Abstract
Objective
The cortisol response during critical illness varies widely among patients. Our objective was to examine single nucleotide polymorphisms (SNPs) in candidate genes regulating cortisol synthesis, metabolism, and activity to determine if genetic differences were associated with variability in the cortisol response among critically ill children.
Design
This was a prospective observational study employing tag SNP methodology to examine genetic contributions to the variability of the cortisol response in critical illness. Thirty-one candidate genes and 31 ancestry markers were examined.
Setting
Patients were enrolled from 7 pediatric critical care units that constitute the Eunice Kennedy Shriver Collaborative Pediatric Critical Care Research Network.
Subjects
Critically ill children (n=92), ages 40 weeks gestation to 18 years of age were enrolled.
Interventions
Blood samples were obtained from all patients for serum cortisol measurements and DNA isolation. Demographic and illness severity data were collected.
Measurements and Main Results
SNPs were tested for association with serum free cortisol (FC) concentrations in context of higher illness severity as quantified by PRISM III score > 7. A SNP (rs1941088) in the MC2R gene was strongly associated (p =0.0005) with a low FC response to critical illness. Patients with the AA genotype were over seven times more likely to have a low FC response to critical illness than those with a GG genotype. Patients with the GA genotype exhibited an intermediate FC response to critical illness.
Conclusions
The A allele at rs1941088 in the MC2R gene, that encodes the ACTH (corticotropin) receptor, is associated with a low cortisol response in critically ill children. These data provide evidence for a genetic basis for a portion of the variability in cortisol production during critical illness. Independent replication of these findings will be important and could facilitate development of personalized treatment for patients with a low cortisol response to severe illness.
Keywords: single nucleotide polymorphism (SNP), single nucleotide variant, serum free cortisol, PRISM III, ACTH (corticotropin) receptor, critical illness, stress response
INTRODUCTION
A variety of noxious stimuli associated with severe illness, such as sepsis and hypotension, are known to increase serum cortisol levels (1). This response is mediated by complex, integrated neurogenic-endocrine-inflammatory signaling networks (2), centered around the activation of the hypothalamic-pituitary-adrenal axis. Because cortisol is such an important adaptive response (3), its role in critical illness, particularly sepsis, has been extensively examined.
The cortisol response varies widely among patients who are subject to the same physiologic challenges. For certain medical conditions such as sepsis, this observation has led to concern that failure to mount an adequate cortisol response may adversely affect survival. In an attempt to improve the outcome of patients with sepsis, steroid supplementation has been advocated. As initially reported, this therapy appeared promising; (4) however, subsequent investigations of both high and low dose steroid supplementation have generally shown equivocal survival benefits (5) and suggest that the incidence of secondary infections may be increased. (6, 7) Nevertheless, interest in cortisol supplementation for critically ill patients remains high (8–10) and efforts are ongoing to identify the subpopulations of patients that will reap the greatest benefit from this intervention. Important components of this undertaking will be to gain a better understanding of tissue level utilization of cortisol and of genetic variation in cortisol regulation.
The concept of corticosteroid resistance at the tissue level, also known as critical illness related corticosteroid insufficiency (CIRCI) is an area of active investigation (11). Free cortisol (FC) represents the biologically active form of the hormone, although total cortisol (TC) measurements are frequently employed in clinical settings. It has been suggested that assessment of FC may provide the most reliable information regarding CIRCI (12, 13).
We recently reported data regarding TC and FC concentrations in a multi-institutional cohort of critically ill children (14). In this group of patients, increasing levels of FC were moderately correlated with increased illness severity. Of interest was the observation that 30% of these critically ill children exhibited FC < 0.8 ug/dL, the suggested cut off value for the diagnosis of CIRCI in adult patients (15); however, none of these subjects demonstrated clinical signs or symptoms of CIRCI (hemodynamic instability, hypoglycemia or hyponatremia). In order to examine genetic influences on the variability of the cortisol response, at the time the cortisol sample was obtained, we collected mononuclear cells from this cohort of children with the intent of undertaking candidate single nucleotide polymorphism (SNP) analysis of genes involved in cortisol metabolism, regulation and activity. Our objective was to determine if any of the genotypes were correlated with the biochemical phenotype of a low cortisol response during critical illness. The long term goal of this research is to identify critically ill children with the greatest benefit/risk ratio for corticosteroid supplementation. An important component of this objective is to identify genotypes that predict which individuals are highly likely to have inadequate cortisol response during critical illness.
MATERIALS AND METHODS
Participating Sites
This study was conducted within the charter Collaborative Pediatric Critical Care Research Network (CPCCRN) (16), that during the first funding cycle encompassed 7 pediatric critical care units (PICUs), and represented a cross-section of critically ill children from geographically diverse regions in the United States.
Institutional Board Review Approval
This project was initially reviewed and approved by the institutional review board at Seattle Children’s Hospital. Subsequently, the individual institutional review boards for each of the 6 other participating CPCCRN performance sites and the data coordinating center reviewed and approved the study protocol. At the time the samples were collected, the parents of all patients enrolled in this investigation provided explicit consent for the study of their children’s DNA.
Patients, Demographics
Samples used in this study were collected from patients participating in the Cortisol Quantification Investigation, which was performed to validate a new laboratory method for rapidly assaying FC concentrations (14). Inclusion criteria for this study were admission to a PICU, age > 40 weeks gestation to 18 years of age, weight > 5 kg and a blood sample obtainable within 24 hours of admission to the PICU. Patients were excluded from the study if they had received systemic steroids within the previous month (including at the time of this ICU admission), or if the reason for admission to the PICU was a requirement for extracorporeal membrane oxygenator support, if the patient was immediately post cardiac surgery requiring cardiopulmonary bypass, if the patient required leukopheresis, plasmapheresis or hemodialysis, if the patient was not expected to survive the PICU admission, or cardiopulmonary resuscitation status was limited. Massive transfusion (> 50% of the total blood volume) was also cause for exclusion from the study. Racial classifications (ancestry) were self-declared. These self-declared classifications were confirmed with a group of ancestry informative SNPs.
