2p-dependent toggling. (A) Histogram of EFRET values from the G53A RNA in buffer without Mg2+ (gray bars, left y-axis) showing predominantly high FRET conformations compared with results obtained with the WT RNA (red outline, right y-axis; from Fig. 1D). (B) Histogram of EFRET values from the G53A RNA with 10 mM Mg2+ (gray bars, left y-axis) compared with results obtained with the WT RNA with 10 mM Mg2+ (red outline, right y-axis; from Fig. 3C). (C) Histogram of EFRET values from the G53A RNA with WT Cus2p (gray bars, left y-axis) compared with results obtained with the WT RNA with WT Cus2p (red outline, right y-axis; from Fig. 3D). (D) Outlines of histogram of EFRET values from the WT RNA in the presence of either WT Cus2p (red and right y-axis; from Fig. 3D), D282N Cus2p (black and left y-axis), or L284F Cus2p (gray and left y-axis). (E) Histogram of EFRET values from the G53A RNA with the D282N Cus2p suppressor (gray bars, left y-axis) compared with results obtained with the WT RNA with WT Cus2p (red outline, right y-axis; from Fig. 3D). (F) Histogram of EFRET values from the G53A RNA with the L284F Cus2p suppressor (gray bars, left y-axis) compared with results obtained with the WT RNA with WT Cus2p (red outline, right y-axis; from Fig. 3D). Each histogram represents data collected from N = 105–112 molecules.
Surprisingly, when we added 10 mM Mg2+ to the G53A RNA, we observed a distribution of EFRET values consistent with a predominant stem IIc conformation (Fig. 5B). That is, while Mg2+ strongly suppresses stem IIc formation in the WT sequence, the G53A mutation allows stem IIc to persist in the presence of Mg2+. Not only does G53A destabilize stem IIa, this mutation also appears to influence Mg2+-dependent toggling.
Given these results, we next wondered how the G53A mutant RNA would respond to addition of Cus2p. EMSA analysis showed that the Cus2p could bind G53A and produce shifts at similar protein concentrations to those observed with other RNA constructs—including the higher gel shift observed upon stem IIc stabilization (Fig. 4). Despite the gel shifts observed by EMSA, addition of WT Cus2p to the G53A RNA in the smFRET assay did not result in suppression of the stem IIc signal or in accumulation of stem IIa (Fig. 5C). Thus, the G53A mutation appears to suppress conformational transitions influenced by both Mg2+ and Cus2p, and Cus2p interactions with the G53A RNA in the smFRET assay do not result in changes to the high EFRET signal.
We also purified mutant D282N and L284F Cus2 proteins and tested their ability to alter the structure of the stem II smFRET reporters. Both suppressor proteins behaved identically to WT Cus2p in the presence of the WT RNA to suppress accumulation of stem IIc (Fig. 5D). Upon addition to the G53A RNA, both suppressors caused a decrease in stem IIc smFRET signals and increase in mid-EFRET states corresponding to stem IIa formation (Fig. 5E,F). Both suppressor proteins restore behaviors to the G53A RNA that we previously observed with the WT RNA sequence and WT Cus2p. Our smFRET assays corroborate previous findings that the G53A mutation destabilizes stem IIa (Ares and Igel 1990) and suggest that this destabilization could occur through disruption of both base-pairing and interactions with Mg2+ and Cus2p. These results also indicate that the function of the Cus2p suppressor mutations may be to restore stem IIc to IIa switching by Cus2p in the presence of IIa-destabilizing mutations.
DISCUSSION
While conformational transitions are implicated in nearly every stage of splicing, many changes in snRNA structure remain uncharacterized due to a lack of structural and biochemical information. We used smFRET to investigate the dynamics of a portion of the U2 snRNA containing the conserved stem II region. Our results indicate that the RNA is dynamic and contains conformations consistent with stem IIc and a mixture of stem IIa conformers. In our model, this equilibrium can be shifted by addition of Mg2+ or Cus2p or by altering the RNA sequence by including a destabilizing G53A mutation (Fig. 6). After addition of Mg2+ or Cus2p to the WT RNA, the RNA remains dynamic, fails to populate the stem IIc conformer, and undergoes a set of conformational transitions also observed when stem IIa is stabilized by mutagenesis. The stem IIa-destabilizing G53A mutation appears to not only change the equilibrium between IIc and IIa but also to alter conformational switching of the RNA by Mg2+ and Cus2p. This interconversion can be restored, however, with Cus2p mutants that suppress G53A in vivo. These results are consistent with Cus2p and Mg2+ facilitating a structural transition within stem II from the IIc to IIa conformers.
