Abstract
The AXL receptor tyrosine kinase is frequently overexpressed in cancers and is important in cancer invasion/metastasis and chemoresistance. Here, we demonstrate a regulatory feedback loop between AXL and microRNA (miRNA) at the post-transcriptional level. Both the GAS6-binding domain and the kinase domain of AXL, particularly the Y779 tyrosine phosphorylation site, are shown to be crucial for this autoregulation. To clarify the role of miRNAs in this regulation loop, approaches using bioinformatics and molecular techniques were applied, revealing that miR-34a may target the 3′ UTR of AXL mRNA to inhibit AXL expression. Interestingly and importantly, AXL overexpression may induce miR-34a expression by activating the transcription factor ELK1 via the JNK signaling pathway. In addition, ectopic overexpression of ELK1 promotes apoptosis through, in part, down-regulation of AXL. Therefore, we propose that AXL is autoregulated by miR-34a in a feedback loop; this may provide a novel opportunity for developing AXL-targeted anticancer therapies.
Keywords: feedback loop regulation, AXL, miR-34a, JNK, ELK1, apoptosis
INTRODUCTION
MicroRNAs (miRNAs) are an abundant class of noncoding RNAs 18–25 nt in length that are involved in post-transcriptional regulation of gene expression and control a wide range of biological processes that represent the hallmarks of cancer, such as apoptosis, invasion, and metastasis (Lujambio and Lowe 2012). MiRNAs post-transcriptionally regulate gene expression to either inhibit mRNA translation or promote its degradation by targeting 3′ UTR (Bartel 2004). Many miRNAs play roles as either oncogenes or tumor suppressors, depending on the nature of their targets, affecting biological functions during cancer initiation and progression. Aberrant expression of miRNAs has been reported in multiple human cancer types (Esquela-Kerscher and Slack 2006).
Receptor tyrosine kinases (RTKs) (e.g., EGFR and MET) may further modulate the strength of their downstream signaling pathways by microRNA-mediated mechanisms in a regulatory feedback manner. For example, miR-7 may down-regulate EGFR signaling (Kefas et al. 2008; Webster et al. 2009) and p21-activated kinase 1 (Reddy et al. 2008) to modulate cell proliferation and invasiveness. On the other hand, EGFR can activate miR-7 expression in lung oncogenesis (Chou et al. 2010). Additionally, MET forms a direct feedback loop with miR-27a in lung cancer (Acunzo et al. 2013). Therefore, RTK expression may be regulated by miRNAs via a feedback mechanism.
AXL, belonging to the TAM family (Tyro3, Mer, AXL) of receptor tyrosine kinases, regulates several aspects of cancer biology. Studies have shown that AXL is highly expressed in many human cancers and it has been implicated in the prognosis of cancer patients, including those with lung (Wimmel et al. 2001; Shieh et al. 2005), renal (Chung et al. 2003), myeloid leukemia (O'Bryan et al. 1991; Rochlitz et al. 1999), breast (Meric et al. 2002), gastric (Wu et al. 2002), prostatic (Wu et al. 2004; Sainaghi et al. 2005), and colon (Craven et al. 1995) cancers. Several pathways have been reported to be downstream from AXL signaling, including PI3K-AKT (Lee et al. 2002; Hasanbasic et al. 2004; Hafizi et al. 2005), MEK-ERK (Fridell et al. 1996), p38 (Wu et al. 2004; Sainaghi et al. 2005), and NF-кB (Georgescu et al. 1999; Demarchi et al. 2001). Moreover, AXL is activated by both ligand-dependent (Demarchi et al. 2001; Gallicchio et al. 2005; Hafizi et al. 2005) and ligand-independent (Bellosta et al. 1995; Heiring et al. 2004) mechanisms. Recent studies have demonstrated that overexpression and/or activation of AXL play a pivotal role in cancer's resistance to chemotherapeutic regimens, in the phenotype of epithelial-to-mesenchymal transition (EMT), and in the promotion of increased metastasis, highlighting the importance of AXL as a therapeutic target (Wu et al. 2014). In addition to its known regulation by some transcription factors, such as EZH2 and MZF1, post-transcriptional regulation of AXL remains largely unknown (Mudduluru et al. 2010; Ott et al. 2012). Therefore, an understanding of the regulation mechanisms related to AXL expression appears important.
The miR-34 family consists of three processed miRNAs that are encoded by two different genes: miR-34a, which is encoded by its own transcript, and miR-34b and miR-34c, which share a common primary transcript (Hermeking 2010). Ectopic expression of miR-34a and miR-34b/c induces apoptosis, senescence, and cell-cycle arrest, as well as inhibits migration and invasion (Hermeking 2010). Their promoters are often silenced by CpG methylation in numerous types of cancers during tumorigenesis and may therefore represent tumor suppressor genes (Lodygin et al. 2008; Vogt et al. 2011). Several studies have reported relationships between AXL and miR-34a in colon carcinomas (Kaller et al. 2011), lung cancer (Lee et al. 2011), breast cancer (Mackiewicz et al. 2011), and leukemia (Boysen et al. 2014). However, how the microRNAs are modulated by the AXL signaling pathway is largely unclear, and the mechanisms underlying how the miR-34 family regulates AXL expression are in need of further study.
In this article, we clarify the involvement of miRNA in the post-transcriptional feedback autoregulation of AXL. We illustrate the negative feedback loop of AXL/ELK1/miR-34a wherein AXL up-regulates ELK1 via the JNK pathway, ELK1 in turn up-regulates miR-34a expression via direct promoter activation, and miR-34a returns to down-regulate AXL mRNA. Indeed, ectopic overexpression of ELK1 significantly decreases AXL expression and induces G1 arrest and apoptosis. In addition, we found that overexpression of miR-34c may down-regulate AXL expression transcriptionally via promoter modulation. This post-transcriptional feedback autoregulation loop of AXL by miR-34a may shed light on how AXL is overexpressed in various cancers and have great therapeutic potential in cancer treatment.
RESULTS
AXL is negatively regulated in a post-transcriptional feedback loop
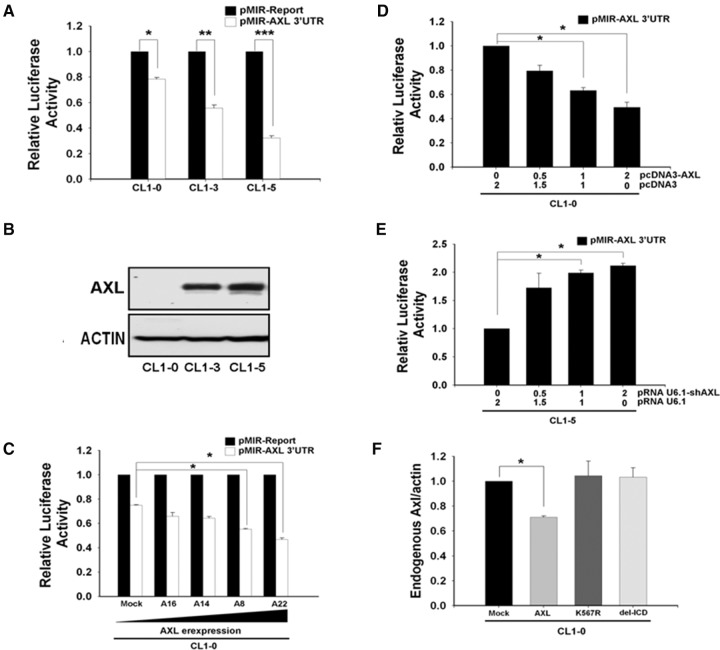
We first found that the expression levels of AXL protein were inversely correlated with its 3′-UTR reporter activity in eight different cancer cell lines (Fig. 1A,B; Supplemental Fig. 1A,B). Expression levels of AXL in different stable clones of CL1-0 were also found to possess an inverse correlation with the AXL 3′-UTR reporter activity (Fig. 1C). These results demonstrate that AXL may regulate its own expression by a previously unknown feedback mechanism; this was further confirmed by performing AXL 3′-UTR reporter assays in CL1-0 cells transiently transfected with AXL expression vector and in CL1-5 cells transfected with AXL shRNA. The results were consistent with expectations (Fig. 1D,E). Next, we designed the AXL 3′-UTR primers specifically to detect endogenous AXL mRNA level and found that it was also down-regulated by overexpression of AXL. Neither the kinase-dead (K567R) nor the intracellular domain deletion (del-ICD) AXL had this effect (Fig. 1F). A similar result was obtained from microarray analysis with the probe targeting the AXL 3′-UTR 4461–4520 region (Supplemental Table 1). Therefore, this feedback regulation may involve AXL kinase activity in suppressing AXL 3′-UTR reporter activity.