Research Procedures
After patients were judged to be eligible for the study, parental permission and patient assent (when applicable) for the study was obtained, and a single blood sample was drawn. Samples were obtained during the day (8:00 AM – 4:00 PM) as soon as the consent process was completed. All samples were collected within 24 hours of admission to the intensive care unit. For this portion of the study, samples were processed differently than those used for the earlier cortisol measurement study. These samples were fractionated by density gradient centrifugation using Cell Preparation Tubes with sodium citrate (Becton, Dickinson and Company, Franklin Lakes, NJ). The mononuclear cell fraction was collected and placed in a 5 mL cryovial containing RNAprotect Cell Reagent (Qiagen, Alameda, CA) to preserve RNA and DNA. These samples were stored at −80°C until the DNA was isolated. DNA and RNA were isolated from the samples using the AllPrep DNA/RNA system (Qiagen). Total and free cortisol were assayed as previously described (14).
Pediatric Risk of Mortality, version III (PRISM III) scores were recorded and used to quantify severity of illness (17).
Candidate genes for this investigation were selected by reviewing glucocorticoid biosynthesis and degradation pathways from the KEGG Pathway Database and GeneGo pathways databases. In addition, the Human Genome Epidemiology (HuGE) Gene Prospector was used to ascertain additional genes that had been identified in the medical literature as having variants that were associated with cortisol synthesis or regulation. SNP genotyping was performed using a custom GoldenGate assay platform (Illumina, San Diego, CA) consisting of 384 SNP loci: 353 SNPs from 33 candidate genes and 31 SNPs as ancestry informative markers (AIMs) (18, 19). We used a tag SNP approach (SNPs in linkage disequilibrium with candidate gene SNPs that are not included on the chip, r2 value ≥ 0.8) to select SNPs for the assay. The 33 candidate genes are known to be important in the metabolism, regulation, and activity of cortisol (Table 1). The complete ancestry marker list consists of 34 SNPs; however, only 31 were available in the GoldenGate assay format.
TABLE 1.
Comparison of all patients in the cortisol measurement study with the subgroup of patients with DNA concentration sufficient analysis for genetic analysis.
| Cortisol Measurement Study |
SNP Genetics Study |
|||
|---|---|---|---|---|
| Frequency | Percent | Frequency | Percent | |
| Native American | 1 | .6 | 1 | 1.1 |
| Asian | 9 | 5.4 | 9 | 9.8 |
| Black | 34 | 20.4 | 12 | 13.0 |
| Other | 10 | 6.0 | 6 | 6.5 |
| Stated unknown | 4 | 2.4 | 3 | 3.3 |
| White | 109 | 65.3 | 61 | 66.3 |
| Female | 80 | 47.9 | 43 | 46.7 |
| Male | 87 | 52.1 | 49 | 53.3 |
| Total | 167 | 100.0 | 92 | 100.0 |
| Mean | Std. Deviation |
Mean | Std. Deviation |
|
| Age | 9.68 | 5.71 | 9.10 | 6.00 |
| PRISM III | 8.06 | 6.97 | 7.49 | 6.99 |
| Total Cortisol | 21.71 | 27.39 | 20.08 | 19.55 |
| Free Cortisol ED | 4.71 | 12.74 | 3.78 | 6.27 |
| Free Cortisol UF | 4.88 | 12.44 | 4.06 | 7.51 |
There were no statistically significant differences between these two groups (Chi square). All ancestry groupings were self-declared by the subjects’ parents. “Other” and “Unknown” are both stated per subjects’ parents. ED, equilibrium dialysis and UF, centrifugal ultrafiltration refer to the fractionation methodology prior to free cortisol analysis.
Sample assays were carried out by the Northwest Genomics Center in the Department of Genome Sciences at the University of Washington (Seattle, WA). Before genotyping, all samples underwent quality control testing to be certain that each sample contained the minimum amount of DNA necessary for successful assay and that the sex matched that of the listed participant. Assay controls were built-in to each oligo pool for allele-specific ligation, PCR uniformity, gender, extension and gap ligation, hybridization, and oligo contamination. After the DNA concentration in the samples was normalized, sample DNA was activated with biotin and captured with streptavidin conjugated paramagnetic beads.
Allele-specific oligonucleotides and linker oligonucleotides were hybridized to the biotinylated DNA. Allele-specific extension and ligation was performed on the hybridized oligonucleotides. Subsequently, PCR incorporating Cy3 and Cy5 dyes was carried out on the extended and ligated products. The fluorescent labeled PCR products were denatured and single-stranded, labeled product was hybridized to microbeads in a 96-well plate format (each well can assay 384 sites). Hybridized microbeads were scanned using the Illumia BeadXpress scanner, a dual-color detection system. Genotypes were called using the Illumina BeadStudio software package, which normalizes and clusters the raw scan data. Of the 353 tag SNPs, 349 were successfully genotyped (fewer than 2% of individuals had low quality/missing calls).
Statistical Analysis
Analyses were performed using the SPSS statistical package (release 18) and R v2.8 (www.r-project.org). All SNPs were tested for Hardy-Weinberg equilibrium as a further quality control measure, and no SNPs were significantly out of equilibrium. Ancestry informative markers were separated from candidate SNPs and decomposed into principal components for use as covariates to adjust for possible confounding by ancestry in the candidate SNP analysis (20, 21). Principal component plots showed good separation of groups according to self-declared ancestry.
