Abstract
Data-independent acquisition (DIA) based proteomics has become increasingly complicated in recent years due to the vast number of workflows described, coupled with a lack of studies indicating a rational framework for selecting effective settings to use. To address this issue and provide a resource for the proteomics community, we compared twelve DIA methods that assay tryptic peptides using various mass-isolation windows. Our findings indicate the most sensitive single injection LC-DIA method uses 6 m/z isolation windows to analyze the densely populated tryptic peptide range from 450–730 m/z, which allowed quantification of 4,465 E. coli peptides. In contrast, using the sequential windowed acquisition of all theoretical fragment-ions (SWATH) approach with 26 m/z isolation windows across the entire 400–1200 m/z range, allowed quantification of only 3,309 peptides. This reduced sensitivity with 26 m/z windows is caused by an increase in co-eluting compounds with similar precursor values detected in the same tandem MS spectra, which lowers the signal-to-noise of peptide fragment-ion chromatograms and reduces the amount of low abundance peptides that can be quantified from 410–920 m/z. Above 920 m/z, more peptides were quantified with 26 m/z windows due to substantial peptide 13C isotope distributions that parse peptide ions into separate isolation windows. Since reproducible quantification has been a long-standing aim of quantitative proteomics, and is a so-called trait of DIA, we sought to determine whether precursor-level chromatograms used in some methods rather than their fragment-level counterparts have similar precision. Our data show extracted fragmention chromatograms are the reason DIA provides superior reproducibility.
Keywords: DIA, SWATH, PAcIFIC, Protalizer, label-free quantification
INTRODUCTION
Liquid chromatography interfaced to tandem mass spectrometers (LC-MS/MS) allows targeted and discovery-based identification of proteins, post-translational modifications, and isoform variants.[1] While peptide identification has become routine, mainly through the increased confidence from high-resolution mass spectrometers, more work remains to be done with regard to developing robust discovery-based quantitative proteomic platforms that are reproducible and broadly used in the proteomics community.
Among the MS acquisition methods that have been described, shotgun acquisition[2] and selected reaction monitoring (SRM)[3] have gained wide acceptance. In shotgun mode, a dynamic process is used to detect precursor-ions in real-time, followed by ranking the most intense precursors, and typically fragmenting the 5–50 most abundant ions. The resulting tandem mass spectra allow peptides to be identified in a discovery-based manner by MS/MS database searching programs that match fragment-ions observed from cleavage of the peptide backbone to peptide sequences in reference proteome databases.[4] For protein quantification across samples in shotgun studies, the number of peptide spectra assigned to a protein is used for relative quantitation,[5] or the area under the curve (AUC) from precursor extracted-ion chromatograms (EICs) associated with peptides.[6]
To measure a targeted list of proteins in a highly reproducible manner, SRM[7–8] is the method of choice due to the elimination of various issues encountered in shotgun assays.[9] Typically in SRM studies, a set of up to ca. 100 peptides is measured, either to confirm protein differences identified in shotgun studies or to measure proteins related to a specific hypothesis. In SRM, a predetermined list of fragment-ions for precursors corresponding to peptides of interest are acquired in a continuous cycle to generate multiple data points as each peptide elutes and is detected. Depending on the instrument used, both the precursor and specific fragment ions are entered for each peptide (e.g. triple quadrupole), or in the product-ion mode when full-scan tandem MS spectra are recorded the precursor information is input for acquisition and the related fragmentions are extracted following LC-MS/MS analysis. Despite procedures to increase the number of peptides that can be measured in SRM studies by acquiring peptides at the approximate times they are expected to elute,[10] the approach allows measuring far fewer proteins than shotgun-based methods that are intended to provide an unbiased analysis of samples.
