Abstract
Translation termination in eukaryotes is mediated by release factors: eRF1, which is responsible for stop codon recognition and peptidyl-tRNA hydrolysis, and GTPase eRF3, which stimulates peptide release. Here, we have utilized ribose-specific probes to investigate accessibility of rRNA backbone in complexes formed by association of mRNA- and tRNA-bound human ribosomes with eRF1•eRF3•GMPPNP, eRF1•eRF3•GTP, or eRF1 alone as compared with complexes where the A site is vacant or occupied by tRNA. Our data show which rRNA ribose moieties are protected from attack by the probes in the complexes with release factors and reveal the rRNA regions increasing their accessibility to the probes after the factors bind. These regions in 28S rRNA are helices 43 and 44 in the GTPase associated center, the apical loop of helix 71, and helices 89, 92, and 94 as well as 18S rRNA helices 18 and 34. Additionally, the obtained data suggest that eRF3 neither interacts with the rRNA ribose-phosphate backbone nor dissociates from the complex after GTP hydrolysis. Taken together, our findings provide new information on architecture of the eRF1 binding site on mammalian ribosome at various translation termination steps and on conformational rearrangements induced by binding of the release factors.
Keywords: human ribosome, translation termination, release factors eRF1 and eRF3, chemical footprinting of rRNAs, rearrangements in rRNAs
INTRODUCTION
Translation termination in eukaryotes is induced by two interacting release factors, eRF1 and eRF3, that bind to ribosome when an mRNA stop codon enters the ribosomal A site (Zhouravleva et al. 1995; Korostelev 2011; Jackson et al. 2012). eRF1 is responsible for stop codon recognition and for triggering hydrolysis of the complex ester bond between the peptidyl moiety and the 3′-terminal ribose of the peptidyl-tRNA located at the P site. Upon binding of eRF1 to ribosome, three highly conserved motifs of eRF1 (YxxCxxxF, TASNIKS, and GTS) located at the apex of the amino-terminal N domain mediate stop codon recognition (Bertram et al. 2000; Song et al. 2000; Chavatte et al. 2002; Frolova et al. 2002; Bulygin et al. 2010, 2011; Conard et al. 2012). Thereafter, the M domain of eRF1 binds to the peptidyl transferase center (PTC) of the ribosome inducing conformational rearrangement of the 28S rRNA and the subsequent release of the polypeptide chain (Frolova et al. 1999; Song et al. 2000). Although the presence of eRF1 alone is sufficient for translation termination in vitro (Kryuchkova et al. 2013; Feng et al. 2014), the eRF1 termination efficiency is considerably reduced in the absence of the translational GTPase eRF3 and GTP (Alkalaeva et al. 2006).
Analysis of cryo-electron microscopy (cryo-EM) models of mammalian pretermination complexes containing the ternary complex of release factors with a nonhydrolysable GTP analog, eRF1•eRF3•GMPPNP, or eRF1 alone (Taylor et al. 2012; des Georges et al. 2014; Muhs et al. 2015) has led to (1) identification of ribosomal proteins and rRNA heliсes involved in interactions with release factors; (2) discovery of conformational changes induced in eRF1 by GTP hydrolysis—namely, translocation of the universally conserved GGQ loop to the PTC to trigger the peptidyl-tRNA hydrolysis; (3) disclosure of ribosomal structural rearrangements (primarily, inward shift of the P stalk base) caused by binding of eRF1•eRF3 complex, which is believed to be required for GTP hydrolysis; and (4) determination of position of eRF1 bound to the pretermination complex in the absence of eRF3. However, cryo-EM structures provided no data on particular rRNA nucleotides involved in interactions with eRF1 and eRF3 in mammalian pretermination complexes. Information concerning rRNA nucleotides that are implicated in the ribosomal conformational rearrangements accompanying translation termination and ensuring proper progression of the events during termination remains unknown as well. A suitable approach to gain this information is chemical footprinting, which is based on identification of rRNA nucleotides whose accessibility to chemical probes changes when a ligand binds to the ribosome. This approach has been fruitfully used for identification of rRNA regions involved in binding of mammalian ribosomes with various ligands (e.g., translation factors [Pisareva et al. 2008], tRNAs [Bulygin et al. 2013], hepatitis C virus [HCV] IRES RNA [Malygin et al. 2013], and ribosomal protein uS2 [p40] [Malygin et al. 2011]). Results obtained by means of chemical footprinting were in general compatible with respective structural data resulted from cryo-EM and X-ray crystallography studies. Utilizing this strategy, the pivotal protein component of the selenoprotein synthesis machinery, the so-called SBP2, which remained unresolved using cryo-EM and X-ray crystallography analysis, was mapped on the 60S subunit structure (Kossinova et al. 2014). Moreover, conformational rearrangements induced by binding of HCV IRES RNA to the 40S subunit (Malygin et al. 2013) and of SBP2 to the 80S or 60S ribosome (Kossinova et al. 2014) have been revealed with the use of chemical footprinting.
In the current work, we have applied the RNA backbone-specific chemical modifications (by hydroxyl radicals and benzoyl cyanide) followed by a footprinting assay to identify 18S and 28S rRNA nucleotides whose accessibility to the probes changes upon association of the tRNA- and mRNA-bound human ribosomes with translation termination factors. Altogether, our results are consistent with cryo-EM data on the eRF1 position on the ribosome, but also reveal additional contacts between rRNAs and eRF1, as well as previously unidentified ribosomal regions that undergo conformational rearrangements induced by eRF1/eRF3 binding before and after GTP hydrolysis. Based on the obtained data, we suggest that eRF3 does not interact with the rRNA ribose-phosphate backbone and that it remains bound in the complex after GTP hydrolysis.
RESULTS
Characterization of the ribosomal termination complexes
In the present work, we studied three types of termination complexes that mimic main states occurring in the course of termination process—namely, a state of stop codon recognition, a state after GTP hydrolysis, and a prerecycling state. In parallel, we examined complexes whose unoccupied A sites were programmed with a sense or stop codon, and a complex where the A site was filled with a tRNA molecule.
Because deacylated tRNA has a higher affinity for the ribosomal P site than for the A and E sites (Graifer and Karpova 2015), the ternary complexes 3 and 5 were obtained by incubation of ribosomes with tRNAPhe and mRNA analogs GAAUUUGACAAA and UUCUAAAAG, respectively (Fig. 1). According to the nitrocellulose filtration test, the mRNA binding levels in these complexes were ∼0.8 mol of mRNA per mole of ribosomes, whereas in the absence of tRNA the binding levels were lower by an order of magnitude, which confirmed strong codon–anticodon interaction at the ribosomal P site. Binding of tRNAAsp with triplet GAC and of eRF1 with stop codon UAA at the A-site resulted in formation of complexes 4 and 8, respectively (Fig. 1). When the ternary complexes eRF1•eRF3•GMPPNP and eRF1•eRF3•GTP were used for binding instead of eRF1, complexes 6 and 7 were formed, respectively (Fig. 1). The eRF1 content in complexes 6–8 was validated by monoclonal antibodies against eRF1 applied to the complexes purified by sucrose gradient centrifugation under conditions optimal for the complex preservation (Fig. 2). Binding of eRF1 under conditions used here is specific (i.e., occurs only if the A site is occupied with a stop codon [Bulygin et al. 2002, 2010, 2011]). Overall, complex 6 mimics a pretermination state, which takes place before GTP hydrolysis, whereas complex 7 corresponds to the post-termination state after GTP hydrolysis. Complex 8 (containing eRF1 alone) represents an intermediate state of the translation termination process that arises before recycling of the post-termination complex after stop codon recognition, GTP hydrolysis, and dissociation of eRF3•GDP.
FIGURE 1.