FIGURE 1.
AXL negatively regulates its own expression through 3′ UTR. (A) Inverse correlation between AXL protein level and AXL 3′-UTR reporter activity. The pMIR-Report-AXL 3′-UTR or p-MIR-Report (mock vector) plasmid was cotransfected with Renilla plasmid, serving as an internal control for transfection efficiency, into CL1-0, CL1-3, and CL1-5. After 24 h, luciferase activity of the cell lysates was measured. Relative fold change in luciferase activity for each cell line was plotted with respect to each pMIR-Report control. (B) AXL protein levels of three cell lines at steady state were analyzed by Western blot, using actin as an internal control. (C) AXL 3′-UTR reporter activity in stable transfectants of CL1-0 overexpressing various levels of AXL. CL1-0/Mock is vector alone control. (D) Ectopic expression of AXL suppresses AXL 3′-UTR reporter activity. The AXL 3′-UTR reporter constructs were cotransfected with various amounts of the AXL expression construct in CL1-0 cells, and luciferase activity was measured 24 h later. Equal amounts of DNA (2 μg) were used for each transfection. (E) Knockdown of AXL expression increased the AXL 3′-UTR reporter activity. The AXL 3′-UTR reporter construct was cotransfected with various amounts of the AXL shRNA construct into CL1-5 cells, and reporter activity was measured as above. (F) Endogenous AXL mRNA levels were down-regulated by AXL in a kinase activity-dependent manner. CL1-0 cells were transfected with various AXL constructs. After 48 h, the endogenous AXL mRNA was measured by real-time RT-PCR using actin as an internal control. The ratios of endogenous AXL to actin were calculated. Mock, vector alone; AXL, full-length wild-type; AXL (K567R), kinase-dead; del-ICD, intracellular domain deletion. In A, C, D, E, and F, the values were derived from three independent experiments. Error bars represent SD (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
The AXL kinase domain and autophosphorylation site Y779 of AXL are crucial to the feedback regulation
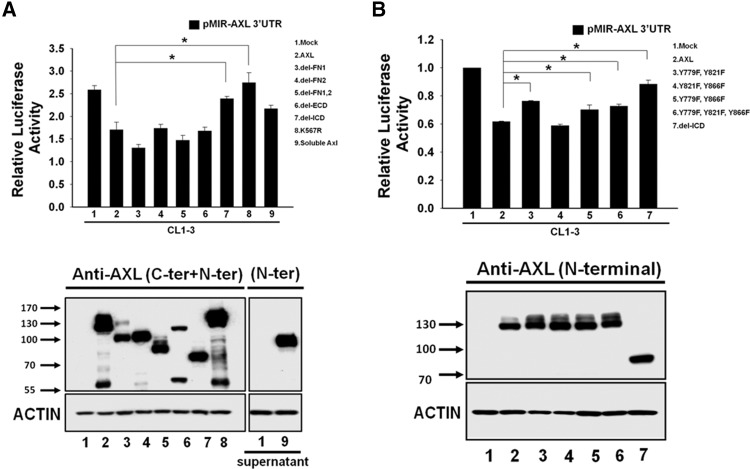
The CL1-3 cell line was originally derived from CL1-0 by selecting for increased invasiveness. Its AXL expression level is higher than CL1-0 and lower than CL1-5. CL1-3 is therefore an appropriate choice for conducting experiments in which both knockdown and overexpression of AXL are required. Among the constructs of various AXL mutations and deletions examined, we found that deletion of the intracellular domain and the kinase-dead point mutation (K567R) could both reverse the feedback regulation in the AXL 3′-UTR reporter assay (Fig. 2A). Among the three putative autophosphorylation sites (Y779, Y821, and Y866) predicted in AXL, only Y779 was required for this feedback regulation (Fig. 2B). Similar results were obtained using CL1-0 cells (Supplemental Fig. 1C,D).
FIGURE 2.
AXL kinase activity and the Y779 phosphorylation site are crucial for feedback regulation. (A) Effects of various AXL mutations on AXL 3′-UTR reporter activity. The AXL 3′-UTR reporter construct was cotransfected with various AXL mutants in CL1-3 cells, and luciferase activity was measured 24 h later. (B) Effects of double or triple mutants in AXL tyrosine phosphorylation sites on AXL 3′-UTR reporter activity. Luciferase reporter activity was assayed for CL-3 cells cotransfected with individual AXL mutants and the AXL 3′-UTR reporter construct. The lower panel shows the expression and molecular weights of various AXL mutant proteins verified by Western blot analysis using a mixture of antibodies against the carboxyl and amino terminal of AXL. Soluble AXL was concentrated from the conditioned medium and detected by Western blotting using AXL amino-terminal antibody. Actin was used as a loading control. The values were derived from three independent experiments. Error bars represent SD (*) P < 0.05.
The AXL ligand GAS6 is important for the negative feedback loop
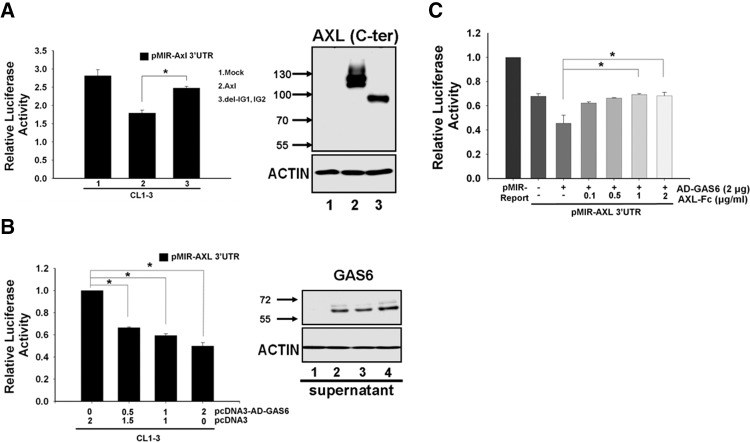
Involvement of the AXL ligand GAS6 in the negative feedback mechanism was tested. Deletion of the immunoglobulin-like domain, which is the domain that contains the GAS6-binding site, significantly reversed the feedback regulation of AXL (Fig. 3A). Meanwhile, when the AXL 3′-UTR reporter plasmid was cotransfected with increasing amounts of GAS6 expression construct, the reporter activity was reduced in a dose-dependent manner in both CL1-3 and MDA-MB-231 cells (Fig. 3B; Supplemental Fig. 2A). In comparison, when GAS6 expression was knocked down by shRNA, increased reporter activity was observed (Supplemental Fig. 2B). Further, addition of recombinant human AXL-Fc, a soluble form of AXL that contains only its extracellular domain that binds GAS6, could antagonize the GAS6-mediated down-regulation of AXL 3′-UTR reporter activity in both CL1-3 and MDA-MB-231 cells (Fig. 3C; Supplemental Fig. 2C). Additionally, R428, a selective small molecule inhibitor of AXL kinase, could block this feedback effect (Fig. 5A; Supplemental Fig. 2D). These results suggest the existence of a feedback loop that is activated by GAS6 to subtly balance the expression of AXL.