Because our primary interest was in SNPs that correspond to low cortisol response during severe illness, we formulated a model to test for differential cortisol concentrations in patients with high PRISM III scores compared to patients with low PRISM III scores. A median PRISM III score of 7 was used to divide the study sample into two groups, one with a lower acuity of illness and another with a higher acuity of illness. We performed a two-stage analysis to test for both differential effects between high and low PRISM III score groups as well as a differential response by allele within the high PRISM III group. The sample median PRISM III score was chosen a priori to define the response groups because equal group sizes provides the highest statistical power (under most genetic scenarios) for this analytic strategy.
Specifically, we first tested each SNP for its association with log2 [FC], including PRISM III score > 7 (the median PRISM III score) as an indicator in the model along with the SNP × PRISM III group interaction. The log2 transformation was used because of the skewed distribution of FC. An additive allele model was used for SNPs: each subject’s genotype for each SNP was scored as 0, 1 or 2 minor alleles. In the second stage, we tested for a differential response by genotype within the high PRISM III score group, again using the additive model. The first and second principal components from the AIMs were tested in each model to adjust for potential confounding by ancestry but then dropped from the models due to lack of effect/significance. Adjustment for multiple testing for top hits (smallest observed p-values) was conducted via Monte Carlo in which the models were run for all SNPs 1000 times under the null hypotheses for both testing stages. The joint distribution of the minimum p-values among the SNPs for the two-stages was determined over the 1000 runs to produce the null distribution for comparison with the observed p-values.
RESULTS
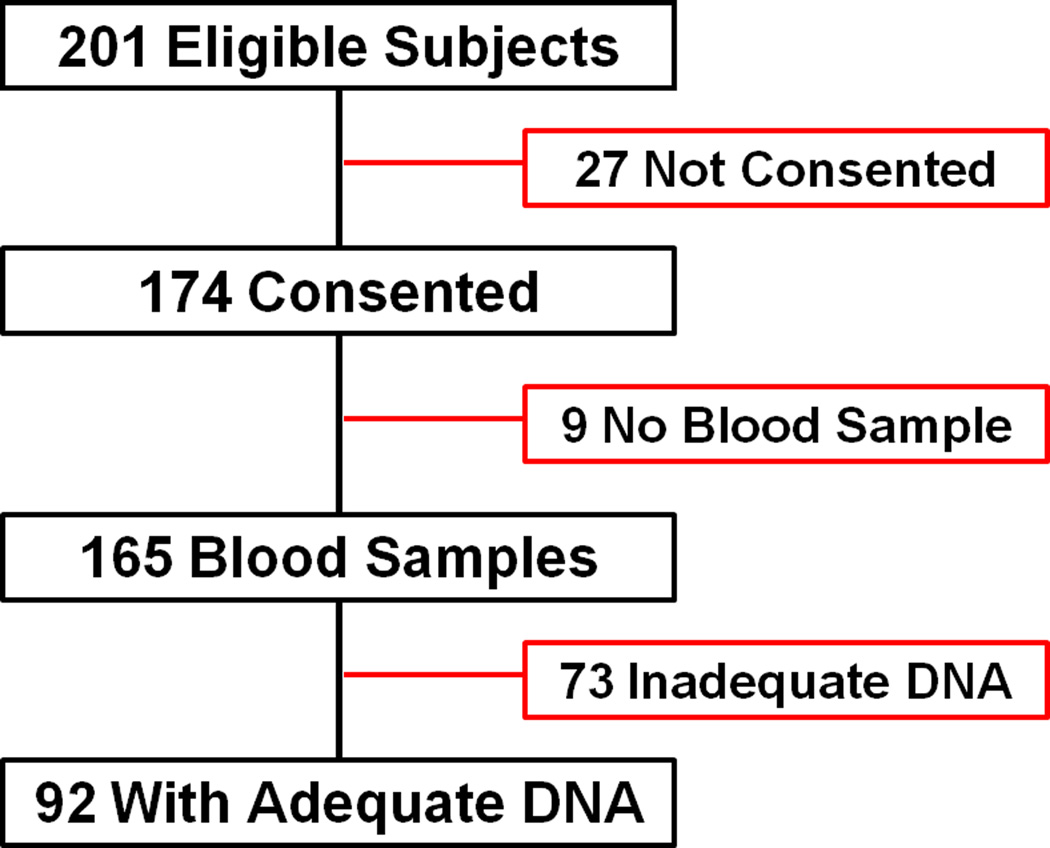
The patients whose samples were used in this investigation are a subset of a larger group of patients who were enrolled in an investigation to validate a new method for measuring FC concentrations. (14) This subset comprised 92 patients with sufficient DNA available for the Illumina platform. Of the 201 patients eligible for cortisol measurement study, parental consent was obtained for 174 patients. Blood samples were available from 165 patients, and samples with DNA of sufficient concentration for analysis was available from 92 of the patients (Figure 1). There were no statistically significant differences between the patients enrolled in the cortisol measurement study and the subpopulation with adequate concentration of DNA in their samples (Table 2).
Figure 1.
Subject and biosample disposition for the investigation.
TABLE 2.
Acute primary PICU admission diagnoses for the patients.
| Diagnosis | Number | Percent |
|---|---|---|
| Postoperative orthopedic surgery | 14 | 15.2% |
| Sepsis/meningitis/pneumonia | 12 | 13.0% |
| DKA, other metabolic disease | 12 | 13.0% |
| Postoperative neurosurgery | 11 | 12.0% |
| Trauma | 10 | 10.9% |
| Postoperative CHD surgery | 7 | 7.6% |
| Seizures, other neurological | 3 | 3.3% |
| Acquired cardiovascular disease | 3 | 3.3% |
| Renal failure | 2 | 2.2% |
| Cancer | 2 | 2.2% |
| Hypox-ischemic-reperfusion | 2 | 2.2% |
| Hematologic disease | 2 | 2.2% |
| Acute abdomen | 2 | 2.2% |
| Hepatic failure | 2 | 2.2% |
| Other diagnoses | 8 | 8.7% |
| Total | 92 | 100.0% |
The median age of the 92 participants was 9.9 years (IQR 25%–75%: 2.8 – 14.6 years), with 43 females and 49 males. The acute primary PICU admission diagnoses for these patients are shown in Table 3. PRISM III scores ranged from 0–39 (Figure 2), with a median value of 7 (IQR 25%-75%: 2 – 10.75). When the study population was divided at the median PRISM III score, gender and ancestry variables were evenly distributed between PRISM III score groups, while mean TC and FC values were higher in the PRISM III >7 group as expected (Table 4).