FIGURE 6.
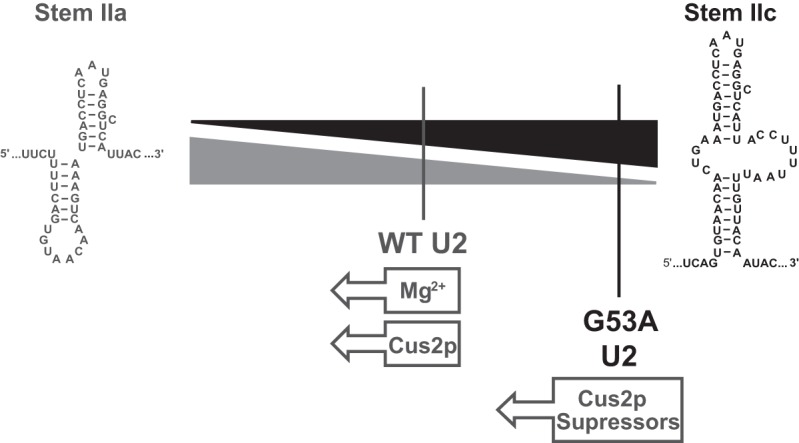
Model for control of U2 stem II toggling with Mg2+ and Cus2p. Stem II exists in dynamic equilibrium between stem IIa and IIc. For the WT RNA, the stem IIc conformer is in approximately equal abundance to a mixture of conformers also observed upon stem IIa stabilization. Addition of either Mg2+ or Cus2p results in suppression of stem IIc and drives the equilibrium toward IIa. The G53A mutant exists predominantly in the stem IIc conformer but can be driven toward IIa by the addition of Cus2p suppressor proteins.
Stem II toggling is spontaneous
Our single molecule data clearly show that the stem II region of the U2 snRNA can interconvert between multiple FRET states that likely correspond to stem IIc and stem IIa base-pairing. While a number of genetic and structural probing experiments have previously indicated that stem II can adopt these conformations, the smFRET experiments reported here provide direct evidence that interconversion between these two conformations can happen spontaneously without the need for an ATP-dependent helicase. Even though these transitions are spontaneous, they are heavily influenced by both metal ions (Mg2+) and proteins (Cus2p) (Fig. 6). Both of these cofactors change stem IIa/IIc equilibrium in a similar manner by increasing stem IIa conformers at the expense of stem IIc. Together these results suggest that components of the U2 snRNP and the spliceosome may be necessary to prevent spontaneous refolding of stem II to an unwanted conformation at an inopportune moment.
smFRET provides a new perspective on Cus2p function
In comparison with EMSAs, smFRET provides additional information when studying RNA/protein interactions by detecting conformational changes that result from these interactions. While our EMSA results suggest that Cus2p can interact with stem II in either the IIa or IIc conformation with an additional shift observed when IIc is stabilized (Fig. 4), the smFRET results reveal that the consequence of interaction between WT stem II and Cus2p is a decrease in IIc FRET signals and an increase in those corresponding to IIa (Figs. 3, 5). Mutations that stabilize stem IIc do not prevent Cus2p from binding (Fig. 4) but do prevent stem IIa formation (Fig. 5; Supplemental Fig. S11). Together the data are consistent with Cus2p facilitating the transition of IIc to IIa and being able to bind both the substrate (IIc) and product (IIa) of that transition. Mutations to Cus2p or the RNA may potentially prevent accumulation of IIa by either stabilizing the IIc substrate against Cus2p action or by destabilizing a IIa/Cus2p product complex.