FIGURE 3.
The AXL ligand GAS6 is crucial for the negative feedback loop. (A) Effects of AXL immunoglobulin domain deletion mutants on AXL 3′-UTR reporter activity. AXL 3′-UTR reporter was cotransfected with wild-type AXL or the immunoglobulin domain deletion mutant into CL1-3 cells, and luciferase activity was measured 24 h later. The expression and molecular weights of various truncated AXL proteins were verified by Western blot analysis (right panel). (B) GAS6 suppresses AXL 3′-UTR reporter activity in a dose-dependent manner. A fixed amount of AXL 3′-UTR reporter construct was cotransfected with various amounts of pcDNA3-AD-Gas6 into CL1-3 cells. The Renilla reporter was used as transfection efficiency control. Expression of GAS6 was concentrated from the conditioned medium and determined by Western blotting (right panel). Actin was used a loading control. (C) GAS6's inhibitory effect on the AXL 3′-UTR reporter activity was neutralized by AXL-Fc. Fixed amounts of the AXL 3′-UTR reporter and the pcDNA3-AD-Gas6 expression construct were cotransfected with various amounts of the AXL-Fc construct into CL1-3 cells. Luciferase activity was measured 24 h post-transfection. Equal amounts of DNA (2 μg) were transfected for each sample in B and C. The values were derived from three independent experiments. Error bars represent SD (*) P < 0.05.
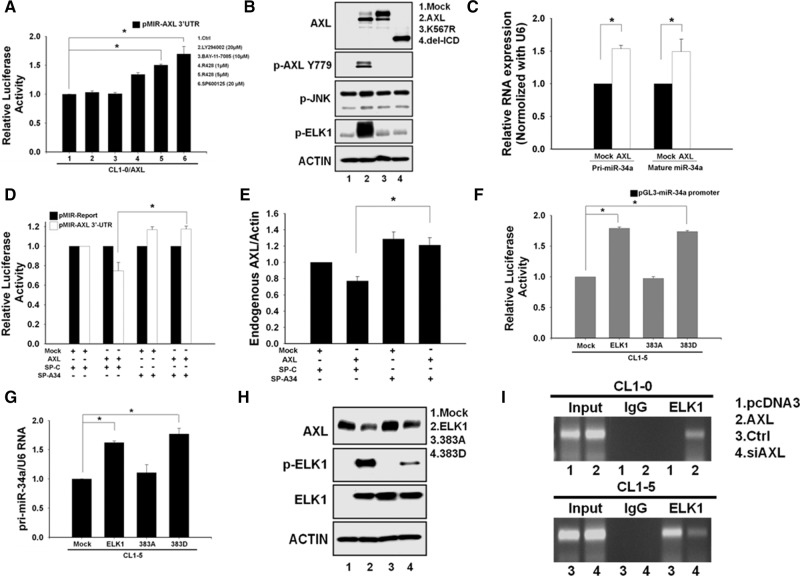
FIGURE 5.
AXL regulates miR-34a via the JNK/ELK1 pathway. (A) The JNK pathway is involved in the negative feedback regulation. After transfection with the AXL 3′-UTR reporter for 24 h, CL1-0/AXL cells were treated with various inhibitors. The luciferase activity was measured 48 h later. An asterisk indicates a significant inhibitory effect on AXL 3′-UTR reporter activity. (B) AXL kinase activity is essential for ELK1 phosphorylation. CL1-0 cells were transfected with the full-length, kinase-dead mutant (K567R) or the intracellular domain deletion construct (del-ICD) of AXL. The protein levels of p-AXL, p-JNK, and p-ELK1 were determined by Western blot. (C) Expression of miR-34a is up-regulated by AXL. After CL1-0 cells were transfected with AXL or the vector-alone control plasmid, the primary transcript and mature forms of miR-34a were measured by real-time PCR. U6 small RNA was used as the internal control. (D) AXL-mediated down-regulation of the reporter activity can be rescued by blocking miR-34a. Luciferase reporter activity was determined in CL1-0 cells after cotransfection with pcDNA3-AXL plus miR-34a sponge (SP-A34) or pcDNA3-AXL plus control sponge (SP-C). The Renilla reporter activity was used to normalize the transfection efficiency. (E) Endogenous AXL expression is up-regulated by blocking miR-34a. CL1-0 cells were cotransfected as in D. The qRT-PCR assay was performed to determine the endogenous AXL mRNA level as described above. (F,G) The promoter activity and primary transcript expression of miR-34a are up-regulated by ELK1. CL1-5 cells were transfected with pcDNA3-ELK1, pcDNA3-ELK1 (S383D), or pcDNA3-ELK1 (S383A) plasmid along with the pGL3-AXL promoter reporter construct. The assay of luciferase reporter activity and primary transcript level of miR-34a were measured as described above. (H) Expression of AXL protein is up-regulated by ELK1. CL1-5 cells were transfected with pcDNA3-ELK1, pcDNA3-ELK1 (S383D), or pcDNA3-ELK1 (S383A) plasmid. The protein levels of AXL, ELK1 and p-ELK1 were determined by Western blot. (I) ELK1 binds to the miR-34a promoter in ChIP. CL1-5 cells were transfected with siAXL to knock down AXL expression. CL1-0 cells were transfected with AXL construct for AXL overexpression. ChIP assay was performed as described in the Materials and Methods section. At least three independent experiments were performed for each assay. Student's t-test was performed to demonstrate statistical significance. (*) P < 0.05.
Prediction of microRNAs that target AXL
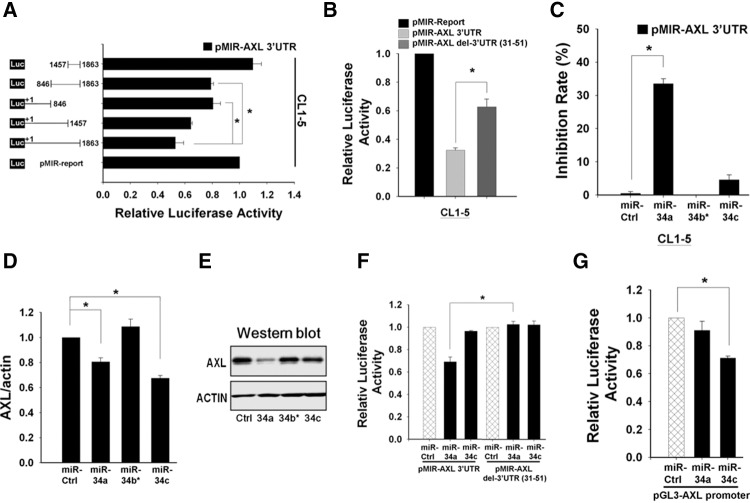
To identify the regions potentially targeted by miRNAs that regulate AXL mRNA in a feedback manner, we used reporter constructs with various lengths of the AXL 3′ UTR. Reporter assays revealed that two regions of the AXL 3′ UTR, 1–846 and 846–1457, may contain putative target sites (Fig. 4A). Similar results were obtained using other cancer cell lines, including MDA-MB-231 and A549 (Supplemental Fig. 3A). By using bioinformatic approaches with four miRNA databases (TargetScan5.1, miRBase, PicTar, miRanda) according to both binding free energy and alignment scores, among the eight miRNAs predicted to have potential binding sites in the AXL 3′-UTR region, only miR-34a, miR-34c, and miR-367 have target sites located within the sequence of the 1–846 region (Supplemental Table 2). The MicroCosm Targets database shows that the 31–56 region was phylogenetically conserved among species (Supplemental Fig. 3C). The AXL 3′-UTR reporter lacking the miR-34 seed region had partially recovered reporter activity in all three cell lines tested (CL1-5, A549, and MDA-MB-MB231) (Fig. 4B; Supplemental Fig. 4A). These results implicate that miR-34 may target this conserved sequence to mediate the feedback regulation of AXL expression.