An emerging acquisition approach termed data-independent acquisition (DIA) offers the ability to assay thousands of proteins in a rapid, discovery-oriented manner with the potential for superior quantification accuracy and precision due to the continuous collection of fragment-ion spectra over a preselected precursor range.[11–16] Multiple variations of the DIA approach have been applied; for a review see Tate et al.[17] For instance, in the MSE method,[12] high and low collision energy is used to fragment all precursors simultaneously from 400–1200 m/z. Whereas, in the sequential windowed acquisition of all theoretical fragment-ions (SWATH)[13] procedure, 26 m/z windows are stepped through every ~3 seconds covering the 400–1200 m/z precursor range in a single analysis. Other methods such as precursor acquisition independent from ion count (PAcIFIC)[14] use isolation windows as small as 2.5 m/z that provide ideal sensitivity and specificity, but require many analyses to assay the 400–1400 precursor m/z range (e.g., in each sample injection a 15 m/z precursor range is analyzed). Although these extreme differences in isolation windows sizes have been applied from 2.5 to 800 m/z in the PAcIFIC and MSE methods, respectively, systematic evaluation of this important parameter in various DIA methods and the precursor m/z ranges analyzed have not been reported.
Just as there is little consensus in the best approach to acquire DIA data, there also is a lack of standardization in the approaches used to identify peptides and their surrogate proteins. Using small isolation windows (e.g., 2.5 m/z), such as those applied in the PAcIFIC method, unprocessed MS/MS spectra can be analyzed by MS/MS searching programs initially developed for shotgun proteomics.[14] In another method, spectral libraries are used for identification whereby peptide fragment-ions are extracted at normalized elution times based on shotgun analyses of the samples analyzed.[13] A drawback to this approach is that additional shotgun runs are needed to generate the spectral library, and peptide standards are generally required to accurately normalize retention time coordinates between the spectral library and actual DIA analyses. Alternatively, spectra deconvolution approaches have been applied to separate co-eluting peptides transmitted through the same mass-isolation window followed by using MS/MS database searching programs for peptide and protein identification. To date, tandem spectra have been deconvoluted by the following procedures: a) matching precursors detected with similar elution time that fall within the mass-isolation window of MS/MS spectra generated – thereby reassigning mass-isolation windows with vast numbers of possible precursor m/z values with accurate mass values;[18] b) eliminating fragment-ions in MS/MS spectra that do not have similar chromatographic behavior (e.g., lift-off, touchdown, or maximum elution time);[19] c) matching fragment-ions with similar elution profiles to precursors with similar elution time that fall within the same isolation window;[20–21] d) a two-stage identification procedure where (b) and (c) are applied and the search results from both rounds of identification are combined.[22]
In this study, we assessed sequential windowed precursor acquisition with three isolation window sizes and various precursor ranges in trypsin digests from E. coli on a Sciex 5600+ QTOF to provide an optimized prototype DIA method for maximizing sensitivity in single injection LC-DIA studies. Although our data were generated from analyzing E. coli tryptic peptides, the results presented from various DIA settings are expected to have general applicability to tryptic peptide samples derived from other species as suggested previously by Scherl et al.[23] We also validate a spectral deconvolution approach for the direct identification of peptides without spectral libraries and benchmark DIA to shotgun sensitivity and assay reproducibility.
EXPERIMENTAL
Preparation of E. coli lysates tryptic digests
E. coli strain BL21-DE3 was grown in 1000 ml of Luria broth (LB) overnight and centrifuged at 4000 x g for 10 min to pellet the cells. The supernatant was decanted and the cells were frozen at −80°C. The following day the cell pellet was resuspended in 25 ml of a lysis buffer containing Lysozyme (Sigma L6876 at 0.5 mg/ml), 50 mM Tris HCl, 10 mM DTT, 1 mM EDTA, Roche complete protease inhibitor (11697498001, 1 tablet for 50 ml) and sigma phosphatase inhibitor cocktail II (P5726, 500 μl for 50 ml). The cell suspension was probe sonicated on ice for 5–8 sec and allowed to cool down. This process was repeated for 8 cycles then the sample was sonicated for 30 min in a water bath at RT. The lysate was centrifuged at 15,000 x g for 30 min and the supernatant was removed. Protein quantification was performed using a colorimetric assay based on dye-metal complexation and monitoring absorbance at 660 nm using a Pierce BCA kit. The E. coli lysate was separated into 1 mg aliquots and stored at −80°C. A one milligram aliquot of the lysate was removed from the −80°C freezer and separated into five 200 μg aliquots for running on a 1D SDS Page gel. The sample volume was reduced by speed vac and the volume of the lysate was brought up to 40 μl with 4X Invitrogen LDS buffer and 10X reducing agent (NP0009). The sample was loaded on a 1.5 mm, 4–12% Bis-Tris Invitrogen NuPage gel (NP0335BOX) and electrophoresed in MOPS buffer (NP0001) until the sample ran 1.5 cm into the gel. Molecular weight markers (Thermo spectra 26623) were run between the samples to indicate the protein-containing region of the gel. The gel was fixed in 50% ethanol/10% acetic acid overnight at RT, then washed in 30% ethanol for 10 min followed by two 10 min washes in MilliQ water (MilliQ Gradient system) and finally scanned on an Epson V700 scanner to record an image of the gel. The lanes were cut out of the gel, cut into small (~2mm) squares and were subjected to in-gel tryptic digestion and subsequent recovery of peptides as described previously.[24] The E. coli peptides from each lane were reconstituted at a concentration of 0.2 μg/μl, combined and stored at −80°C.