TABLE 3.
Demographics, illness severity, and serum cortisol concentrations for the study population.
| PRISM ≤ 7 | PRISM > 7 | ||||
|---|---|---|---|---|---|
| Number | Percent | Number | Percent | P Value | |
| Gender | |||||
| Male | 23 | 46.0 | 20 | 47.6 | NS |
| Female | 27 | 54.0 | 22 | 52.4 | NS |
| Ancestry | |||||
| Native Am | 1 | 2.0 | 0 | 0.0 | NS |
| Asian | 6 | 12.0 | 3 | 7.1 | NS |
| Black | 4 | 8.0 | 8 | 19.0 | NS |
| Other | 3 | 6.0 | 3 | 7.1 | NS |
| Unknown | 3 | 6.0 | 0 | 0.0 | NS |
| Caucasian | 33 | 66.0 | 28 | 66.7 | NS |
| Mean | SD | Mean | SD | P Value | |
| Age (years) | 8.68 | 5.59 | 9.60 | 6.49 | NS |
| PRISM III | 2.60 | 2.14 | 13.31 | 6.26 | NA |
| Total Cortisol (µg/dL) | 13.48 | 10.40 | 27.93 | 24.56 | 0.0003 |
| Free Cortisol (µg/dL) | 2.12 | 3.26 | 6.36 | 10.13 | 0.0063 |
PRISM III, Pediatric Risk of Mortality, version III; Am, American; under Ancestry, “Other” and “Unknown” are both stated per subjects’ parents. The racial differences between the PRISM III ≤ 7 group and the PRISM III > 7 group are not statistically significant (Fisher exact test). NS, not significant; NA, not applicable. Total and free cortisol concentrations were determined per methodology summarized in reference 14.
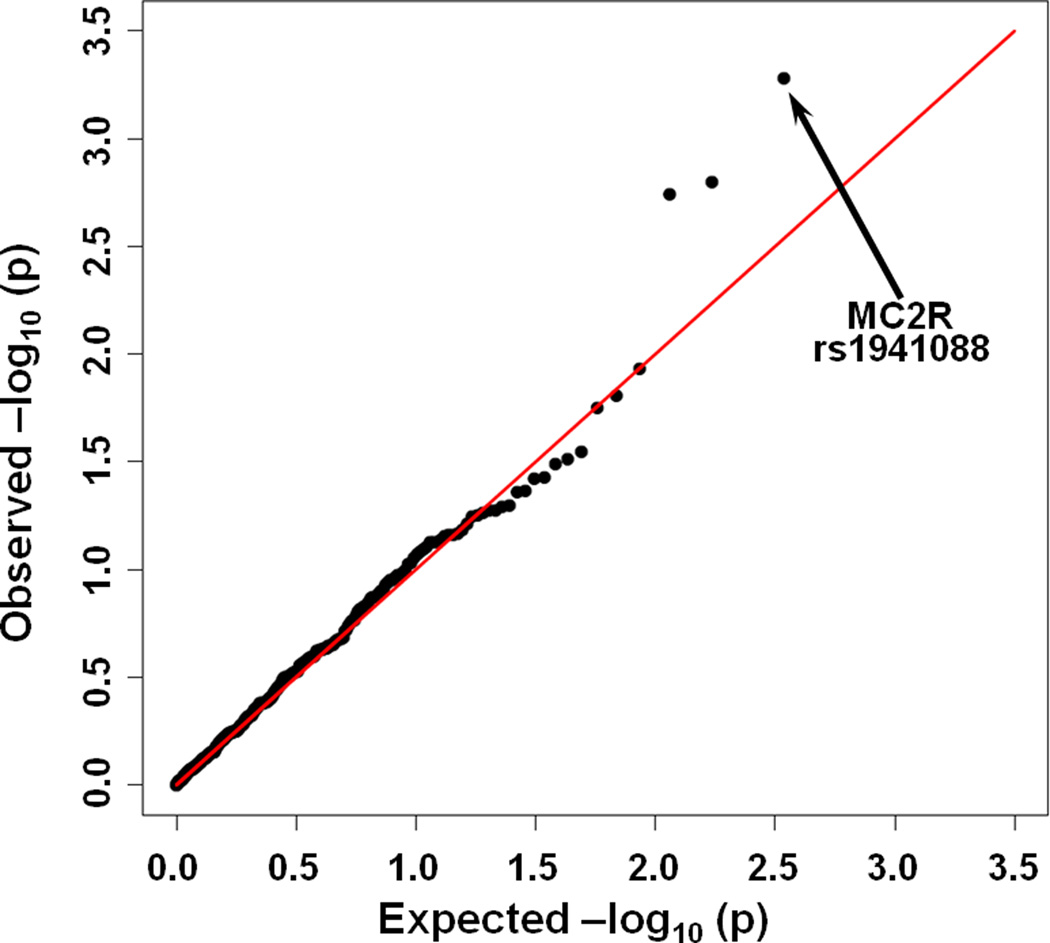
Figure 2.
Expected versus observed log10 (p-values) for association of SNP genotypes with differential response to PRISM III scores > 7. SNP rs1941088 is highly associated with a differential cortisol response (p =0.00052).