FIGURE 4.
AXL expression is regulated by microRNAs and through different mechanisms. (A) Identification of the microRNA-binding region of AXL 3′ UTR. The cotransfection reporter assay was performed in the CL1-5 cells using constructs of a series of AXL 3′-UTR deletion mutants (0.5 μg), with Renilla luciferase plasmid serving as an internal control for transfection efficiency. Then the luciferase activity was measured 48 h post-transfection. The schematic presentation on the left shows the deleted regions of various constructs in the AXL 3′ UTR. Reporter activity obtained from the pMiR vector (bottom bar) was set to 1.0 and the activity of the other constructs was calculated and plotted. Relative luciferase activity is shown on the right and is presented as fold values relative to the basal activity. (B) Confirmation of target site (seed region) in AXL 3′ UTR. Luciferase reporter vectors pMIR-Report-AXL 3′ UTR (intact) or pMIR-Report-AXL 3′ UTR (seed region deletion mutant) were transfected into CL1-5 cells, and luciferase activity was measured 24 h later. (C) Inhibition of AXL 3′ UTR by miR-34a/b/c. The AXL 3′-UTR luciferase reporter activities of CL-5 cells transfected with miR-34a/b/c mimics or control oligos were determined. Luciferase activity of control oligos (siRNA) was set as 100% for normalization. An asterisk indicates a significant difference between miR-34a and the control. (D) Expression of the AXL mRNA was determined, in triplicate, by real-time RT-PCR. Fold changes between cells transfected with miR-34a/b/c mimics compared with control oligos are shown. (E) Western blot shows the inhibition of AXL protein levels by miRNAs in CL1-5 cells. Actin serves as a loading control. (F) Discrimination of the target site for miR-34a, but not miR-34c, in AXL 3′ UTR. CL1-5 cells were cotransfected with miR-34a or miR-34c mimic, along with pMIR-Report-AXL 3′ UTR (intact) or pMIR-Report-AXL del 3′ UTR (seed region deletion mutant). An asterisk indicates a significant difference between miR-34c and the control. (G) MiR-34c, but not miR-34a, affects AXL promoter activity. CL-5 cells were transfected with miR-34a or miR-34c mimic along with the pGL3-AXL promoter luciferase construct. Luciferase reporter activity was assayed 48 h later. An asterisk indicates a significant difference between miR-34c and the control. All reporter assays were performed in triplicate and the Renilla reporter activity was used to normalize the transfection efficiency. The error bars represent SD (*) P < 0.05.
miR-34a targets the 3′ UTR of AXL mRNA and inhibits its expression
To identify the miR-34 family that regulates AXL mRNA, we examined the effects of miR-34a, miR-34b*, and miR-34c mimics on the AXL 3′-UTR reporter activity in CL1-5 cells (Supplemental Fig. 3B). We found that only miR-34a could down-regulate AXL 3′-UTR reporter activity (Fig. 4C). Similar results were obtained in MDA-MB-231 cells (Supplemental Fig. 4B). However, as shown, both miR-34a and miR-34c mimics could significantly down-regulate the expression of AXL mRNA and protein (Fig. 4D,E). When the putative miR-34 target site of the AXL 3′ UTR was deleted from the reporter, miR-34a could no longer reduce the AXL 3′-UTR reporter activity (Fig. 4F). In contrast, miR-34c changed neither AXL reporters’ activities (Fig. 4F). In examining these miRNAs’ effects on the AXL promoter's reporter activity, we found that miR-34c significantly down-regulated the AXL promoter's reporter activity, but miR-34a did not (Fig. 4G). These findings implicate that, in contrast to miR-34a, miR-34c may regulate AXL expression through an indirect mechanism, probably by targeting other molecules that modulate AXL promoter activity.
JNK-ELK1 signal pathway is involved in the regulation of miR-34a by AXL
To investigate what pathway is involved in this AXL feedback regulation, we used various inhibitors in the AXL 3′-UTR reporter assay, including PD98059 and U0126 (MEK-ERK), LY294002 (PI3K-AKT), SP600125 (JNK), PP2 (SRC), AG490 (JAK), BAY-11-7086 (NF-κB), and SB203580 (P38). Only JNK inhibitor could significantly recover the AXL 3′-UTR reporter activity suppressed by AXL overexpression in CL1-0 and CL1-5 cells (Fig. 5A; Supplemental Fig. 5A,B). The AXL kinase inhibitor R428 had a similar effect (Fig. 5A; Supplemental Fig. 2D). Because ELK1 is downstream from the JNK pathway (Cavigelli et al. 1995; Whitmarsh et al. 1995), we wondered whether AXL kinase activity is required to activate ELK1. We examined the effects of overexpression of various AXL mutants in CL1-0 cells and observed that wild-type AXL could up-regulate p-AXL, p-JNK, and p-ELK1 expression but not the AXL kinase-dead (K567R) or the intracellular domain deletion (del-ICD) mutants of AXL (Fig. 5B). These results demonstrate that AXL kinase may activate ELK1 via the JNK pathway. To identify the AXL-inducible miRNAs, we compared the miRNA expression arrays between the CL1-0 cells and cells stably overexpressing AXL and found that miR-34a was among the AXL-inducible miRNAs (Supplemental Table 3). To confirm that AXL may up-regulate miR-34a, we transfected the constructs of AXL into CL1-0 cells and analyzed the primary transcript and mature form of miR-34a using quantitative real-time PCR. As a result, expression of miR-34a was indeed found to be up-regulated by AXL overexpression (Fig. 5C). The reporter activity of miR-34a promoter was also increased by AXL in a dose-dependent manner (Supplemental Fig. 5C). These results indicate that activation of AXL is essential for up-regulating the expression of miR-34a.
To clarify whether AXL decreases its own 3′-UTR activity in the presence of a miR-34 inhibitor, we designed two sets of miRNA sponges for anti-miR-34a usage. We used the miR-34a targeting sequence of AXL to synthesize a single-strand RNA oligonucleotide with 2′-Ome modification (SP-A34). The second miR-34a sponge has a sequence perfectly complementary to that of miR-34a (CM-A34). Delivery of the miRNA sponge can specifically neutralize and inhibit mature miRNA from target recognition and translational repression. AXL, MET, and BCL2 are known targets of miR-34a. Our results demonstrate that treatment of CM-A34 or SP-A34 resulted in a significant up-regulation of AXL, MET and BCL2 protein expression in CL1-0, CL1-5, and A549 cells, especially SP-A34 (Supplemental Fig. 6A). We also found that AXL-mediated down-regulation of its 3′-UTR reporter activity and the endogenous AXL mRNA level was significantly increased upon SP-A34 treatment (Fig. 5D,E).
A recent study found that ELK1 may regulate miR-34a expression via promoter targeting during oncogene-induced senescence (Christoffersen et al. 2010). To investigate the effect of ELK1-S383 phosphorylation on its ability to function as an activator for the miR-34a gene promoter, the S383 residue was mutated to aspartate (S383D) to mimic phosphorylation status or to alanine (S383A) to mimic nonphosphorylation status as a control. The miR-34a promoter activity and primary transcript expression were both increased by transfection with wild-type ELK1 and ELK1-S383D, but not ELK1-S383A (Fig. 5F,G; Supplemental Fig. 6B). To further validate the existence of an ELK1-mediated feedback loop for AXL regulation, we transfected ELK1 or ELK1 siRNA into CL1-5 cells and found that overexpression and knockdown of ELK1 resulted in down- and up-regulation of AXL protein expression, respectively (Fig. 5H; Supplemental Fig. 6C). ChIP assay was performed to determine whether ELK1 directly binds to the miR-34a promoter when AXL expression was knocked down by siRNA in CL1-5 and when AXL was ectopically overexpressed in CL1-0 cells. The results show that there is a direct binding between ELK1 protein and the miR-34a promoter (Fig. 5I), demonstrating that ELK1 is a transcription activator of the miR-34a promoter. Indeed, ELK1-mediated down-regulation of AXL 3′-UTR reporter activity was significantly reversed when cells were treated with miR-34a sponge SP-A34 (Supplemental Fig. 6D). Furthermore, AXL-mediated up-regulation of miR-34a and down-regulation of its own mRNA expression could also be reversed by ELK1 siRNA (Supplemental Fig. 6E,F). In summary, this feedback loop is activated by AXL overexpression through JNK-mediated ELK1 activation, and this subsequently leads to an up-regulated expression of miR-34a that in turn down-regulates AXL protein expression.