It has been previously shown that Cus2p can coimmunoprecipitate more U2 snRNA containing a mutation (G53A) that destabilized stem IIa than when the snRNA was rendered incapable of forming stem IIc due to the ΔCC mutation (Perriman and Ares 2007). In light of our smFRET and EMSA data, increased co-IP of the G53A mutant may have resulted from Cus2p binding to the stem IIc conformer of that U2 but failing to toggle the RNA and release stem IIa. Lower amounts of U2 snRNA being pulled down with the ΔCC/stem IIa snRNA may indicate that Cus2p has lower avidity for binding stem IIa or that the ΔCC mutation promotes dissociation of Cus2p. Since co-IP assays often do not have a kinetic component, it is also possible that ΔCC/stem IIa RNAs may have already progressed beyond the point of Cus2p action and those U2 snRNPs may be already primed for branchsite association by Prp5p.
One consequence of our model is that Cus2p may also function to convert stem II from IIc to IIa within the intact U2 snRNP. The ability of Cus2p suppressor proteins to restore IIc to IIa toggling in a G53A mutant RNA (Fig. 5) and to suppress the G53A mutation in vivo (Yan et al. 1998) suggests that features observed in our model system may also be present in U2. However, it is also possible that specific interactions that occur within the snRNP are not being recapitulated within the model RNA and with recombinant protein. While we have shown that Cus2p appears to switch stem II from IIc to IIa, this should be interpreted with caution, and we cannot yet exclude other models for Cus2p function or predict the influence of other U2 components (e.g., the SF3 complex). Understanding how these components influence stem II smFRET dynamics and Cus2p binding will be critical for dissecting transitions occurring within intact U2.
Implication for the roles of Cus2p and Prp5p during prespliceosome formation
In the absence of Cus2p, ATP hydrolysis by Prp5p is no longer required to form the prespliceosome (Perriman and Ares 2000), and this result has led to a model in which Prp5p activates U2 by first removing Cus2p (Liang and Cheng 2015). In light of our model that Cus2p may function to directly form stem IIa (which in turn is required for prespliceosome formation), an alternate proposal is that Cus2p and Prp5p work in tandem during U2 activation. The RNA structure-specific RRM domain of Cus2p could be used to facilitate stem II conformational rearrangements in preparation for loading of Prp5p. The DEAD-box Prp5p could then displace Cus2p and provide proofreading for snRNA/branchsite duplex formation via regulated ATP hydrolysis (Xu and Query 2007) or by timing of its release from the prespliceosome (Liang and Cheng 2015). This concept of Cus2p and Prp5p working in tandem is particularly attractive if stem II conformational change, removal of Cus2p, Prp5p loading, and BSL opening are a highly coupled series of events. Efficient loading of Prp5p onto U2 and timing of BSL opening may be aided by optimization of the stem II region by Cus2p. This interpretation is similar to one previously proposed by Perriman and Ares (2000) and is supported by genetic interactions between Cus2p and some alleles of Prp5p. Overexpression of Cus2p or Cus2 suppressor proteins can eliminate the temperature sensitivity of a mutation in Prp5p (prp5-1), while deletion of Cus2p results in the prp5-3 mutation becoming lethal (Perriman and Ares 2000). Neither of these results would necessarily be expected if Cus2p and Prp5p operated independently of one another or if Cus2p functioned solely as a block to prespliceosome formation that is removed by Prp5p.
Magnesium-dependent transitions within the splicing machinery
Our smFRET data show that a helicase such as Prp5p is not required for disruption of stem IIc or IIa. Stem II interconverts spontaneously between different structures, and stem IIc can be suppressed merely by addition of Mg2+. In the spliceosome it is unclear how Mg2+ accessibility could be regulated within the stem II region during specific stages of splicing or during transitions between different spliceosomal complexes. In a recent cryo-electron microscopy structure of a spliceosomal complex, the stem II region is both solvent accessible and disordered, likely due to dynamic exchange between different conformations and the absence of the SF3 complex (Yan et al. 2015). It is possible that the Mg2+-dependent transitions we observe here could be acting as a surrogate for other interactions present in the spliceosome but not in our model system (e.g., positively charged amino acid side chains). Switching stem II conformation from IIc to IIa during catalysis could therefore rely on collaboration between Mg2+ and proteins. On the other hand, our results could indicate that the spliceosome may need to make specific interactions with the stem IIc conformer to prevent spontaneous switching to IIa by cellular Mg2+. In this case, Mg2+ could facilitate IIc to IIa toggling once stabilizing interactions between the spliceosome and IIc have been disrupted.