In the SNP regression models, we were able to analyze 330 of the 349 successfully genotyped SNPs; 19 SNPs could not be analyzed in the primary regression model due to too few patients with particular genotypes. Assessment of p-values and coefficients for the ancestry principal components in the regression models showed no effect for the adjustment (i.e. no confounding between ancestry and low FC response to more severe illness). Therefore the ancestry principal components were dropped from the models in order to increase power.
In the primary analysis, the SNP most highly associated with a different cortisol response in the high and low PRISM III groups was rs1941088 in a non-coding region of the melanocortin 2 receptor gene (MC2R; p=0.00052), followed by rs1800630 (TNF; p=0.0016) and rs1313379 (NR3C2, nuclear receptor subfamily 3, group C, member 2; p=0.0018) (Figure 3).
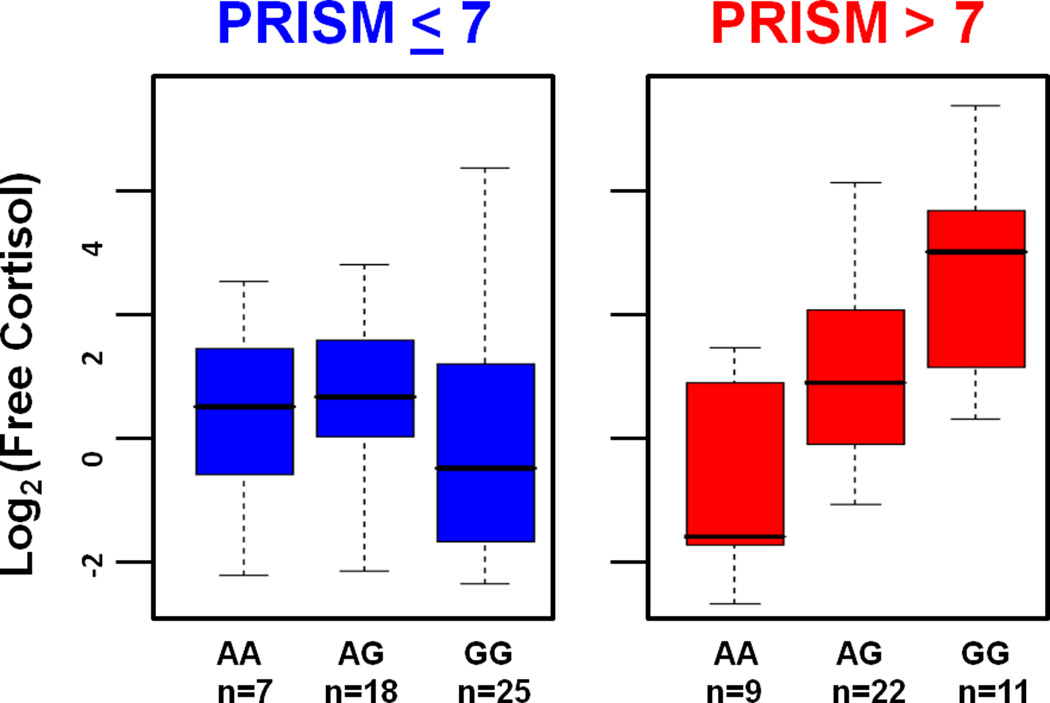
Figure 3.
Log2 of free cortisol (FC) concentration in relation genotype for the SNP rs1941088 for subjects with PRISM scores ≤ 7 or > 7. Greater stressed subjects (PRISM III scores > 7) with the AA genotype failed to respond to physiological stress with a vigorous FC response. In the less stressed state (PRISM III scores of ≤ 7), the AA genotype is indistinguishable from alternative genotypes.
The above association for rs1941088 was stronger among self-declared Caucasians (p=0.000032). In stage two of the analysis, rs1941088 was again the most significant SNP and showed a strong, significant relationship between FC response and number of minor alleles among children in the higher PRISM III score group (p=0.0007; Figure 4). Children with PRISM III >7 showed a ~4-fold reduction in FC for each A allele at this site, with the AA genotype children exhibiting a median FC concentration of only 0.34 ug/dL. Again, the effect was stronger among Caucasians (p=0.00007). SNPs rs1800630 and rs1313379 showed weaker associations with FC among children in the high (>7) PRISM III score group (p=0.008 and p=0.048 respectively; graphical data not shown), but only rs1941088 remained statistically significant after the correction for multiple testing via Monte Carlo determination of the null distributions of the most significant SNPs in both stages of the analysis (p=0.016, p=0.34, p=0.39, for MC2R-rs1941088, TNF-rs1800630 and NR3C2-rs1313379, respectively).
Of interest, in the second stage analysis, the second most significant SNP was rs1940907 (p=0.007), also an MC2R SNP (3-prime-untranslated region). The genotype scores for these two MC2R SNPs are highly correlated (ρ=0.67, p<10−12), consistent with the similar findings for each.
DISCUSSION
We have identified a SNP, rs1941088, in the non-coding region of the MC2R gene (melanocortin 2 receptor, also known as the ACTH receptor) that is significantly associated with a low FC response among children whose PRISM III scores were above the median in this study. Among patients with PRISM III scores >7, the FC response was proportional to the number of minor alleles at this locus, consistent with an additive causal genetic effect (Figure 4). In order to conduct an analysis comparing less severely ill patients to more severely ill patients, we used the median PRISM III score of 7 as a point to partition patients into groups with greater and lesser severity of illness. As expected, children with a PRISM III score of > 7 showed a higher cortisol response than those with PRISM III scores ≤ 7. Children with a PRISM III score exceeding 7 and with the AA genotype generated significantly less FC than children with AG and GG genotypes. Alternatively children with the GG genotype were 7.4 times more likely to have FC above the median value than children with the AA genotype among all those with PRISM III scores exceeding 7. No genotype dose-response was apparent for children with PRISM III scores ≤ 7, patients in whom illness severity is expected to be relatively low. We elected to use a PRISM III score of 7 to partition patients into groups of more severe and less severe illness. Our rationale for this selection was that the PRISM III score of 7 was the median PRISM III score for the patients in this study, thus allowing us to partition patients into equal size groups according to severity of illness.