ELK1 promotes apoptosis in part through inhibition of AXL kinase activity
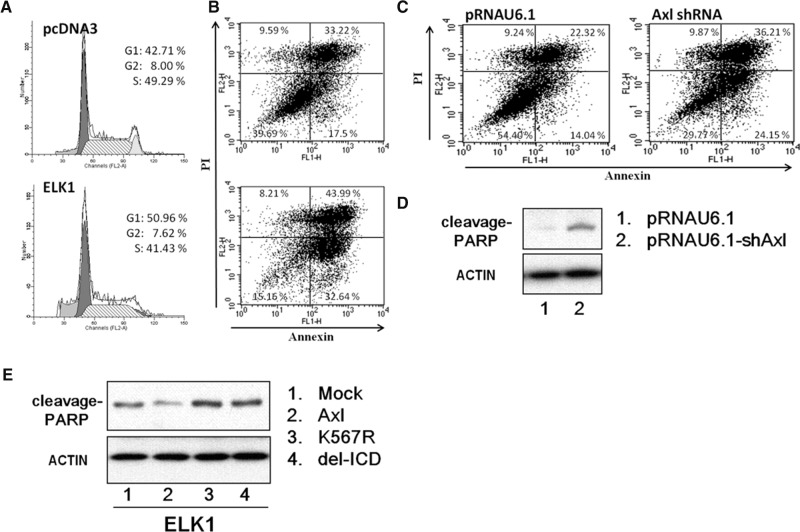
To explore the effect of ELK1 on the cell cycle, we analyzed the cell-cycle distribution in ELK1-transfected cells by flow cytometry. ELK1 caused significant G0/G1 arrest in CL1-5 cells (Fig. 6A). To further elucidate the mechanism underlying this cell-cycle arrest effect, we analyzed apoptosis of transiently ELK1-transfected cells by flow cytometry and found that overexpression of ELK1 induced apoptosis (Fig. 6B). Similar results were obtained using AXL shRNA (Fig. 6C). Double transfection experiments were conducted to clarify that AXL is one of the main mediators of ELK1-induced apoptosis. CL1-5 cells were first transfected with full-length AXL construct, AXL kinase-dead mutant (K567R), or AXL intracellular domain deletion construct (del-ICD), the next day, all were transfected with an ELK1 expression vector. We found that only the wild-type AXL (full-length AXL), but not the kinase-dead AXL or the intracellular domain deletion mutant AXL, decreases poly-ADP-ribose polymerase (PARP) expression; this suggests that AXL kinase activity could attenuate ELK1-induced apoptosis (Fig. 6E). Under conditions with knockdown expression of AXL, the protein level of cleaved PARP was drastically increased (Fig. 6D), further supporting that inhibition of AXL could trigger apoptosis.
FIGURE 6.
ELK1 protein induces G1 arrest and apoptosis. (A) Cell-cycle progression of CL1-5 cells overexpressing ELK1. Cells were transfected with ELK1 or mock vector, harvested, fixed with ethanol, and stained with propidium iodide. DNA content was measured by flow cytometry. The percentage of the cell population in each of the G1, G2–M, and S phases was calculated. Representative plots of one set of triplicate experiments are shown. (B) Overexpression of ELK1 promotes apoptosis. Cells were transfected with ELK1 or mock vector for 48 h. Apoptosis was determined by annexin V-FITC/PI double staining and flow cytometry analysis. The percentage in the lower right quadrant indicates the percentage of early apoptotic cells. The percentage in the upper right quadrant indicates the percentage of late apoptotic cells. (C) Knockdown of AXL expression promotes apoptosis. CL1-5 cells were transfected with AXL shRNA or mock vector for 48 h. Apoptosis was determined by annexin V-FITC/PI double staining and flow cytometry analysis. The percentage in the lower right quadrant indicates the percentage of early apoptotic cells. The percentage in the upper right quadrant indicates the percentage of late apoptotic cells. (D) Western blot analysis of poly (ADP-ribose) polymerase (PARP) cleavage. Whole cell lysates were subjected to Western blotting using antibody against PARP. Actin was used as an internal control for protein loading. (E) Western blot analysis of poly (ADP-ribose) polymerase (PARP) cleavage. After transfection with various constructs of AXL for 24 h, cells were allowed to express ELK1 for 2 days prior to harvest. Total cell lysates were subjected to Western blot analysis for PARP protein. Actin was used as an internal control for protein loading. Mock, vector alone; AXL, full-length wild-type; K567R, kinase dead; delICD, deletion of the entire intracellular domain.
DISCUSSION
Regulation of AXL is of great clinical interest because of AXL's important relationships with cancer invasion and chemotherapy resistance. In this study, we found not only that AXL could up-regulate miR-34a expression but also that miR-34a could turn back to down-regulate AXL by directly targeting its 3′ UTR. We further identified an AXL regulation feedback loop that involves ELK1-mediated up-regulation of miR-34a expression via the JNK pathway and that subsequently represses AXL expression to induce G1 arrest and apoptosis. Although several studies have shown that miR-34a negatively regulates AXL expression (Mackiewicz et al. 2011; Mudduluru et al. 2011; Boysen et al. 2014), here we report that AXL can regulate miR-34a expression through the JNK/ELK1 pathway. Our findings suggest that the relationship between miR-34a and AXL may play a critical role in the maintaining of balanced AXL expression and may provide new insights into its role in tumor progression.
AXL affects multiple cellular behaviors including chemoresistance, epithelial-to-mesenchymal transition, and invasion/metastasis process of cancer cells via different signaling pathways (Wu et al. 2014). AXL can be activated either by Gas6, the biological ligand for AXL, or self-dimerization upon overexpression (Wu et al. 2014). Our results demonstrate that the AXL kinase domain is necessary for this feedback mechanism by using a series of AXL mutants in the reporter experiments (Fig. 2). There are three putative autophosphorylation sites; only one of them, Y779, is required for the feedback loop regulation (Fig. 2B; Supplemental Fig. 1D). In addition, residual reporter activities remained for the double and triple mutations of the Y779, Y821, and Y866 sites, when compared with that of the whole intracellular domain deletion mutation (ICD) (Fig. 2B). Therefore, we assume that there may be other potential phosphorylation sites of AXL that regulate other factors involved in the feedback mechanism. Additionally, deletion of AXL's IG domain, which is required for Gas6 binding, significantly compromised its ability to inhibit AXL 3′-UTR reporter activity (Fig. 3A). Supporting this, the AXL ligand GAS6 was also found to be involved in the negative feedback regulation of AXL expression (Fig. 3B,C). Taken together, this feedback regulation of AXL may involve both ligand-dependent and ligand-independent pathways.