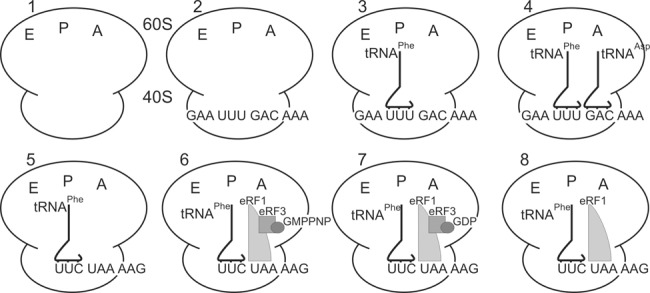
Composition of complexes used in the study. Note, in complex 7 GTP is converted to GDP by eRF3 GTPase activity. Because it is not known exactly at which step of termination eRF3•GDP dissociates from the ribosomal complex, the figure shows one of possible states after GTP hydrolysis, where eRF3•GDP remains bound to the ribosome.
FIGURE 2.
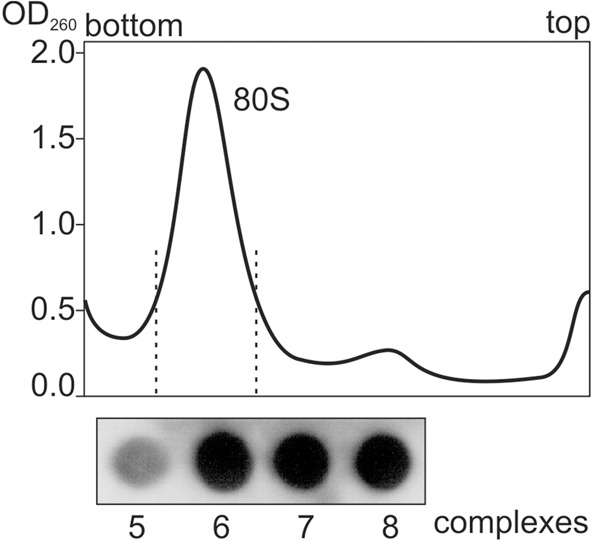
Dot blot measurement of eRF1 content in complexes 6–8 with the use of monoclonal antibodies against eRF1. Complexes 5–8 were purified by sucrose gradient and indicated fractions were loaded onto the membrane.
Identification of rRNA nucleotides that are protected from chemical attack in termination complexes and their mapping on the secondary structure of 28S and 18S rRNAs
To determine sites of rRNA backbones that are protected from chemical attack by release factor binding, as well as the sites that increase their accessibility when the factors are bound to ribosomes, we performed rRNA footprinting in the translation termination complexes using probes with an exquisite sensitivity to local RNA conformation, hydroxyl radicals, and benzoyl cyanide. Hydroxyl radicals induce strand scissions in RNA by attacking riboses at the exposed C4′ atoms, whereas benzoyl cyanide selectively forms covalent ribose 2′-O-adducts (Latham and Cech 1989; Pogozelski et al. 1995; Mortimer and Weeks 2008). Although both probes are specific for the ribose, very small-sized hydroxyl radicals cleave solvent accessible RNA regions, whereas massive benzoyl cyanide modifies preferentially flexible nucleotides (Weeks 2010). Thus, structural data obtained with use of these probes are mutually complementary.
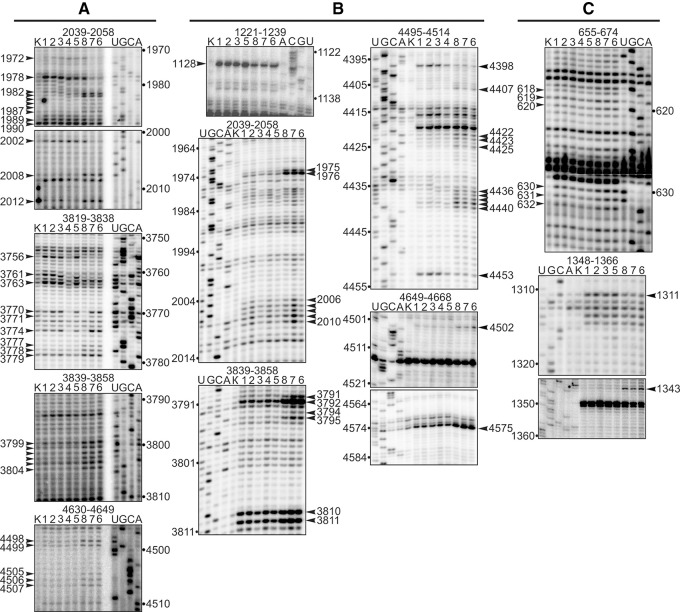
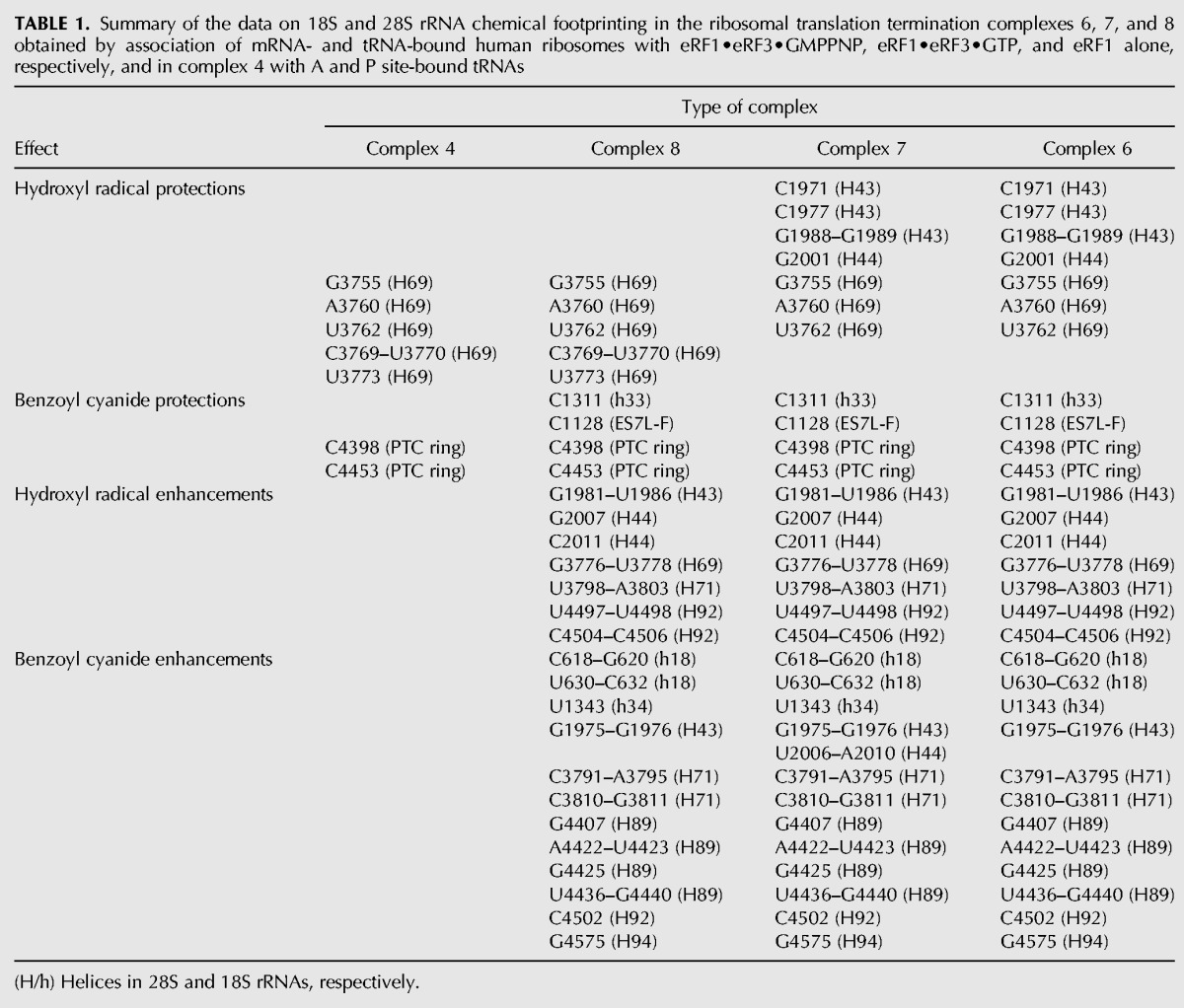
Positions of the scissions in rRNA chains and of nucleotides modified with benzoyl cyanide were determined by primer extension of 5′ labeled oligodeoxyribonucleotides assuming that stops of the reverse transcription occurred at 3′-nucleotides proximal to the cleavage sites, and nucleotides bearing a bulk group at the ribose moiety caused pauses. Comparison of footprints for complex 5 and complexes 6–8 allowed identification of nucleotides whose accessibility to the chemical probes changes upon termination factor binding, whereas comparison of those for complexes 3 and 4 enabled determination of residues changing the reactivity toward the probes after tRNA binding at the A site (Fig. 3). The identified positions of all these nucleotides were mapped on the 18S and 28S rRNA secondary structures (Fig. 4; Anger et al. 2013) and the results are summarized in Table 1. The most prominent protections from hydroxyl radicals were found in helix H69 in the complex 4 (where the A site was occupied by tRNA) and in the eRF1-containing complex 8 (Fig. 4A), whereas protections from benzoyl cyanide were detected in the PTC ring in both complexes (Fig. 4B). The same footprinting results were obtained with benzoyl cyanide for the eRF3-containing complexes 6 and 7 with the addition of nucleotide C1128 in the expansion segment ES7L-F, which was protected in complex 8 as well (Fig. 4B). Treatment with hydroxyl radicals revealed in complexes 6 and 7 protections in H69 as well as in H43 and H44 of the GTPase-associated center (GAC, the universally conserved center of the ribosome including also H42 and sarcin–ricin loop of H95) (Fig. 4A). As for the 18S rRNA, the single nucleotide C1311 in h33 was protected against benzoyl cyanide in complexes 6–8 (Fig. 4C).