We elected to use the PRISM III scoring system to quantitate the physiologic variables associated with critical illness. Although other scoring systems such as the PIM or PELODS could have been used, we selected the PRISM III system because it has been serially validated and it is in wide use throughout the United States. Importantly, for purposes of this investigation, the PRISM III is the illness severity metric employed by the CPCCRN group and the research coordinators have been trained to collect these data accurately.
The genotyping of multiple SNPs necessarily involves the testing of multiple hypotheses, which means that a 0.05 level p-value for any one test among these has an increased probability of being a false positive. Accordingly, we adjusted the results of our analyses to correct for multiple hypothesis testing. Even after these adjustments, the genotype of SNP rs1941088 remained significantly associated with a low cortisol response to more severe illness (PRISM III score >7) (p=0.016). Furthermore, a closely linked MC2R SNP, rs1940907, was next most strongly associated with FC response among children with high PRISM III scores. Consistent with our finding that rs1941088 is tagging a variant that is important in cortisol regulation, several studies have shown that polymorphisms in and near the promotor region of MC2R are associated with lowered sensitivity to ACTH (high ACTH/cortisol ratios). These conditions include phenotypically normal adults (22, 23), infantile spasms resistant to ACTH treatment (24), early adrenarche (25), and heroin addiction (26). Together, these findings strongly suggest that a portion of variability of the cortisol response to illness severity is attributable to genetic variants affecting the expression of MC2R.
Given the importance of the MC2R protein in transducing the cortisol response to signal ACTH signaling originating from the hypothalamus, it is biologically plausible that variation in the MC2R gene, that encodes the ACTH receptor, could exert an effect on serum cortisol concentrations. The ACTH receptor, is a 297 amino acid transmembrane protein, expressed on the surface of cells in the zona fasciculata of the adrenal gland, the tissue responsible for cortisol synthesis. Expression of this receptor is necessary for the adrenal cortex to synthesize cortisol in response to ACTH stimulation (27). When the MC2R gene is knocked out in experimental animals, it is frequently lethal (28). Mutations in the MC2R gene are responsible for the disorder known as familial, type 1 glucocorticoid deficiency (29–31). These data are consistent with the hypothesis that polymorphisms in MC2R are associated with variation in the cortisol response to critical illness.
Because our sample size was small, we felt that the candidate gene study design was superior to the genome wide association study design. The relatively small sample precludes testing of thousands of SNPs, because statistical power would be low in the face of adjustment for multiple testing. While one could perform genome-wide SNP genotyping and select only candidate SNPs for statistical testing, the additional cost of such genotyping was not warranted given the uncertain utility of the additional data. In general, small genetic studies are discouraged because they tend to be underpowered and to have significant risk of Type II error (failing to identify a significant association when one is present) (32). Fortunately, small studies do not pose any additional risk of Type I error (false positive), as long as proper corrections for multiple testing are implemented. Type I error can be affected by hidden confounders and/or biased sampling, but these issues can affect large studies as well as smaller studies. Our sample included balanced numbers of children from multiple centers to ensure broad representation of children admitted for critical care, and the study procedures were centrally coordinated (via the CPCCRN) to provide consistent data collection and application of study procedures. Consequently, great care has been taken to eliminate confounding factors, and the significance value assigned to the association between rs1941088 and lower cortisol response during more severe illness (PRISM III score >7) should be valid.
Because of the relatively small sample size, we were unable (due to lack of statistical power) to use more than two PRISM categories for the outcome and did not attempt to estimate the non-linear functional relationship between the PRISM score and cortisol. However, the fact that we have obtained a signal with a straightforward and uncomplicated statistical model can be seen as a strength of the result: the signal does not depend on ability to estimate the best cut-points for the PRISM score nor does it depend on a precise model for the PRISM vs. cortisol relationship. With a larger sample size in future studies, more complex models can be fitted to help determine the PRISM vs. cortisol and PRISM vs. genotype relationships with more precision to help elucidate the underlying pathophysiology.
We used a tag SNP approach in this investigation, meaning rather than sample every frequently occurring SNP in the MC2R gene, we examined a subset of SNPs that are in high linkage disequilibrium (correlation) with other frequently occurring (>5%) SNPs in the MC2R gene. Although this approach is efficient, it does not definitively identify the SNP that is responsible for the observed effect (low cortisol response). Instead, any combination of variants (haplotype) that is in linkage disequilibrium with rs1941088 could be causal. Of note, rs1941088 is in linkage disequilibrium with SNPs rs1893219 and rs1893220 (correlation 0.50 to 0.93) in the MC2R gene. These two SNPs are located in the promoter region of MC2R and have been previously shown to be associated with higher ACTH/cortisol ratios. (24, 33) All known non-synonymous variants in the coding region of MC2R are rare and highly deleterious [per dbSNP, (34) making it unlikely that our results are due to linkage disequilibrium with coding variants. Causal variants in the single intron for MC2R cannot be ruled out at this stage, however, and rs1941088 also tags other intronic SNPs.