In this study, bioinformatic approaches were used to identify the miR-34 family as potential candidates that may target the AXL 3′ UTR (Fig. 4C; Supplemental Table 2). Both miR-34a and miR-34c may down-regulate the AXL mRNA and protein levels (Fig. 4D,E). By cotransfecting the miR-34 family and the AXL 3′-UTR reporters containing or not containing the seed sequences targeted by the respective miRNAs, our results have shown that AXL is a real target of miR-34a (Fig. 4F), a finding consistent with the results of Mudduluru et al. (2011). However, miR-34c apparently suppressed AXL expression through its promoter rather than its 3′ UTR (Fig. 4F,G), indicating that miR-34c may down-regulate AXL expression in an indirect manner. In mammalian cells, microRNAs primarily modulate gene expression by targeting 3′ UTR of mRNAs. However, recent studies have indicated an alternative way of microRNAs in regulating gene expression through promoter targeting (Li et al. 2006; Place et al. 2008; Younger and Corey 2011). However, we identified no miR-34c targeting sequences in the AXL promoter region (+1 to −1726). We believe that miR-34c most likely may target a transcription factor crucial for the AXL promoter activity and thus deserves to be further investigated. Taken together, our results suggest that miR-34a directly binds on the seed region of AXL 3′ UTR in controlling AXL expression, whereas miR-34c down-regulates AXL expression through indirect modulation of its promoter activity.
The results of our miRNA microarray analyses show that AXL-overexpressing cells have higher expression of miR-34a and miR-34b* (Supplemental Table 3). In addition, the miR-34a promoter reporter activity was also increased by AXL in a dose-dependent manner (Supplemental Fig. 5C). Although AXL kinase activity is required for up-regulation of ELK1 and expression of the primary transcript and mature miR-34a (Fig. 5B,C), ectopic overexpression of ELK1 in CL1-5 cells dramatically suppressed the reporter activity of AXL 3′ UTR (Supplemental Fig. 6D). By overexpression or knockdown of ELK1, we demonstrate that ELK1 expression levels may influence ELK1's own phosphorylation, subsequently resulting in up- or down-regulation of AXL protein expression (Fig. 5H; Supplemental Fig. 6C). Moreover, down-regulation of both the AXL- and ELK1-mediated AXL 3′-UTR reporter activity could be rescued by blocking miR-34a (Fig. 5D,E; Supplemental Fig. 6D), suggesting the existence of an AXL/ELK1/miR-34a regulation loop.
It is clear that miR-34a affects cell proliferation, induces apoptosis (Chang et al. 2007; Yamakuchi et al. 2008; Lize et al. 2010), and targets multiple oncogenes to down-regulate their expression and inhibit invasion ability in various cancers (Migliore et al. 2008; Li et al. 2009a,b; Yan et al. 2009; Corney et al. 2010; Mudduluru et al. 2011), acting like a tumor suppressor. More interestingly, a recent study reveals that miR-34a and miR-199a/b could inhibit AXL expression in solid tumors via epigenetic mechanisms (Mudduluru et al. 2011); the importance of this observation is further strengthened by several studies reporting the close relationships between AXL and miR-34a in colon carcinoma (Kaller et al. 2011), lung cancer (Lee et al. 2011), breast cancer (Mackiewicz et al. 2011), and leukemia (Boysen et al. 2014). However, recent studies also show that miR-34a is highly expressed in papillary thyroid carcinoma (PTC) and functions as an oncogenic miRNA by regulating GAS1 expression via the PI3K/Akt/Bad pathway to promote cell proliferation and prevent apoptosis (Cahill et al. 2006; Ma et al. 2013). Additionally, expression of miR-34a may also commonly be high in many human cancers (Dutta et al. 2007). As either an onco-molecule or as a tumor suppressor, miR-34a is important, and its mutual regulation with AXL as demonstrated in this study highlights its potential in cancer therapeutic opportunities. These results also implicate that the tumor-suppressive roles played by microRNAs are not as simple as they might seem. In different types of cancers, these microRNAs may be under the control of different yet complicated fine-tuning mechanisms and may target multiple genes; some may even possess opposite functions in certain different conditions. Therefore, the detailed regulatory mechanism of AXL modulated by miRNA remains to be further clarified.
Although p53 mutation rate is >50% in many human cancers, up-regulation of miR-34a may promote the cancer cells toward apoptosis regardless of the p53 status (Raver-Shapira et al. 2007; Christoffersen et al. 2010; Hermeking 2010). A recent study has shown that wild-type p53 activation negatively regulates AXL expression via miR-34a and may offer therapeutic opportunities for chronic lymphocytic leukemia B cells (Boysen et al. 2014). According to our data, phosphorylated ELK1 not only is essential for up-regulating miR-34a expression (Fig. 5F,G) but also directly binds to the miR-34a promoter in vivo (Fig. 5I). Because these CL1 sublines have a p53 status of R248W mutation, the regulatory mechanism described in this report is likely to be p53-independent (Tsai et al. 2014). In this study we emphasize that ELK-1 could also directly induce mir-34a expression via promoter targeting, a result consistent with that observed during oncogene-induced senescence (Christoffersen et al. 2010). Although overexpression of ELK1 led to significant G1 arrest and apoptosis (Fig. 6A,B), knockdown of AXL expression with shRNA produced a similar effect (Fig. 6C,D). We further found that AXL kinase activity could overcome the ELK1-induced apoptosis (Fig. 6E). In line with these results, overexpression of ELK1 significantly decreased AXL expression and subsequently induced G1 arrest and apoptosis via a miR-34a-mediated feedback loop. Recently, several reports have indicated that miR-34a expression is silenced in various cancers because of aberrant CpG methylation on its promoter (Lodygin et al. 2008; Vogt et al. 2011). Once the miR-34a promoter is methylated, phosphorylated ELK1 may not bind to its promoter to inhibit AXL expression, and the feedback loop will be consequently interrupted. Dysfunction of the AXL/ELK1/miR-34a feedback loop may presumably shift the equilibrium of AXL expression from a proto-oncogene into a bona fide oncogene by overexpression during cancer progression. In the future, we plan to verify the correlation between AXL-mediated ELK1 phosphorylation and the methylation status of miR-34a promoter, especially the role of AXL phosphorylation at Y779 in this feedback regulation.
The present study shows that AXL up-regulates miR-34a expression via the JNK/ELK1 signal pathway and that miR-34a consequently returns to inhibit AXL expression via binding to AXL’s mRNA 3′-UTR. These results demonstrate that this negative regulatory loop might play an important role in maintaining a balanced AXL expression and that malfunction of this regulatory loop may contribute to resisting apoptosis and promoting invasion and metastasis in cancer cells. For example, cancers may be more malignant because of AXL overexpression caused by miR-34a inactivation, whereas miR-34a activation by ELK1 may slow down cancer progression because of reduced AXL expression. Therefore, if we could better understand the molecular mechanism of this negative feedback loop regulation, we would be able to develop drugs that induce the expression of miR-34a and/or ELK1 to improve anticancer efficacy in clinical applications. Therefore, we believe that the insights into this feedback loop autoregulation of AXL may help to enhance the treatment efficacy of using AXL inhibitors along with chemotherapies for AXL-associated cancers.
MATERIALS AND METHODS
Cell lines, transfection, and siRNA
The CL1 sublines (i.e., CL1-0, CL1-3 and CL1-5) were established by selection for increasingly invasive cancer cells from a human lung adenocarcinoma cell line, CL1-0, as described previously (Lay et al. 2007). CL1 sublines, MCF-7, and MDA-MB-231 were cultured in RPMI; A549, HeLa, and PANC-1 were cultured in DMEM. Both media are supplemented with 10% fetal bovine serum. AXL 3′-UTR reporter (0.5 μg), AXL promoter, Renilla reporter (0.05 μg), with or without other plasmids or miRNA mimics, were transfected into 5 × 105 cells by using Lipofectamine 2000 (Invitrogen) for reporter assays. miRNA mimics (50 nM) (MDBio Inc.) and various plasmids (2 μg) were transfected into 1 × 106 cells for qRT-PCR or Western blot analysis. The siRNA sequence for ELK1: 5′-GCGGCCAGAAGUUCGUCUACAAGUU-3′ (MDBio Inc.). The anti-miR-34a sequences were shown in Supplemental Table 4.