FIGURE 3.
Autoradiograms obtained after chemical footprinting of the ribosomal complexes shown in Figure 1. (A,B) 28S rRNA footprints after treatment with hydroxyl radicals and benzoyl cyanide, respectively; (C) 18S rRNA footprints after treatment with benzoyl cyanide. rRNA positions are indicated on the top, lanes 1–8 correspond to complexes shown on Figure 1, and lanes K represent the results of primer extension on rRNAs isolated from untreated ribosomes. Positions of reverse transcription stops are indicated with arrows.
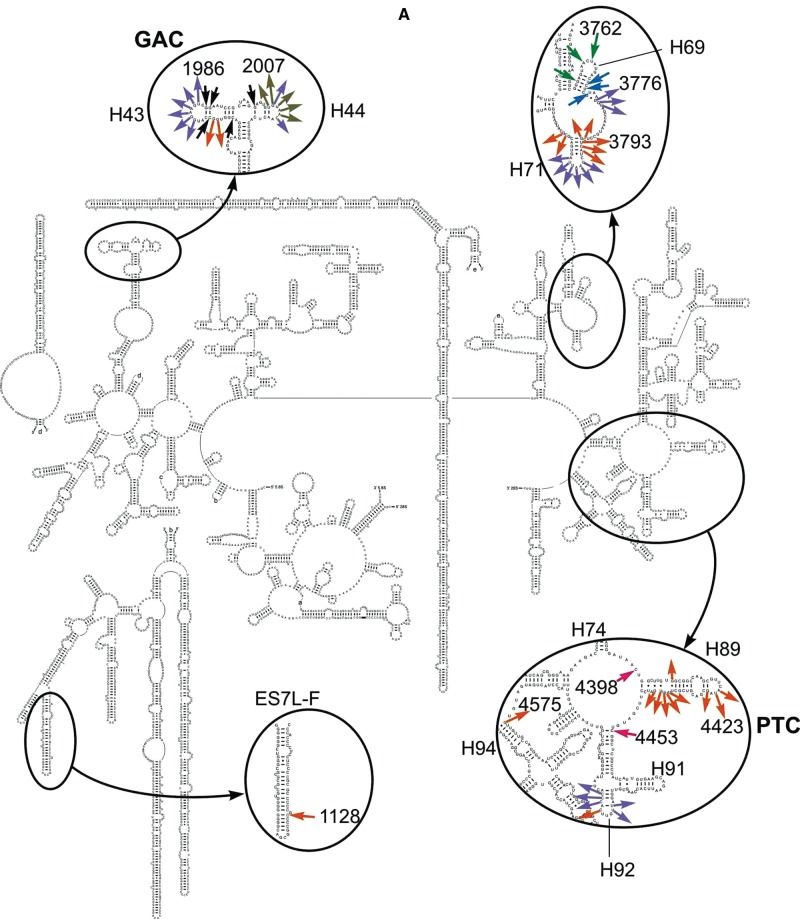
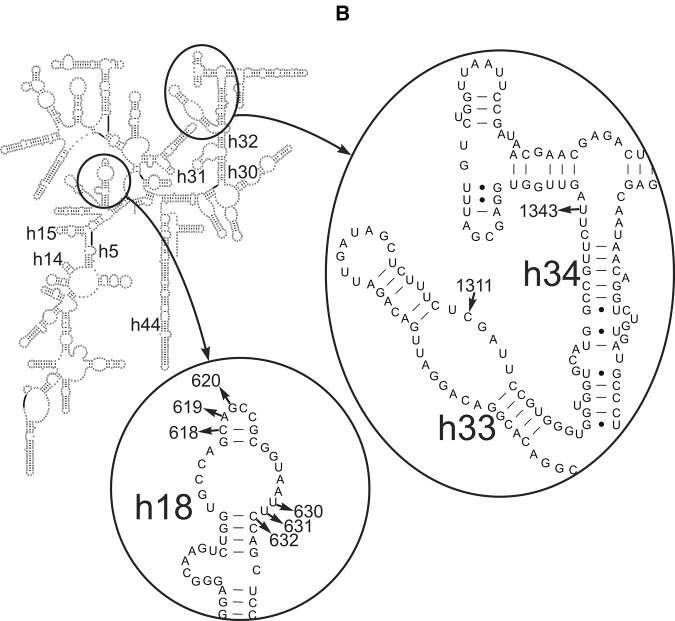
FIGURE 4.
Schematic diagrams of the human 28S (A) and 18S (B) rRNA secondary structures showing locations of nucleotides with changed accessibility to chemical probes. Here and thereafter, 28S rRNA and 18S rRNA helices are designated with uppercase (H) and lowercase (h) letters, respectively. Arrows directed to and from nucleotides designate protections and enhancements, respectively. In A, nucleotides identified after treatment with hydroxyl radicals are shown in violet (complexes 6–8), black (6 and 7), green (4 and 6–8), and blue (4 and 8) and after treatment with benzoyl cyanide in orange (6–8), olive (7), and magenta (4 and 6–8). In B, benzoyl cyanide enhancements and protections are shown in black.
TABLE 1.