Although glucocorticoids alter the expression of over 1300 genes (35), the regulation and synthesis of cortisol, the endogenous glucocorticoid, is governed by a modest number of genes. We selected 33 candidate genes related to cortisol synthesis and that we believed had the highest likelihood of association with the cortisol response to severe illness. Among these were a small number of inflammatory cytokine genes, because some of the inflammatory cytokines have a role in regulating the synthesis of cortisol. For example, TNFα and IL1β alter the activity of 11β-HSD1 and 11β-HSD2 (hydroxysteroid dehydrogenases) to favor the conversion of cortisone to cortisol, the biologically active mediator of the stress response (36–39). Among the group of inflammatory cytokine genes that were examined, we did not detect any SNPs that were significantly associated with altered cortisol response to severe illness. It is possible that cytokine influences on cortisol synthesis were not detectable because of the power limitations in this study. Additionally, the effects of pro-inflammatory cytokines may be most dramatic during illnesses with a strong inflammatory component such as sepsis. Because most of our subjects were admitted to PICUs for reasons other than sepsis, effects caused by genetic variations in the cytokine system might be much less evident in this population. Given the current evidence for a genetic component in cortisol response to severe illness, future studies could be designed to refine these findings to determine whether the genetic association varies with the type of illness (i.e. sepsis vs. non-sepsis) and thus further refine future treatments.
In conclusion this investigation demonstrates that the genotype of the rs1941088 SNP in the MC2R gene is associated with a low FC response in critically ill children with a PRISM III score of > 7. Our investigation measured these findings at a single point in time, just after patients were admitted to a pediatric intensive care unit. We did not attempt to correlate these findings with outcome, which will be important to do in future studies. This finding expands our understanding of the variability of cortisol response during critical illness and provides a plausible biological explanation for a portion of the cortisol variation that is observed. As in all genotype/phenotype association studies, it will be important to validate these findings in an independent investigation. It will also be important to identify the causative SNP(s), which will reveal the mechanism underlying these observations. Confirmation of our findings would indicate MC2R genotyping is a potential tool for identifying one subset of critically ill patients who might benefit from corticosteroid supplementation in critical illness.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Karen Cornell for her technical assistance and the CPCCRN performance site research coordinators for their assistance with conduct of the study.
This work was supported, in part, by cooperative agreements (U10HD050096, U10HD049981, U10HD500009, U10HD049945, U10HD049983, U10HD050012 and U01HD049934) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Department of Health and Human Services as well as a Recovery Act Administrative Supplement 3U10HD049945-05S1.
REFERENCES
- 1.Cuesta JM, Singer M. The stress response and critical illness: A review. Crit Care Med. 2012;40:3283–3289. doi: 10.1097/CCM.0b013e31826567eb. [DOI] [PubMed] [Google Scholar]
- 2.Molina PE. Neurobiology of the stress response: contribution of the sympathetic nervous system to the neuroimmune axis in traumatic injury. Shock. 2005;24:3–10. doi: 10.1097/01.shk.0000167112.18871.5c. [DOI] [PubMed] [Google Scholar]
- 3.Chrousos GP. The hypothalamic-pituitary-adrenal axis and immune-mediated inflammation. New Eng J Med. 1995;332:1351–1362. doi: 10.1056/NEJM199505183322008. [DOI] [PubMed] [Google Scholar]
- 4.Schumer W. Steroids in the treatment of clinical septic shock. Ann Surg. 1976;184:333–341. doi: 10.1097/00000658-197609000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefering R, Neugebauer EA. Steroid controversy in sepsis and septic shock: a meta-analysis. Crit Care Med. 1995;23:1294–1303. doi: 10.1097/00003246-199507000-00021. [DOI] [PubMed] [Google Scholar]
- 6.Cronin L, Cook DJ, Carlet J, et al. Corticosteroid treatment for sepsis: a critical appraisal and meta-analysis of the literature. Crit Care Med. 1995;23:1430–1439. doi: 10.1097/00003246-199508000-00019. [DOI] [PubMed] [Google Scholar]
- 7.Sprung CL, Annane D, Keh D, et al. Hydrocortisone therapy for patients with septic shock. N Engl J Med. 2008;358:111–124. doi: 10.1056/NEJMoa071366. [DOI] [PubMed] [Google Scholar]
- 8.Patel GP, Balk RA. Systemic steroids in severe sepsis and septic shock. Am J Respir Crit Care Med. 2012;185:133–139. doi: 10.1164/rccm.201011-1897CI. [DOI] [PubMed] [Google Scholar]
- 9.Contrael KM, Killian AJ, Gregg SR, et al. Prescribing patterns of hydrocortisone in septic shock: a single-center experience of how surviving sepsis guidelines are interpreted and translated into bedside practice. Crit Care Med. 2013;41:2310–2317. doi: 10.1097/CCM.0b013e31828cef29. [DOI] [PubMed] [Google Scholar]
- 10.Venkatesh B, Myburgh J, Finfer S, et al. The ADRENAL study protocol: adjunctive corticosteroid treatment in critically ill patients with septic shock. Crit Care Resusc. 2013;15:83–88. [PubMed] [Google Scholar]
- 11.Marik PE. Critical illness-related corticosteroid insufficiency. Chest. 2009;135:181–193. doi: 10.1378/chest.08-1149. [DOI] [PubMed] [Google Scholar]
- 12.Arafah BM. Hypothalamic pituitary adrenal function during critical illness: limitations of current assessment methods. J Clin Endocrinol Metab. 2006;91:3725–3745. doi: 10.1210/jc.2006-0674. [DOI] [PubMed] [Google Scholar]