Identification of miRNA binding sites
We used miRBase, PicTar, TargetScan5.1 (Lewis et al. 2003), and miRanda (John et al. 2004) software for prediction of potential AXL 3′-UTR sites targeted by micro-RNAs. The sequence of AXL 3′ UTR is shown in Supplemental Table 2.
qRT-PCR for microRNAs quantification
To detect miR-34a expression, total RNA was isolated with TRIzol reagent (Invitrogen). The Mir-X miRNA First-Strand Synthesis Kit (BD Clontech) was used for cDNA synthesis from miRNAs. To detect endogenous AXL mRNA and primary miRNA expression, total RNA was prepared from CL1-0 cells transfected with various AXL constructs using the RNeasy Mini Kit (QIAGEN). All primer sequences used for AXL mRNA and miR-34a detection are listed in Supplemental Table 4. Actin and U6 RNA were used as the internal control for normalization of AXL and pri-miR-34a, respectively. PCR was performed in a 20 μL reaction mixture containing 10 μM forward and reverse primers, and 1× SYBER GREEN reaction mix (Takara). All reactions were performed in an ABI 7500 sequence detection system.
Plasmids construction
The AXL 3′-UTR reporters were constructed in the pMIR-Report vector by inserting full-length and various mutation or deletion forms of the AXL 3′-UTR sequences into the SpeI/SacI restriction site of the pMIR-Report vector. The miR-34a promoter reporters were constructed by insertion of the PCR-amplified sequences into the XhoI/HindIII site of the pGL3 vector. All primers used for these constructs are listed in Supplemental Table 4. AD-Gas6 is a constitutively active form of Gas6 with its amino-terminal Gla domain deleted. Constructs of AXL shRNA, AXL promoter, and wild-type and various mutants of AXL were as described previously (Lay et al. 2007; Hong et al. 2008; Huang et al. 2013).
The mutations of ELK1 on the S383 residue and the double/triple mutations of the AXL putative autophosphorylation sites were generated by site-directed mutagenesis. All primers used for these constructs are listed in Supplemental Table 4.
Microarray analysis of AXL-responsive miRNAs
CL1-0 cells and their stable transfectants overexpressing AXL were compared by using the Agilent human miRNA microarray of 723 human and 76 human viral miRNAs. Data were analyzed using the GeneSpring 7.3.1 program (Agilent Technologies).
Immunoblots
Primary antibodies to AXL (amino and carboxyl terminals), Gas6, ELK1, p-ELK1, and PARP1 were from Santa Cruz Biotechnology. HRP-conjugated anti-goat, anti-mouse, and anti-rabbit secondary antibodies were from Santa Cruz Biotechnology. Anti-phospho-AXL (Y779) was made-to-order from Genetex. Anti-phospho-JNK was from Cell Signal, Inc. Antibody to actin was from Sigma-Aldrich.
Reagents
The MEK/ERK inhibitors (PD98059 and U0126) and PI3K/AKT inhibitors (Wortmannin and LY294002) were from Cell Signaling Technology. The JNK inhibitor (SP600125), SRC inhibitor (PP2), JAK inhibitor (AG490), and P38 inhibitor (SB203580) were from Calbiochem. The NF-kB inhibitor (BAY-11-7086) was from Santa Cruz. AXL-Fc was from R&D Systems. R428 was kindly provided by Rigel, Inc.
Chromatin immunoprecipitation (ChIP) assay
An EZ-ChIP kit and reagents from Millipore were used, according to the manufacturer's instructions, to detect the in vivo binding between DNA and proteins. Briefly, anti-ELK1 was incubated with the nuclear extract overnight at 4°C with rotation. After immunoprecipitation, DNA was recovered and subjected to PCR amplification by using the primers specific for detection of the +335/+125 region of the miR-34a promoter that contains the putative ELK1-binding site. The primer sequences used for PCR detection of the ELK promoter region are listed in Supplemental Table 4.
Cell-cycle analysis
At harvest, cells were collected and washed twice with PBS, resuspended in 1 mL of PBS, fixed by adding 4 mL of 100% ethanol at −20°C for 2–24 h, and then stained with a propidium iodide (PI) solution containing 50 μg/mL PI and 50 μg/mL RNase (Sigma-Aldrich) in PBS at room temperature for 30 min in the dark. Stained cells were passed through a nylon mesh sieve to remove cell clumps and were then analyzed by flow cytometry (BD). Data were collected and analyzed by CellQuest and ModFit LT software.
Apoptosis analysis by flow cytometry
Apoptosis was also determined using the Annexin V-FITC/propidium iodide (PI) detection kit from BD Pharmingen. At harvest, cells were washed in cold PBS, stained with Annexin V-FITC and PI, following the manufacturer's instructions. The cells were then analyzed by flow cytometry.
Statistical analysis
Quantitative PCR and reporter assays data are shown as mean ± SD. Differences between control and treatment groups were analyzed by Student's t-test. All experiments were performed in triplicate and repeated at least three times and P-values <0.05 were considered statistically significant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Health Research Institutes (CA-104-PP-14, CA-104-SP-01), the Ministry of Health and Welfare (MOHW104-TDU-B-212-124-008), and the Ministry of Science and Technology (MOST-103-2320-B-400-009, NSC 99-2314-B-400-001-MY3), Taiwan. We thank Rigel Pharmaceuticals (South San Francisco, CA) for providing the AXL inhibitor R428. We are also grateful to Dr. Shen-Liang Chen (National Central University, Taiwan) for help during the revision.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.052571.115.
REFERENCES
- Acunzo M, Romano G, Palmieri D, Lagana A, Garofalo M, Balatti V, Drusco A, Chiariello M, Nana-Sinkam P, Croce CM. 2013. Cross-talk between MET and EGFR in non–small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci 110: 8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bellosta P, Costa M, Lin DA, Basilico C. 1995. The receptor tyrosine kinase ARK mediates cell aggregation by homophilic binding. Mol Cell Biol 15: 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen J, Sinha S, Price-Troska T, Warner SL, Bearss DJ, Viswanatha D, Shanafelt TD, Kay NE, Ghosh AK. 2014. The tumor suppressor axis p53/miR-34a regulates Axl expression in B-cell chronic lymphocytic leukemia: implications for therapy in p53-defective CLL patients. Leukemia 28: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill S, Smyth P, Finn SP, Denning K, Flavin R, O'Regan EM, Li J, Potratz A, Guenther SM, Henfrey R, et al. 2006. Effect of ret/PTC 1 rearrangement on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer 5: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavigelli M, Dolfi F, Claret FX, Karin M. 1995. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J 14: 5957–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, Lin SC, Chang YC, Lin SY, Chen SJ, et al. 2010. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res 70: 8822–8831. [DOI] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. 2010. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 17: 236–245. [DOI] [PubMed] [Google Scholar]
- Chung BI, Malkowicz SB, Nguyen TB, Libertino JA, McGarvey TW. 2003. Expression of the proto-oncogene Axl in renal cell carcinoma. DNA Cell Biol 22: 533–540. [DOI] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, et al. 2010. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res 16: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Xu LH, Weiner TM, Fridell YW, Dent GA, Srivastava S, Varnum B, Liu ET, Cance WG. 1995. Receptor tyrosine kinases expressed in metastatic colon cancer. Int J Cancer 60: 791–797. [DOI] [PubMed] [Google Scholar]
- Demarchi F, Verardo R, Varnum B, Brancolini C, Schneider C. 2001. Gas6 anti-apoptotic signaling requires NF-κB activation. J Biol Chem 276: 31738–31744. [DOI] [PubMed] [Google Scholar]
- Dutta KK, Zhong Y, Liu YT, Yamada T, Akatsuka S, Hu Q, Yoshihara M, Ohara H, Takehashi M, Shinohara T, et al. 2007. Association of microRNA-34a overexpression with proliferation is cell type-dependent. Cancer Sci 98: 1845–1852. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. 2006. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269. [DOI] [PubMed] [Google Scholar]