Summary of the data on 18S and 28S rRNA chemical footprinting in the ribosomal translation termination complexes 6, 7, and 8 obtained by association of mRNA- and tRNA-bound human ribosomes with eRF1•eRF3•GMPPNP, eRF1•eRF3•GTP, and eRF1 alone, respectively, and in complex 4 with A and P site-bound tRNAs
Protections of rRNA nucleotides by ligands in the ribosomal 80S complexes imply probable participation of the respective rRNA regions in binding to these ligands, although one cannot exclude that several protections arise from long distance conformational changes caused by the ligand binding. In this line, protections in helices H43, H44, and H69 (Fig. 4A) correlate well with cryo-EM data on eRF1 positioning within the mammalian ribosome associated with eRF1•eRF3•GMPPNP or eRF1 alone (des Georges et al. 2014; Muhs et al. 2015). In the mentioned cryo-EM studies, eRF3 positioning in the pretermination complex has been also determined, and eRF3 has been shown to be located in close proximity to H95. The lack of eRF3-specific footprints in the H95 region in complex 6 (containing eRF1•eRF3•GMPPNP) can be considered as an indication that the H95 backbone has no contacts with eRF3. Of course, the lack of these footprints is not the exact proof of the absence of the respective contacts of ribosome with eRF3 because one cannot rule out a possibility that eRF3 replaces existing intraribosomal contacts implicated H95 by displacing ribosomal components interacting with it in the absence of the factor. All other footprints found in the current work with termination complexes are located in those regions of 28S rRNA (3′ fragment of H69, PTC ring, and ES7L-F) that have no contacts with eRF1 according to cryo-EM data (Muhs et al. 2015). Protections in the 3′ side of H69 observed with complex 8 could arise because of the possible flexibility of eRF1 in the termination complex (Fig. 4A). As for the contact of helix ES7L-F with eRF1/eRF3, it could not be detected by cryo-EM because the position of this helix had not yet been visualized in the mammalian 80S ribosome. Although no contacts of eRF1 with the PTC ring have been detected by cryo-EM, interaction of eRF1with nucleotides located immediately in the PTC was observed in the pretermination complex with eRF1 alone (Muhs et al. 2015). Notably, contacts of eRF1 with 18S rRNA nucleotides (except for the contact with h33 [Fig. 4C]) that were revealed by cryo-EM (des Georges et al. 2014; Muhs et al. 2015) were not detected in this study, indicating that they mainly involved rRNA bases.
The majority of rRNA nucleotides identified by footprinting of the termination complexes exhibit enhanced reactivity toward chemical probes (Fig. 4) that could indicate conformational rearrangements at the respective regions induced by eRF1/eRF3 binding. In complexes 6–8, the most pronounced enhancements in reactivity of rRNA nucleotides toward hydroxyl radicals were observed in the GAC region (H43 and H44, nucleotides 1981, 1982, 2007, and 2011), in the apical loop of H71 (nucleotides 3798–3803), and in H92 (nucleotides 4497 and 4498) (Fig. 4A). About the same number of enhancements in 28S rRNA was revealed after treatment of the complexes with benzoyl cyanide (Fig. 4B) and nearly half of them were located in a vicinity of enhancements detected with hydroxyl radicals (H43, H71, and H92), whereas enhancements in H89 and H94 were specific to benzoyl cyanide. Moreover, treatment with benzoyl cyanide revealed GTP hydrolysis-dependent enhancements in the GAC (H44) in the complex 7 (Fig. 4B).
In complexes 6–8 treated with benzoyl cyanide, the 18S rRNA sites with increased accessibility comprised nucleotide U1343 in h34 and nucleotides C618–C620 and U630–C632 in the apical and internal loops of h18, respectively (Fig. 4C).
Spatial mapping of changes in rRNA accessibility in termination complexes
The results of chemical footprinting were mapped on the three-dimensional cryo-EM models of the pretermination complexes that contained eRF1 or eRF1•eRF3•GMPPNP (Fig. 5; Muhs et al. 2015). Analysis revealed two groups of protected nucleotides in the 28S rRNA of complex 8: four nucleotides (A3760, U3762, G4398, and G4453) were located within or in a close proximity to the eRF1 binding site, whereas four others were located at a distance (G3755, C3769, U3770, and U3773) (Fig. 5B). The same was true for complexes 6 and 7 (nucleotides A3760 and G3762 vs. G3755, G4398, and G4453) (Fig. 5A). Unresolved nucleotide G1128 in helix ES7L-F that is protected in complexes 6–8 seems to be distant from the eRF1 binding site but it could interact with eRF1 because of possible flexibility of this helix. The GAC including H43 and H44, where nucleotides protected against hydroxyl radicals in eRF3-containing complexes 6 and 7 are located, is not resolved in the cryo-EM models (Muhs et al. 2015) as well. However, a contact of this center with eRF1 in the pretermination complex containing eRF1•eRF3•GMPPNP has been observed (Taylor et al. 2012). We tried to locate the GAC in the published models (Muhs et al. 2015) analyzing the cryo-EM structure of the human 80S ribosome (Anger et al. 2013), in which the GAC is resolved. Taking into account the arrangement of this center in the human ribosome, we determined its placement in the above-mentioned models (see Fig. 5A,B) and found that nucleotides C1977, G1988, and G1989 of the GAC, which were protected against hydroxyl radicals in complexes 6 and 7, were located close to the eRF1 binding site (Fig. 5A).
FIGURE 5.
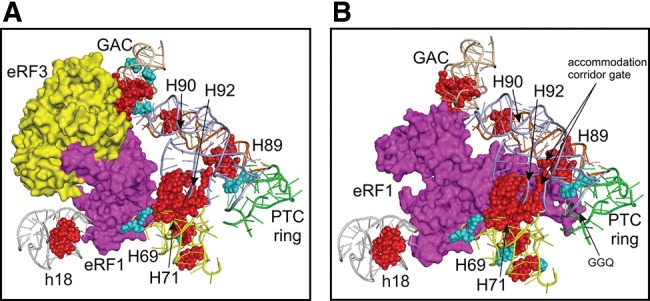
Three-dimensional representation of rRNA regions undergoing changes in accessibility upon binding of eRF1•eRF3•GMPPNP (A) (PDB 4D61 and 4D67) or eRF1 alone (B) (PDB 4D5N and 4D5Y). Release factors eRF1 and eRF3 are shown in magenta and yellow, respectively, and nucleotides with decreased or enhanced accessibility are designated by cyan or red, respectively. Regions in 28S rRNA where changes take place are shown in wheat (for H43 and H44), yellow (H69 and H71), orange (H89), blue (H90 and H92), and green (PTC ring). A fragment of 18S rRNA h18 containing enhancements close to the eRF1 binding site is shown in gray.
Remarkably, the majority of the 28S rRNA nucleotides that enhance their accessibility in complex 8 also reside near the eRF1 position on the model of the ribosomal complex with eRF1 alone (Fig. 5B), although several nucleotides of this kind (G1976, G1985, and U1986 in H43, and U3798–A3800 in H71) are located at some distance from the eRF1 binding site. As for enhancements found with eRF3-containing complexes 6 and 7, a part of them including enhancements in the GAC, as well as those in regions of H71 (at positions 3793–3795, 3798, and 3799) and H92 (4504–4506), is close enough to the eRF1 binding site on the model of ribosomal complex with eRF1•eRF3•GMPPNP (Fig. 5A). Noteworthy, enhancements residing in H89, which are observed in all types of termination complexes, are distant from eRF1/eRF3 binding sites in complexes 6 and 7 (Fig. 5A), but in complex 8 with eRF1 alone those turn out to be close to the location of eRF1 on the ribosome (Fig. 5B). Nucleotides U2006–A2010 of the GAC, which display enhanced accessibility to benzoyl cyanide only in complex 7, where eRF1 is bound in the complex using eRF3•GTP, are located not far from the eRF1 binding site by analogy with nucleotides of H43 and H44 protected against hydroxyl radicals in complexes 6 and 7 (Fig. 5A). Notably, on both models, 18S rRNA nucleotides that increase their accessibility to benzoyl cyanide in complexes 6–8 are away from eRF1/eRF3 binding sites.