The Mg2+-dependent transitions we observe with stem II are reminiscent of those previously observed with other spliceosomal components, namely conformational dynamics of a U2/U6 RNA duplex (Guo et al. 2009) and the RNase H domain of the spliceosomal protein, Prp8p (Abelson 2013; Schellenberg et al. 2013). It is unclear whether Mg2+-dependent structural transitions represent tools used by the spliceosome or are merely diagnostic of more flexible conformations of spliceosomal snRNAs and proteins. Nonetheless, control of toggling of spliceosomal components by metals and proteins could represent an important strategy for how catalytic RNPs may have evolved to regulate conformational transitions.
MATERIALS AND METHODS
Preparation of RNAs
RNAs for smFRET experiments were prepared by splinted ligation of two synthetic fragments (IDT) comprising nucleotides 31–78 and 79–126 of the yeast U2 snRNA. The 5′ fragment contained an amino allyl incorporated at position U56 for fluorophore derivatization (AGUGUAGUAUCUGUUCUUUUCAGUG-/iAmMC6T/AACAACUGAAAUGACCUCAAUG). The 3′ fragment contained both an amino allyl incorporated at position U101 and a 3′ biotin for surface immobilization (AGGCUCAUUACCUUUUAAUUUG/iAmMC6T/UACAAUACACAUUUUUUGGCACCCA-BIO). RNAs stabilized by mutation into stem IIa or IIc were created by purchasing oligos containing U50G/A52G/U63C/A65C or G53A/A95C/U97G mutations, respectively. Additional RNAs containing a deoxy substitution at U73 in stem IIb, the G53A mutation in stem IIa, or the full 5′ region of U2 were prepared by purchasing 5′ fragments containing those mutations.
All oligos were labeled with fluorophores by reaction with a single vial Cy3- or Cy5-NHS monoreactive dye (GE Healthcare Life Sciences) according to the manufacturer's directions. The labeling buffer used was 100 mM Na2B4O7 (pH 8.8) and labeling was carried out in the dark at room temperature overnight. Excess dye was removed using a G-25 MicroSpin column (GE Healthcare Life Sciences) prior to gel purification.
Labeled RNA fragments were joined using splinted ligation (Crawford et al. 2013) with a DNA splint (AAAAGGTAATGAGCCTCATTGAGGTCATTTCA) and the double-stranded T4 RNA ligase 2 (NEB). Ligated RNAs were purified using a denaturing polyacrylamide gel electrophoresis, eluted from the gel fragments, resuspended in water, and stored as concentrated stocks (1 µM) at −20°C.
smFRET experimental conditions
RNAs were immobilized onto PEG/PEG-biotin derivatized quartz slides as previously described (Crawford et al. 2013). smFRET experiments were carried out in 1× imaging buffer (20 mM HEPES at pH 7.9, 1 mM EDTA, 125 mM KCl, 5% glycerol, 0.1% Triton X-100, 1 mM DTT) using protocatechuate dioxygenase as the oxygen scavenger. Experiments with the G53A RNA were carried out in the same buffer except trolox (2 mM) was added in place of DTT and glucose oxidase/catalase was used as the oxygen scavenger, neither buffer modification affects stem II conformational switching. RNasin (Promega) was added to the buffers at a concentration of 0.2 U/µL. When present, Cus2p was added to a final concentration of 3 or 6 µM (for WT RNA and G53A RNA experiments, respectively). Buffers containing Mg2+ did not contain EDTA.