- Fridell YW, Jin Y, Quilliam LA, Burchert A, McCloskey P, Spizz G, Varnum B, Der C, Liu ET. 1996. Differential activation of the Ras/extracellular-signal-regulated protein kinase pathway is responsible for the biological consequences induced by the Axl receptor tyrosine kinase. Mol Cell Biol 16: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicchio M, Mitola S, Valdembri D, Fantozzi R, Varnum B, Avanzi GC, Bussolino F. 2005. Inhibition of vascular endothelial growth factor receptor 2-mediated endothelial cell activation by Axl tyrosine kinase receptor. Blood 105: 1970–1976. [DOI] [PubMed] [Google Scholar]
- Georgescu MM, Kirsch KH, Shishido T, Zong C, Hanafusa H. 1999. Biological effects of c-Mer receptor tyrosine kinase in hematopoietic cells depend on the Grb2 binding site in the receptor and activation of NF-κB. Mol Cell Biol 19: 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafizi S, Ibraimi F, Dahlbäck B. 2005. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J 19: 971–973. [DOI] [PubMed] [Google Scholar]
- Hasanbasic I, Cuerquis J, Varnum B, Blostein MD. 2004. Intracellular signaling pathways involved in Gas6-Axl-mediated survival of endothelial cells. Am J Physiol Heart Circ Physiol 287: H1207–H1213. [DOI] [PubMed] [Google Scholar]
- Heiring C, Dahlback B, Muller YA. 2004. Ligand recognition and homophilic interactions in Tyro3: structural insights into the Axl/Tyro3 receptor tyrosine kinase family. J Biol Chem 279: 6952–6958. [DOI] [PubMed] [Google Scholar]
- Hermeking H. 2010. The miR-34 family in cancer and apoptosis. Cell Death Differ 17: 193–199. [DOI] [PubMed] [Google Scholar]
- Hong CC, Lay JD, Huang JS, Cheng AL, Tang JL, Lin MT, Lai GM, Chuang SE. 2008. Receptor tyrosine kinase AXL is induced by chemotherapy drugs and overexpression of AXL confers drug resistance in acute myeloid leukemia. Cancer Lett 268: 314–324. [DOI] [PubMed] [Google Scholar]
- Huang JS, Cho CY, Hong CC, Yan MD, Hsieh MC, Lay JD, Lai GM, Cheng AL, Chuang SE. 2013. Oxidative stress enhances Axl-mediated cell migration through an Akt1/Rac1-dependent mechanism. Free Radic Biol Med 65: 1246–1256. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Roh S, Hoffmann R, Warscheid B, Hermeking H. 2011. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, et al. 2008. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68: 3566–3572. [DOI] [PubMed] [Google Scholar]
- Lay JD, Hong CC, Huang JS, Yang YY, Pao CY, Liu CH, Lai YP, Lai GM, Cheng AL, Su IJ, et al. 2007. Sulfasalazine suppresses drug resistance and invasiveness of lung adenocarcinoma cells expressing AXL. Cancer Res 67: 3878–3887. [DOI] [PubMed] [Google Scholar]
- Lee WP, Wen Y, Varnum B, Hung MC. 2002. Akt is required for Axl-Gas6 signaling to protect cells from E1A-mediated apoptosis. Oncogene 21: 329–336. [DOI] [PubMed] [Google Scholar]
- Lee JH, Voortman J, Dingemans AM, Voeller DM, Pham T, Wang Y, Giaccone G. 2011. MicroRNA expression and clinical outcome of small cell lung cancer. PLoS One 6: e21300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. 2006. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci 103: 17337–17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. 2009a. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett 275: 44–53. [DOI] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, et al. 2009b. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 69: 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizé M, Pilarski S, Dobbelstein M. 2010. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ 17: 452–458. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. 2008. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7: 2591–2600. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. 2012. The microcosmos of cancer. Nature 482: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Qin H, Cui Y. 2013. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun 441: 958–963. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. 2011. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat 130: 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric F, Lee WP, Sahin A, Zhang H, Kung HJ, Hung MC. 2002. Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res 8: 361–367. [PubMed] [Google Scholar]
- Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. 2008. MicroRNAs impair MET-mediated invasive growth. Cancer Res 68: 10128–10136. [DOI] [PubMed] [Google Scholar]
- Mudduluru G, Vajkoczy P, Allgayer H. 2010. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol Cancer Res 8: 159–169. [DOI] [PubMed] [Google Scholar]
- Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. 2011. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene 30: 2888–2899. [DOI] [PubMed] [Google Scholar]
- O'Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, Prokop C, Espinosa R III, Le Beau MM, Earp HS, Liu ET. 1991. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol 11: 5016–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Litzenburger UM, Sahm F, Rauschenbach KJ, Tudoran R, Hartmann C, Marquez VE, von Deimling A, Wick W, Platten M. 2012. Promotion of glioblastoma cell motility by enhancer of zeste homolog 2 (EZH2) is mediated by AXL receptor kinase. PLoS One 7: e47663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. 2008. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci 105: 1608–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743. [DOI] [PubMed] [Google Scholar]
- Reddy SD, Ohshiro K, Rayala SK, Kumar R. 2008. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 68: 8195–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlitz C, Lohri A, Bacchi M, Schmidt M, Nagel S, Fopp M, Fey MF, Herrmann R, Neubauer A. 1999. Axl expression is associated with adverse prognosis and with expression of Bcl-2 and CD34 in de novo acute myeloid leukemia (AML): results from a multicenter trial of the Swiss Group for Clinical Cancer Research (SAKK). Leukemia 13: 1352–1358. [DOI] [PubMed] [Google Scholar]
- Sainaghi PP, Castello L, Bergamasco L, Galletti M, Bellosta P, Avanzi GC. 2005. Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol 204: 36–44. [DOI] [PubMed] [Google Scholar]
- Shieh Y-S, Lai C-Y, Kao Y-R, Shiah S-G, Chu Y-W, Lee H-S, Wu C-W. 2005. Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia 7: 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Chen PM, Cheng YW, Chen CY, Sheu GT, Wu TC, Lee H. 2014. LKB1 loss by alteration of the NKX2-1/p53 pathway promotes tumor malignancy and predicts poor survival and relapse in lung adenocarcinomas. Oncogene 33: 3851–3860. [DOI] [PubMed] [Google Scholar]
- Vogt M, Munding J, Grüner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. 2011. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch 458: 313–322. [DOI] [PubMed] [Google Scholar]
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. 2009. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284: 5731–5741. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Shore P, Sharrocks AD, Davis RJ. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269: 403–407. [DOI] [PubMed] [Google Scholar]
- Wimmel A, Glitz D, Kraus A, Roeder J, Schuermann M. 2001. Axl receptor tyrosine kinase expression in human lung cancer cell lines correlates with cellular adhesion. Eur J Cancer 37: 2264–2274. [DOI] [PubMed] [Google Scholar]
- Wu CW, Li AF, Chi CW, Lai CH, Huang CL, Lo SS, Lui WY, Lin WC. 2002. Clinical significance of AXL kinase family in gastric cancer. Anticancer Res 22: 1071–1078. [PubMed] [Google Scholar]
- Wu YM, Robinson DR, Kung HJ. 2004. Signal pathways in up-regulation of chemokines by tyrosine kinase MER/NYK in prostate cancer cells. Cancer Res 64: 7311–7320. [DOI] [PubMed] [Google Scholar]
- Wu X, Liu X, Koul S, Lee CY, Zhang Z, Halmos B. 2014. AXL kinase as a novel target for cancer therapy. Oncotarget 5: 9546–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci 105: 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Zhou X, Chen X, Hu DN, Dong XD, Wang J, Lu F, Tu L, Qu J. 2009. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci 50: 1559–1565. [DOI] [PubMed] [Google Scholar]
- Younger ST, Corey DR. 2011. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res 39: 5682–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.