DISCUSSION
We have found that eRF1 bound alone to the pretermination complex comprising the 80S ribosome, tRNA (P site), and mRNA (stop codon at the A site) protects the backbone of 28S rRNA in the region of H69 from attack by ribose-specific probes. The 5′ part of H69 is also protected in eRF1•eRF3-containing complexes 6 and 7. The backbone of H43 and H44 in the GAC becomes protected only in complexes with eRF3. The 28S rRNA nucleotides, C1128 located in the specific to vertebrates ES7L-F helix, G4398, and G4453 in the PTC ring, as well as C1311 in h33 of 18S rRNA, were protected in all types of termination complexes. Of note, the same nucleotides of the PTC ring together with the region of H69 were protected by A site–bound tRNA. Along with the protections, we have revealed rRNA regions with increased accessibility to probes when the ribosomal complexes contained either both release factors or eRF1 alone (Table 1).
All this allowed us to obtain new information regarding eRF1/eRF3 binding sites on the mammalian ribosome and the sites where conformational rearrangements induced by release factors binding could occur. In particular, our findings displayed the sites in H43 and H44 of the GAC, the region of H69, and the PTC ring of 28S rRNA and in h33 of 18S rRNA that could be directly involved in interactions with the termination factors. Furthermore, our results indicate the sites in H43, H44, H71, H89, H92, and H94 of 28S rRNA, as well as in h18 and h34 of 18S rRNA, that become more accessible because of conformational rearrangements induced by the factors binding.
Regions of rRNA involved in binding to release factors
The same sets of protected nucleotides (G3755, A3760, and U3762 in H69 as well as C4398 and C4453 in the PTC ring) of 28S rRNA in all types of termination complexes allow us to refer these nucleotides to the eRF1 binding site. This site could also comprise nucleotides of the 3′-area of H69 (C3769, U3770, and U3773) that decreased their accessibility upon eRF1 binding to ribosomes alone. However, the eRF1 binding site derived from our chemical probing data only partially overlaps with the eRF1 positions on the cryo-EM models of termination complexes of mammalian 80S ribosome containing eRF1 alone or eRF1•eRF3•GMPPNP (Fig. 5; Muhs et al. 2015). For all of that, eRF1 binding site in complex 8 displays more similarity with eRF1 position on the respective model as compared with those in complexes 6 and 7. The above-mentioned nucleotides of the region of H69 and the PTC ring seem to belong to a universal core of the A site responsible for binding of both the elongator tRNAs and class 1 release factors, as evidenced by protections of the same nucleotides with A site–bound tRNA in complex 4 (Table 1). The PTC ring is known to harbor the CCA ends of tRNAs and the universally conserved GGQ motif of eRF1 responsible for triggering peptidyl-tRNA hydrolysis (Frolova et al. 1999). Therefore, data on protections of C4398 and C4453 of the PTC ring in complexes 4 and 6–8 suggest that these nucleotides are capable of binding to eRF1 independently of the mode of its entering the A site. However, this assumption does not agree with the model of the pretermination complex with eRF1•eRF3•GMPPNP (Muhs et al. 2015), where C4398 and C4453 nucleotides of the PTC ring are located apart from eRF1 binding site (Fig. 5A). Moreover, in the cryo-EM study (Preis et al. 2014) performed with a similar termination complex containing eRF1 and eRF3 from yeast it has been shown that the M domain of eRF1 contacts the 40S subunit, so that h14 is in direct contact with the GGQ motif.
Protection patterns in the PTC ring observed in complex 6 (containing eRF1•eRF3•GMPPNP and deacylated tRNA at the P site) might be caused by displacement of eRF1 toward the PTC, facilitated by the lack of the peptidyl moiety in the P site–bound tRNA. Remarkably, the location of eRF1 close to the PTC in complex 6 is consistent with the data on cross-linking of the P site–bound periodate-oxidized tRNA to eRF1 in the analogous ribosomal complex containing eRF1•eRF3•GMPPNP (Hountondji et al. 2014). This cross-linking also takes place in complexes similar to those 7 and 8. It has been supposed (Hountondji et al. 2014) that in all these complexes tRNA could adopt both P/P and P/E configurations and that tRNA in the P/P state is involved in the cross-linking with eRF1, whereas tRNA in the P/E state is cross-linked to the ribosomal protein eL44 (eL36AL). On the contrary, peptidyl-tRNA occupying the P site of the pretermination complex seemed to prevent eRF1 bound to eRF3 and GMPPNP from extending to the PTC and contacting the PTC ring (Taylor et al. 2012). Something similar could take place when the pretermination complex was obtained by association of the ternary complex eRF1•eRF3•GMPPNP with the binary 80S.CrPV-STOP mRNA complex; this pretermination complex corresponds to the canonical POST state of 80S ribosome with P/P- and E/E-tRNAs (Muhs et al. 2015). However, in the pretermination complex assembled on the 80S.CrPV-STOP mRNA with eRF1 alone, eRF1 was capable of extending toward the PTC, probably because of its flexibility (Muhs et al. 2015).
The same sets of nucleotides protected against hydroxyl radicals in H43 and H44 of the GAC in eRF3-containing complexes 6 and 7 and the lack of the respective protections in complex 8 with eRF1 alone enable assignment of these nucleotides to the eRF3 binding site (i.e., consistent with the GAC involvement in the interaction with translational GTPases [e.g., see Rodnina et al. 2000; Qin et al. 2009; Li et al. 2011]). However, on the cryo-EM model of the pretermination complex containing eRF1•eRF3•GMPPNP (Muhs et al. 2015) with a fitted GAC region taken from the cryo-EM structure of the mammalian ribosome (Fig. 5A; Anger et al. 2013), these helices are located closer to eRF1 rather than to eRF3. This is in accordance with the previous results showing contacts of the GAC to eRF1 in the pretermination complex with eRF1•eRF3•GMPPNP (Taylor et al. 2012). Therefore, one can conclude that helices H43 and H44 of GAC are involved in interaction with the ribosome-bound eRF1 within the complex with eRF3, whereas eRF3 itself has no contacts with the GAC.
It is still a subject of discussion whether eRF3 leaves the ribosome after GTP hydrolysis. In particular, taking into account the data on tighter binding of eRF1 to post-termination complexes in the presence of eRF3 (Pisarev et al. 2010), it has been proposed (Taylor et al. 2012) that eRF3 release is mediated by a recycling factor ABCE1, whose binding site on the ribosome overlaps with that of eRF3 (Becker et al. 2012). In contrast, eRF3 dissociation from the ribosome immediately after GTP hydrolysis and before ABCE1 binding is suggested (Preis et al. 2014). Our data on the same sets of rRNA nucleotides protected from attack by the probes in complexes 6 and 7 give us the grounds to assume that eRF3 does not dissociate from the ribosome after GTP hydrolysis. This assumption explains the lack of protections in the 3′-area of H69 in complex 7 compared with complex 8. If eRF3 is still present in complex 7 after GTP hydrolysis, it can prevent eRF1 from the occupation of the same position on the ribosome as in complex 8. Although protection patterns for complexes 6 and 7 are the same, the enhancement patterns look different. In particular, the accessibility of several nucleotides of the GAC is much more enhanced in the GDP-bound complex 7 than in the GMPPNP-bound complex 6. This does not contradict our assumption on the presence of eRF3 in complex 7, and it relates to conformational rearrangements in the GAC region caused by GTP hydrolysis (see the next subsection). Overall, further studies are required to obtain direct evidence that eRF3 does not dissociate (or, on the contrary, dissociates) from the ribosomal termination complex after GTP hydrolysis.
As for protection of the nucleotide G1128 in the helix ES7L-F of 28S rRNA observed in all termination complexes, we suggest that it is not concerned with the involvement of G1128 in eRF1 binding because this nucleotide is located far away from the eRF1 binding site and is not connected to ribosomal components implicated in its formation (see Anger et al. 2013). Most likely, this protection reflects flexibility of the large expansion segment ES7L comprising this helix in the ribosome, allowing its transient placement in a protected position in the termination complexes.