Data acquisition protocol for smFRET experiments
smFRET data were collected on a prism-based total internal reflection fluorescence (TIRF) microscope with laser excitation at 532 and 640 nm. The 640 nm laser (12–14 mW, measured prior to entry into the prism) excitation was used to localize the RNA at the beginning and the end of each experiment. The 532 nm laser (10 mW, measured prior to entry into the prism) was used to detect FRET with a frame rate of 200 msec for a time span of 3 min with continuous data collection. Donor and acceptor emissions were collected simultaneously using a DualView apparatus (DV2, Photometrics) and an EM-CCD camera (Andor). Images were recorded using Metamorph software (Molecular Devices).
Single-molecule data analysis
Raw microscope images were analyzed using custom MATLAB (Mathworks) software. Integrated fluorescence intensities for donor (ICy3) and acceptor (ICy5) spots of fluorescence were then used to calculate apparent FRET efficiencies by EFRET = ICy5/(ICy3 + ICy5). Hidden Markov modeling was performed using HaMMy software (Mckinney et al. 2006). Transition occupancy density plots (TODPs) were prepared as previously described (Abelson et al. 2010; Blanco and Walter 2010).
Cloning, expression, and purification of recombinant Cus2p
Cus2p was cloned from S. cerevisiae genomic DNA by PCR and placed into the p11 expression plasmid (DNASU [Seiler et al. 2014]) at the NdeI and BamHI restriction sites using standard procedures. Protein was produced in Escherichia coli BL21(DE3) Gold cells by growing the bacteria in LB media at 37°C until an OD600 of ∼0.6 was reached. At that time, protein production was induced by addition of 1 mM IPTG, and cells were incubated for an additional 3 h at 37°C. Cells (typically from a 1 L culture) were collected by centrifugation and stored at −80°C.
Cus2p was purified by first resuspending the cell pellet in 15 mL of 1× IMAC binding buffer (Immobilized Metal Affinity Chromatography; 25 mM HEPES at pH 7.6, 500 mM NaCl, 10% glycerol, 0.5 mM DTT, 25 mM imidazole). Protease inhibitors (Pierce Protease Inhibitor Mini Tablets, EDTA-free), 100 ng/µL lysozyme, and DNase (Turbo DNase [Life Technologies], 15 µL) were then added and incubated with the cell suspension for 30 min at room temperature. Cell lysis was then continued by sonication on ice (40% power level, 3 cycles of 3 sec on/2 sec off for 1 min with a 1-min rest between cycles). Cell debris was removed by centrifugation for 30 min at 4°C at 18,000 rpm in a Beckman J-20 rotor. The resulting supernatant was then filtered through a 0.4 µm filter before being loaded onto a 1 mL HisTrap column (GE Healthcare Life Sciences) attached to an AKTA Prime protein purification system. Cus2p was eluted using a gradient of IMAC buffer against IMAC buffer with 500 mM imidazole and 1 mL fractions were collected. Cus2p containing fractions were identified by SDS-PAGE, combined, and dialyzed against 10 mM HEPES (pH 7.9), 250 mM NaCl overnight at 4°C. After dialysis, the protein was aliquoted and stored at −80°C. Removal of the his-tag was accomplished by incubation of Cus2p with TEV protease (purified in laboratory) during dialysis at a mass ratio of Cus2p:protease of 50:1. The his-tag, remaining his-tag-Cus2p, and his-tag-TEV were separated from the cleaved protein using a HisTrap column as before. Cleaved Cus2p fractions were dialyzed, aliquoted, and stored following procedures for the uncleaved protein.
Preparation of Cus2p mutant proteins
D282N and L284F mutant Cus2 proteins were prepared by PCR and inclusion of the point mutations in the reverse primers. Mutant proteins were expressed using procedures described for WT Cus2p and purified by IMAC.
Preparation of RNAs for EMSAs
For EMSA and structure probing assays, different model T7-U2 snRNA constructs were prepared by in vitro transcription using T7 RNA polymerase. Transcripts were either internally labeled with [α-32P]-UTP during transcription or post-transcriptionally 5′ end labeled with [γ-32P]-ATP using T4 polynucleotide kinase. After transcription or labeling, RNAs were gel-purified, suspended in water and stored at 1 µM at −20°C.