- 13.Hamrahian AH, Oseni TS, Arafah BM. Measurements of serum free cortisol in critically ill patients. N Engl J Med. 2004;350:1629–1638. doi: 10.1056/NEJMoa020266. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman JJ, Donaldson A, Barker RM, et al. Real-time free cortisol quantification among critically ill children. Pediatr Crit Care Med. 2011;12:525–531. doi: 10.1097/PCC.0b013e3181fe4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annane D, Maxime V, Ibrahim F, et al. Diagnosis of adrenal insufficiency in severe sepsis and septic shock. Am J Respir Crit Care Med. 2006;174:1319–1326. doi: 10.1164/rccm.200509-1369OC. [DOI] [PubMed] [Google Scholar]
- 16.Willson DF, Dean JM, Newth C, et al. Collaborative Pediatric Critical Care Research Network (CPCCRN) Pediatr Crit Care Med. 2006;7:301–307. doi: 10.1097/01.PCC.0000227106.66902.4F. [DOI] [PubMed] [Google Scholar]
- 17.Pollack MM, Patel KM, Ruttimann UE. PRISM III: an updated Pediatric Risk of Mortality score. Crit Care Med. 1996;24:743–752. doi: 10.1097/00003246-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Phillips C, Salas A, Sanchez JJ, et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Gen. 2007;1:273–280. doi: 10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Fondevila M, Phillips C, Santos C, et al. Revision of the SNPforID 34-plex forensic ancestry test: Assay enhancements, standard reference sample genotypes and extended population studies. Forensic Sci Int Gen. 2013;7:63–74. doi: 10.1016/j.fsigen.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Seldin MF, Price AL. Application of ancestry informative markers to association studies in European Americans. PLoS Genet. 2008;4:e5. doi: 10.1371/journal.pgen.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassir R, Kosoy R, Tian C, et al. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC Genetics. 2009;10:39. doi: 10.1186/1471-2156-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reisch N, Slawik M, Zwermann O, et al. Genetic influence of an ACTH receptor promoter polymorphism on adrenal androgen secretion. Eur J Endocrinol. 2005;153:711–715. doi: 10.1530/eje.1.02015. [DOI] [PubMed] [Google Scholar]
- 23.Slawik M, Reisch N, Zwermann O, et al. Characterization of an adrenocorticotropin (ACTH) receptor promoter polymorphism leading to decreased adrenal responsiveness to ACTH. J Clin Endocrinol Metab. 2004;89:3131–3137. doi: 10.1210/jc.2003-032010. [DOI] [PubMed] [Google Scholar]
- 24.Liu ZL, He B, Fang F, et al. Genetic polymorphisms of MC2R gene associated with responsiveness to adrenocorticotropic hormone therapy in infantile spasms. Chin Med J (Engl) 2008;121:1627–1632. [PubMed] [Google Scholar]
- 25.Lappalainen S, Utriainen P, Kuulasmaa T, et al. ACTH receptor promoter polymorphism associates with severity of premature adrenarche and modulates hypothalamo-pituitary-adrenal axis in children. Pediatr Res. 2008;63:410–414. doi: 10.1203/PDR.0b013e3181659c14. [DOI] [PubMed] [Google Scholar]
- 26.Proudnikov D, Hamon S, Ott J, et al. Association of polymorphisms in the melanocortin receptor type 2 (MC2R, ACTH receptor) gene with heroin addiction. Neurosci Lett. 2008;435:234–239. doi: 10.1016/j.neulet.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forti FL, Dias MH, Armelin HA. ACTH receptor: ectopic expression, activity and signaling. Mol Cell Biochem. 2006;293:147–160. doi: 10.1007/s11010-006-9237-0. [DOI] [PubMed] [Google Scholar]
- 28.Chida D, Nakagawa S, Nagai S, et al. Melanocortin 2 receptor is required for adrenal gland development, steroidogenesis, and neonatal gluconeogenesis. Proc Natl Acad Sci U S A. 2007;104:18205–18210. doi: 10.1073/pnas.0706953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metherell LA, Chan LF, Clark AJ. The genetics of ACTH resistance syndromes. Best Pract Res Clin Endocrinol Metab. 2006;20:547–560. doi: 10.1016/j.beem.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Chung TT, Webb TR, Chan LF, et al. The majority of adrenocorticotropin receptor (melanocortin 2 receptor) mutations found in familial glucocorticoid deficiency type 1 lead to defective trafficking of the receptor to the cell surface. J Clin Endocrinol Metab. 2008;93:4948–4954. doi: 10.1210/jc.2008-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung TT, Chan LF, Metherell LA, et al. Phenotypic characteristics of familial glucocorticoid deficiency (FGD) type 1 and 2. Clin Endocrinol (Oxf) 2010;72:589–594. doi: 10.1111/j.1365-2265.2009.03663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall IP, Blakey JD. Genetic association studies in Thorax. Thorax. 2005;60:357–359. doi: 10.1136/thx.2005.040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding YX, Zou LP, He B, et al. ACTH receptor (MC2R) promoter variants associated with infantile spasms modulate MC2R expression and responsiveness to ACTH. Pharmacogenet Genomics. 2010;20:71–76. doi: 10.1097/FPC.0b013e328333a172. [DOI] [PubMed] [Google Scholar]
- 34.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phuc Le P, Friedman JR, Schug J, et al. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaattela M, Carpen O, Stenman UH, et al. Regulation of ACTH-induced steroidogenesis in human fetal adrenals by rTNF-alpha. Mol Cell Endocrinol. 1990;68:R31–R36. doi: 10.1016/0303-7207(90)90196-f. [DOI] [PubMed] [Google Scholar]
- 37.Jaattela M, Ilvesmaki V, Voutilainen R, et al. Tumor necrosis factor as a potent inhibitor of adrenocorticotropin-induced cortisol production and steroidogenic P450 enzyme gene expression in cultured human fetal adrenal cells. Endocrinology. 1991;128:623–629. doi: 10.1210/endo-128-1-623. [DOI] [PubMed] [Google Scholar]
- 38.Cai TQ, Wong B, Mundt SS, et al. Induction of 11beta-hydroxysteroid dehydrogenase type 1 but not -2 in human aortic smooth muscle cells by inflammatory stimuli. J Steroid Biochem Mol Biol. 2001;77:117–122. doi: 10.1016/s0960-0760(01)00041-3. [DOI] [PubMed] [Google Scholar]
- 39.Tomlinson JW, Moore J, Cooper MS, et al. Regulation of expression of 11beta-hydroxysteroid dehydrogenase type 1 in adipose tissue: tissue-specific induction by cytokines. Endocrinology. 2001;142:1982–1989. doi: 10.1210/endo.142.5.8168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.