Thus, our data on protections of 28S rRNA nucleotides in ribosomal termination complexes allow the proposal of the location of eRF1 alone or in concert with eRF3 on the ribosome. In these complexes eRF1 is located close to nucleotides of the PTC ring, where the acceptor ends of tRNAs are bound as well as nucleotides of the GAC, if the factor is associated with the ribosome within the ternary complex eRF1•eRF3•GDP (or GMPPNP). After GTP hydrolysis, eRF3 does not dissociate from the ribosome, and this is the reason why the eRF1 binding site in the complex containing eRF1•eRF3•GDP does not differ from that in the complex with eRF1•eRF3•GMPPNP.
Regions of rRNAs implicated in the conformational rearrangements induced by release factors binding
Our data show that binding of release factors to the ribosome induces conformational changes in both 40S and 60S ribosomal subunits. In the 18S rRNA of the small subunit, structural rearrangements primarily take place in h18 and, to a lesser extent, in h34 in all three types of termination complexes (see Fig. 4C; Table 1), implying that these changes are induced by eRF1 irrespective of whether it enters the A site alone or within the complex with eRF3. Remarkably, the group of nucleotides in h18 that displays increased reactivity toward chemical probes includes nucleotide U630, which has been identified as one of the key structural elements of the mRNA binding channel of human ribosome (Graifer et al. 1994a,b) and the nucleotides surrounding it. Contacts between h18 and the N-domain of eRF1 taking place upon mRNA stop codon recognition by eRF1 (des Georges et al. 2014) and involving nucleotide bases rather than ribose residues (see above) apparently require rearrangement of the backbone of 18S rRNA in the region of the mRNA binding channel. Notably, nucleotides U630–C632 enhancing their accessibility to benzoyl cyanide adjoin nucleotides U627 and A628, which, in their turn, face h34 and so may be involved in the formation of the mRNA entry channel latch (Lomakin and Steitz 2013; Hussain et al. 2014). Therefore, one can suggest that the above-mentioned structural rearrangement occurs because of the opening of the latch caused by the binding of eRF1 at the A site.
In the 28S rRNA of the large subunit, rearrangements take place in helices H43, H44, H71, H89, H92, and H94 and in the stretch between H69 and H71 in all termination complexes (see Fig. 4A,B; Table 1), indicating that structural changes in these helices are caused by eRF1 binding. Noteworthy, a large number of nucleotides involved in the structural rearrangements belong to H89, whose secondary structure is universally conserved in the 23S-like RNAs of all three domains of life (Cannone et al. 2002). The helix H89 resides at the 60S subunit surface facing the 40S subunit and extends from the PTC ring (Anger et al. 2013). According to numerous data obtained with bacterial ribosomes (e.g., see O'Connor and Dahlberg 1995; La Teana et al. 2001; Burakovsky et al. 2011), H89 is involved in different steps of the translation process. In eukaryotes, this helix is implicated in interaction of the ribosomes with eIF5B (Unbehaun et al. 2007). Molecular dynamics studies show that H89 forms a wall of an accommodation corridor, which is used by acceptor stem of aa-tRNA on its way from the A/T state to A/A state. At the same time, H90 and H92 helices, in which nucleotides enhancing their accessibility to probes as a result of eRF1/eRF3 binding are also found (Table 1), constitute the opposite wall of the corridor (Sanbonmatsu et al. 2005). The universally conserved nucleotides U4438 (U2492 in E. coli 23S rRNA) in H89 and the opposite C4502 (C2556 in E. coli 23S rRNA) in H92 act as a three-dimensional gate to the PTC for aminoacyl-tRNA (aa-tRNA), and deletions of the respective nucleotides in the rRNA of Saccharomyces cerevisiae, as well as any substitutions at C4502, are lethal for the cells (Rakauskaite and Dinman 2011). In our study, the increase of accessibility of C4502 (H92) and the stretch U4436–G4440, which includes U4438 (H89), to benzoyl cyanide and the enhanced reactivity of U4497–U4498 and C4504–C4506 (H92) neighboring to C4502 toward hydroxyl radicals (Table 1) obviously reflect rearrangements of the accommodation corridor gate caused by eRF1 binding alone or in complex with eRF3 to the ribosome. It seems surprising that the enhancement profiles are similar for all three termination complexes, whereas location of eRF1 in complex 8 and those in complexes 6 and 7 are distinct (Fig. 5). In these complexes, H69 is the common structural element of the eRF1 binding site, and enhancements in H69 were observed with all three termination complexes (Table 1). Analyzing the structure of the human 80S ribosome (Anger et al. 2013), one can propose that rearrangements of the accommodation corridor gate are the consequence of allosteric transitions in the 28S rRNA structure induced by binding of eRF1 with H69 and propagated from H69 to H89 through H69–H71, H71–H92, and H92–H89 interactions (Fig. 5).
It is possible to presume that these rearrangements lead to the opening of the accommodation corridor, enabling the eRF1 GGQ motif to pass into the PTC. In this line, the increase of accessibility of nucleotides A4422−U4423 and C4425, located in the apex of H89, to benzoyl cyanide (Table 1) might relate to rearrangements of the apex providing an initial stage of opening of the corridor.
Notably, the apical loop of H89 undergoes conformational rearrangements as well when the ribosome binds the SelenoCysteine Insertion Sequence (SECIS) binding protein 2 (SBP2) (Kossinova et al. 2014), a key player in selenoprotein synthesis utilizing recoding of the UGA stop codon as a selenocysteine (Sec) codon (e.g., see Bulteau and Chavatte 2015). The functions of the protein are based on its ability to interact with SECIS present in the 3′ UTR of all eukaryotic selenoprotein mRNAs and with the ribosome—in particular, with helix ES7L-E of the 28S rRNA (Kossinova et al. 2014 and references therein). SBP2 bound to SECIS of selenoprotein mRNA has been suggested to provide delivery of Sec-tRNASec (within its ternary complex with GTP and a specific elongation factor eEFSec) to the ribosomal A site programmed with UGA codon and its subsequent accommodation to this site (Kossinova et al. 2013). The rearrangements caused by SBP2 binding to the ribosome involve H89 apex sites (riboses of U4419 and C4421 [Caban and Copeland 2012] and nucleotide bases of A4414 and A4422 [Kossinova et al. 2014]) that are not implicated in structural changes induced by release factors binding (Table 1). One can assume that SPB2-induced rearrangements in the H89 apex prevent the accommodation corridor from opening by eRF1. Because eRF1 and SPB2 binding sites do not overlap, eRF1 should be able to associate with SPB2-bound ribosome, and, accordingly, SPB2 might block the placement of the eRF1 GGQ-motif into the PTC enabling the UGA stop codon to be read through as a codon for selenocysteine.
Another region that contains a large number of nucleotides enhancing their accessibility to probes in the presence of release factors is the GAC (Fig. 4A,B; Table 1). By studying the interaction of the GAC with the aa-tRNA•EF-Tu•GTP ternary complex using molecular dynamics simulation (Li et al. 2006), it has been suggested that nucleotide A1067 (G1981 in the human 28S rRNA) in H43 loop has to flip out to interact with the D loop of the A/T aa-tRNA bound to the ribosome within the ternary complex. Enhancement at nucleotide G1981 in the apical loop of H43 observed with complexes 6–8 supports the assumption of reorientation of this nucleotide in the presence of the A/T site bound ligand and indicates that eRF1 alone being bound to the ribosome is in the A/T state similar to eRF1 bound within the ternary complex with eRF3 and GDP (GMPPNP). Moreover, the reorientation of G1981 was accompanied by rearrangements in the neighbor region 1982–1986, as argued by enhancements observed at these nucleotides with complexes 6–8. By analogy with study of Li et al. (2006), it was reasonable to conclude that all these rearrangements provide interactions of eRF1 with the nucleotide G1981 and, probably, with several bases in the region 1982–1986, which could not be detected with the use of the backbone-specific probes. Moreover, the data obtained with complex 7 showed that additional structural changes in the GAC could occur in the region 2006–2010 located in the apical loop of H44 due to eRF3-dependent GTP hydrolysis, which took place in this complex.