RNA mobility shift assays
RNA–protein interactions using His-tagged Cus2p variants and model U2 snRNAs were studied essentially as previously described (Yan et al. 1998). Various amounts of Cus2p (0.4–2 µM) were incubated with 20 fmol of 32P-labeled model U2 RNAs in 1× EMSA buffer (20 mM HEPES at pH 7.9, 1 mM EDTA, 125 mM KCl, 5% v/v glycerol, 0.1% v/v Triton X-100, 1 mM DTT), 2 pmol yeast total tRNA per 10 µL reaction, and RNasin RNase inhibitor (0.3 U per 10 µL reaction, Promega) for 30 min at 30°C. Reaction products were separated in a 5% nondenaturing polyacrylamide gel (60:1) buffered with 0.5× TBE for 4 h at 150 V at 4°C, also using 0.5× TBE as running buffer. RNA-protein complexes were visualized by phosphorimaging and data analyzed using Image Quant software (GE Healthcare Life Sciences).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Manny Ares, Clara Kielkopf, Charles Query, Sam Butcher, Dave Brow, Marv Wickens, and members of the Hoskins laboratory for helpful discussions. We also acknowledge the reviewers of the manuscript for making many helpful suggestions. We thank the laboratories of Taekjip Ha and Nils Walter for smFRET analysis software. We thank Fatemehsadat Jamalidinan for computer software development. We acknowledge support from startup funding from the University of Wisconsin-Madison, Wisconsin Alumni Research Foundation (WARF), and the Department of Biochemistry. All authors are supported by awards from the National Institutes of Health (R00 GM086471 and R01 GM112735), the Arnold and Mabel Beckman Foundation, and the Shaw Scientist Program of the Greater Milwaukee Foundation. M.L.R. is supported by the Molecular Biophysics Training Program (NIH T32-GM08293).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.052233.115.
REFERENCES
- Abelson J. 2013. Toggling in the spliceosome. Nat Struct Mol Biol 20: 645–647. [DOI] [PubMed] [Google Scholar]
- Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, et al. 2010. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol 17: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M Jr, Igel AH. 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev 4: 2132–2145. [DOI] [PubMed] [Google Scholar]
- Blanco M, Walter NG. 2010. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol 472: 153–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA. 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. 2013. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. Proc Natl Acad Sci 110: 6783–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Karunatilaka KS, Rueda D. 2009. Single-molecule analysis of protein-free U2-U6 snRNAs. Nat Struct Mol Biol 16: 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. 2011. Ordered and dynamic assembly of single spliceosomes. Science 331: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC. 2006. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 21: 543–553. [DOI] [PubMed] [Google Scholar]
- Kosowski TR, Keys HR, Quan TK, Ruby SW. 2009. DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle. RNA 15: 1345–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. 2013. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat Struct Mol Biol 20: 1450–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W-W, Cheng S-C. 2015. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev 29: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM. 2007. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol 14: 519–526. [DOI] [PubMed] [Google Scholar]
- Mckinney SA, Joo C, Ha T. 2006. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J 91: 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr. 2000. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev 14: 97–107. [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr. 2010. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell 38: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Barta I, Voeltz GK, Abelson J, Ares M Jr. 2003. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci 100: 13857–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM. 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–354. [DOI] [PubMed] [Google Scholar]
- Schellenberg MJ, Wu T, Ritchie DB, Fica S, Staley JP, Atta KA, LaPointe P, MacMillan AM. 2013. A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation. Nat Struct Mol Biol 20: 728–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler CY, Park JG, Sharma A, Hunter P, Surapaneni P, Sedillo C, Field J, Algar R, Price A, Steel J, et al. 2014. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res 42: D1253–D1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. 2008. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-K, Cheng S-C. 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784. [DOI] [PubMed] [Google Scholar]
- Tseng C-K, Cheng S-C. 2013. The spliceosome catalyzes debranching in competition with reverse of the first chemical reaction. RNA 19: 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Xu Y-Z, Query CC. 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M Jr. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol 18: 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CCL, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Zavanelli MI, Ares M Jr. 1991. Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev 5: 2521–2533. [DOI] [PubMed] [Google Scholar]
- Zavanelli MI, Britton JS, Igel AH, Ares M Jr. 1994. Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol Cell Biol 14: 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.