All other enhancements (in particular those observed in H71, H94, and the stretch between H69 and H71, as well as in h34) were located far away from the eRF1 binding site in complexes 6–8 (Fig. 5) and could reflect allosteric long-range rearrangements upon the factor binding to the ribosome.
Overall, the results of chemical footprinting of the rRNA backbone in termination complexes containing eRF1•eRF3•GMPPNP, eRF1•eRF3•GDP, or eRF1 alone led us to identification of one region in 18S rRNA and two regions in 28S rRNA where significant conformational rearrangements induced by binding of release factors to human ribosome could occur. These rearrangements include (1) structural changes in the 18S rRNA in region of mRNA binding channel during the eRF1-mediated stop codon recognition, (2) opening of the accommodation corridor ensuring placement of the eRF1 GGQ-motif into the PTC region of the 23S rRNA, and (3) reorientation of H43 in the GAC region of 23S rRNA required for eRF1 binding and reorganizations in the apical loop of H44 in the GAC related to eRF3-dependent GTP hydrolysis.
Thus, our findings provide new information on the architecture of the eRF1 binding site at different translation termination steps on mammalian ribosome and on conformational rearrangements induced by binding of the release factors to the ribosome.
MATERIALS AND METHODS
Purification of ribosomes and release factors
40S and 60S ribosomal subunits were isolated from unfrozen human placenta according to Matasova et al. (1991), reactivated by incubation in buffer A (50 mМ HEPES-KOH, рН 7.5, 100 mМ KCl, 10 mМ MgCl2, and 0.1 mМ EDTA) for 10 min at 37°C, and reassembled, taken in equimolar ratios. Activity of the obtained 80S ribosomes in poly(U)-directed binding of [14C]Phe-tRNAPhe from Escherichia coli (kindly provided by Dr. V.I. Katunin, B.P. Konstantinov's St. Petersburg Institute of Nuclear Physics RAS, Gatchina, Russia) was ∼80% (maximum binding level 1.6 mol of Phe-tRNAPhe per mole of the ribosomes). Yeast tRNAAsp transcript was obtained by in vitro T7 transcription. Oligoribonucleotides used as mRNAs were from Sigma-Aldrich. Generation of DNA constructs containing sequences coding for human eRF1 (full length) and eRF3 (lacking the nonessential amino-terminal 138 amino acids) and the preparation and purification of the respective recombinant proteins were carried out as described (Frolova et al. 1998, 2000, 2002); measurement of the factor’s activity was performed according to Frolova et al. (1994, 1996).
Formation and analysis of ribosomal complexes
Ribosomal complexes containing mRNAs, tRNAs, and release factors were obtained in buffer A at 20°C; the extent of mRNA binding was examined by nitrocellulose filtration with the use of the oligoribonucleotides labeled at the 5′-termini by [γ-32P]ATP and polynucleotide kinase (Graifer et al. 1997). Binary complex 2 (lacking tRNA) was obtained by incubation of ribosomes (0.5 μM) with oligoribonucleotide UUC UAA AAG (5 μM) for 40 min. Ternary complexes 3 and 5 (with tRNAPhe in the P site and vacant A site) were obtained by incubation of 80S ribosomes (0.5 μM) with tRNAPhe (5 μM) and oligoribonucleotide GAA UUU GAC AAA or UUC UAA AAG (5 μM), respectively. To obtain complex 4 (with tRNAAsp in the A site), complex 3 was incubated with tRNAAsp (5 μM) for 40 min at 20°C. To obtain complexes 6, 7, and 8, complex 5 was mixed with eRF1•eRF3•GMPPNP, eRF1•eRF3•GTP, or eRF1 alone, respectively, and incubated for 40 min at 20°C. Both eRF1 and eRF3 were taken in the eightfold excess over the ribosomes, and the final concentrations of GTP and GMPPNP were 1 mМ. The extent of mRNA analogs binding to 80S ribosomes was examined with the use of 5′-32P-labeled analogs by nitrocellulose filtration; the filters (Millipore, pore size 0.45 μm) were pretreated with 0.6 M KOH for 25 min at 25°C to decrease unspecific sorption of the mRNA analogs (Demeshkina et al. 2003). For validation of eRF1 content in the termination complexes 6–8, aliquots containing 20 pmol of ribosomes were layered onto 15%–30% sucrose (w/w) linear gradient in buffer A with subsequent centrifugation (rotor SW40, 17 h, 21,800 rpm, 4°C). After ethanol precipitation of pooled ribosomal complexes, the obtained pellets were dissolved in minimal amounts of water and blotted onto a nitrocellulose membrane. To determine eRF1 content, the membrane was incubated with rabbit polyclonal anti-eRF1 antibodies (1/2000 dilution) (Novus Biologicals) and then treated with anti-rabbit HRP-conjugated secondary antibody (1/10000 dilution) (Sigma-Aldrich), developed with the ECL-plus kit (Thermo Scientific) and exposed to X-ray film (GE Healthcare).
Chemical probing of rRNAs
Ribosomal complexes obtained in buffer A as described above were supplied with benzoyl cyanide (Sigma-Aldrich) at a final concentration of 80 mM and incubated for 2 min at 20°C; the reaction was stopped by addition of 0.8 volume of ice-cold ethanol. Hydroxyl radical cleavage of 18S and 28S rRNAs within ribosomal complexes assembled in the same buffer was carried out by adding to the reaction mixtures of 1/10 v of freshly prepared solution containing 12 mM Fe(NH4)2(SO4)2, 62 mM ascorbic acid, 25 mM EDTA-KOH (pH 7.5), and 0.6% H2O2 according to Malygin et al. (2013). A typical reaction mixture contained 10 pmol of 80S ribosomes and all other components in the respective amounts. The rRNA was isolated from the treated complexes by phenol deproteination and was used for reverse transcription with 5′-32P-labeled primers complementary to the human 28S rRNA regions 1221–1239, 2039–2058, 3839–3858, 4495–4514, and 4649–4668 and to the human 18S rRNA regions 161–180, 506–525, 655–674, 1348–1366, and 1537–1556 as described (Malygin et al. 2013). Each reaction mixture containing 1 pmol of rRNA and 2 pmol of a primer in 5 μL of water was heated at 90°C for 1 min; cooled down in ice; supplied with 5 μL of solution containing 100 mM Tris–HCl (pH 8.3), 150 mM KOAc, 16 mM Mg(OAc)2, 20 mM DTT, 0.5 mM dNTPs, and 1 U of AMV reverse transcriptase (Fermentas); and then incubated for 30 min at 37°C. Reaction was stopped by adding 1 μL of 3 M KOAc (pH 5.0), and the reaction products were ethanol precipitated.
ACKNOWLEDGMENTS
We thank Nicolay Lisitsin for help in editing the manuscript. This work was supported by the Russian Foundation for Basic Research (grant 14-04-00709 to G.G.K.) and by the Program “Molecular and Cell Biology” of the Presidium of the Russian Academy of Sciences (to G.G.K. and L.Y.F.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.053801.115.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau A-L, Chavatte L. 2015. Update on selenoprotein biosynthesis. Antioxid Redox Signal 23: 775–794. [DOI] [PubMed] [Google Scholar]
- Bulygin KN, Repkova MN, Ven'yaminova AG, Graifer DM, Karpova GG, Frolova LY, Kisselev LL. 2002. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett 514: 96–101. [DOI] [PubMed] [Google Scholar]
- Bulygin KN, Khairulina YS, Kolosov PM, Ven'yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Kisselev LL, Karpova GG. 2010. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA 16: 1902–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulygin KN, Khairulina YS, Kolosov PM, Ven'yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Karpova GG. 2011. Adenine and guanine recognition of stop codon is mediated by different N domain conformations of translation termination factor eRF1. Nucleic Acids Res 39: 7134–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulygin K, Malygin A, Hountondji C, Graifer D, Karpova G. 2013. Positioning of CCA-arms of the A- and the P-tRNAs towards the 28S rRNA in the human ribosome. Biochimie 95: 195–203. [DOI] [PubMed] [Google Scholar]
- Burakovsky DE, Sergiev PV, Steblyanko MA, Konevega AL, Bogdanov AA, Dontsova OA. 2011. The structure of helix 89 of 23S rRNA is important for peptidyl transferase function of Escherichia coli ribosome. FEBS Lett 585: 3073–3078. [DOI] [PubMed] [Google Scholar]
- Caban K, Copeland PR. 2012. Selenocysteine Insertion Sequence (SECIS)-binding protein 2 alters conformational dynamics of residues involved in tRNA accommodation in 80S ribosomes. J Biol Chem 287: 10664–10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Mueller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J 21: 5302–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conard SE, Buckley J, Dang M, Bedwell GJ, Carter RL, Khass M, Bedwell DM. 2012. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA 18: 1210–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Laletina E, Meschaninova M, Ven'yaminova A, Graifer D, Karpova G. 2003. Positioning of mRNA codons with respect to 18S rRNA at the P and E sites of human ribosome. Biochim Biophys Acta 1627: 39–46. [DOI] [PubMed] [Google Scholar]
- des Georges A, Hashem Y, Unbehaun A, Grassucci RA, Taylor D, Hellen CU, Pestova TV, Frank J. 2014. Structure of the mammalian ribosomal pre-termination complex associated with eRF1.eRF3.GDPNP. Nucleic Acids Res 42: 3409–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Yamamoto A, Wilkins SE, Sokolova E, Yates LA, Müenzel M, Singh P, Hopkinson RJ, Fischer R, Cockman ME, et al. 2014. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol Cell 53: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341. [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Simonsen JL, Merkulova TI, Litvinov DY, Martensen PM, Rechinsky VO, Camonis JH, Kisselev LL, Justesen J. 1998. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur J Biochem 256: 36–44. [DOI] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Merkulova TI, Kisselev LL. 2000. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Seit-Nebi A, Kisselev L. 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graifer D, Karpova G. 2015. Interaction of tRNA with eukaryotic ribosome. Int J Mol Sci 16: 7173–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graifer DM, Juzumiene DI, Wollenzien P, Karpova GG. 1994a. Cross-linking of messenger-RNA analogs containing 4-thiouridine residues on the 3′- or 5′-side of the coding triplet to the messenger-RNA binding center of the human ribosome. Biochemistry 33: 3878–3884. [DOI] [PubMed] [Google Scholar]
- Graifer DM, Juzumiene DI, Karpova GG, Wollenzien P. 1994b. Messenger-RNA binding track in the human 80S ribosome for messenger-RNA analogs randomly substituted with 4-thiouridine residues. Biochemistry 33: 6201–6206. [DOI] [PubMed] [Google Scholar]
- Graifer DM, Malygin AA, Matasova NB, Mundus DA, Zenkova MA, Karpova GG. 1997. Studying functional significance of the sequence 980–1061 in the central domain of human 18S rRNA using complementary DNA probes. Biochim Biophys Acta 1350: 335–344. [DOI] [PubMed] [Google Scholar]
- Hountondji C, Bulygin K, Créchet JB, Woisard A, Tuffery P, Nakayama J, Frolova L, Nierhaus KH, Karpova G, Baouz S. 2014. The CCA-end of P-tRNA contacts both the human RPL36AL and the A-site bound translation termination factor eRF1 at the peptidyl transferase center of the human 80S ribosome. Open Biochem J 8: 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2012. Termination and post-termination events in eukaryotic translation. Adv Prot Chem Struct Biol 86: 45–93. [DOI] [PubMed] [Google Scholar]
- Korostelev AA. 2011. Structural aspects of translation termination on the ribosome. RNA 17: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossinova O, Malygin A, Krol A, Karpova G. 2013. A novel insight into the mechanism of mammalian selenoprotein synthesis. RNA 19: 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossinova O, Malygin A, Krol A, Karpova G. 2014. The SBP2 protein central to selenoprotein synthesis contacts the human ribosome at expansion segment 7L of the 28S rRNA. RNA 20: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryuchkova P, Grishin A, Eliseev B, Karyagina A, Frolova L, Alkalaeva E. 2013. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucleic Acids Res 41: 4573–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A, Gualerzi CO, Dahlberg AE. 2001. Initiation factor IF 2 binds to the α-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7: 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Cech TR. 1989. Defining the inside and outside of a catalytic RNA molecule. Science 245: 276–282. [DOI] [PubMed] [Google Scholar]
- Li W, Sengupta J, Rath BK, Frank J. 2006. Functional conformations of the L11-ribosomal RNA complex revealed by correlative analysis of cryo-EM and molecular dynamics simulations. RNA 12: 1240–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Trabuco LG, Schulten K, Frank J. 2011. Molecular dynamics of EF-G during translocation. Proteins 79: 1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA. 2013. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500: 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Babaylova ES, Loktev VB, Karpova GG. 2011. A region in the C-terminal domain of ribosomal protein SA required for binding of SA to the human 40S ribosomal subunit. Biochimie 93: 612–617. [DOI] [PubMed] [Google Scholar]
- Malygin AA, Kossinova OA, Shatsky IN, Karpova GG. 2013. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res 41: 8706–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasova NB, Myltseva SV, Zenkova MA, Graifer DM, Vladimirov SN, Karpova GG. 1991. Isolation of ribosomal subunits containing intact rRNA from human placenta: estimation of functional activity of 80S ribosomes. Anal Biochem 198: 219–223. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2008. Time-resolved RNA SHAPE chemistry. J Am Chem Soc 130: 16178–16180. [DOI] [PubMed] [Google Scholar]
- Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol Cell 57: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M, Dahlberg AE. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol 254: 838–847. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5′-UTRs requires DExH-box protein DHX29. Cell 135: 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogozelski WK, Mcneese TJ, Tullius TD. 1995. What species is responsible for strand scission in the reaction of [Fe(II)EDTA]2- and H2O2 with DNA. J Am Chem Soc 117: 6428–6433. [Google Scholar]
- Preis A, Heuer A, Barrio-Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R. 2014. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep 8: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Grigoriadou C, Cooperman BS. 2009. Interaction of IF2 with the ribosomal GTPase-associated center during 70S initiation complex formation. Biochemistry 48: 4699–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakauskaite˙ R, Dinman JD. 2011. Mutations of highly conserved bases in the peptidyltransferase center induce compensatory rearrangements in yeast ribosomes. RNA 17: 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Stark H, Savelsbergh A, Wieden HJ, Mohr D, Matassova NB, Peske F, Daviter T, Gualerzi CO, Wintermeyer W. 2000. GTPases mechanisms and functions of translation factors on the ribosome. Biol Chem 381: 377–387. [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung C-S. 2005. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci 102: 15854–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. 2000. The crystal structure of human eukaryotic release factor eRF1−mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321. [DOI] [PubMed] [Google Scholar]
- Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci RA, Pestova TV, Frank J. 2012. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc Natl Acad Sci 109: 18413–18418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Marintchev A, Lomakin IB, Didenko T, Wagner G, Hellen CU, Pestova TV. 2007. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J 26: 3109–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM. 2010. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 20: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guillec R, Inge-Vechtomov S, Kisselev L, Philippe M. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14: 4066–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]