Nano liquid chromatography coupled to electrospray tandem mass spectrometry (nLC-ESI-MS/MS)
nLC-ESI-MS/MS analyses were performed on a 5600+ QTOF mass spectrometer (Sciex, Toronto, On, Canada) interfaced to an Eksigent (Dublin, CA) nanoLC.ultra nanoflow system. Each acquisition method was analyzed in technical triplicate. An amount corresponding to 500 nanograms of total protein was loaded (via an Eksigent nanoLC.as-2 autosampler) onto an IntegraFrit Trap Column (outer diameter of 360 μm, inner diameter of 100, and 25 μm packed bed) from New Objective, Inc. (Woburn, MA) at 2 μl/min in formic acid/H2O 0.1/99.9 (v/v) for 15 min to desalt and concentrate the samples. For the chromatographic separation of peptides, the trap-column was switched to align with the analytical column, Acclaim PepMap100 (inner diameter of 75 μm, length of 15 cm, C18 particle sizes of 3 μm and pore sizes of 100 Å) from Dionex-Thermo Fisher Scientific (Sunnyvale, CA). The peptides were eluted using a variable mobile phase (MP) gradient from 95% phase A (Formic acid/H2O 0.1/99.9, v/v) to 40% phase B (Formic Acid/Acetonitrile 0.1/99.9, v/v) for 70 min, from 40% phase B to 85% phase B for 5 min and then keeping the same mobile phase composition for 5 additional min at 300 nL/min. The nLC effluent was ionized and sprayed into the mass spectrometer using NANOSpray® III Source (Sciex). Ion source gas 1 (GS1), ion source gas 2 (GS2) and curtain gas (CUR) were respectively kept at 8, 0 and 35 vendor specified arbitrary units. The mass spectrometer method was operated in positive ion mode and the interface heater temperature and ion spray voltage were kept at 150°C, and at 2.6 kV respectively. The data was recorded using Analyst-TF (version 1.7) software.
We used 12 DIA methods to compare various isolation windows and precursor m/z range results. The details of each of these methods are provided below and in Table 1. Method 1) the mass spectrometer method was set to go through 1757 cycles for 99 minutes, where each cycle performed one TOF-MS scan type (0.25 sec accumulation time, from the 410.0 to 690.0 precursor m/z range) followed by 56 sequential DIA windows of 6 Daltons each across the precursor range. Note that the Analyst software automatically added 1 Dalton to each DIA window to provide overlap between adjacent isolation windows, thus an input of 5 Da in the method set up window results in an overlapping 6 Da collection window width (e.g. 410–416 m/z, then 415–421 m/z, followed by 420–426 m/z, etc.). Within the DIA windows a charge state of +2, high sensitivity mode, and rolling collision energy with a collision energy spread (CES) of 15 V was selected. Methods 2–4) these are the same as method 1 except that the TOF-MS scan range was 550 to 830 m/z, 690.0 to 970.0 m/z, and 970.0 to 1250.0 m/z, respectively. Method 5) the mass spectrometer method was set to go through 1358 cycles, where each cycle performed one TOF-MS scan type (0.25 sec accumulation time, from 550.0 to 750.0 precursor m/z) followed by 40 sequential DIA windows of 6 Daltons each. Method 6) the mass spectrometer method was set to go through 1523 cycles, where each cycle performed one TOF-MS scan type (0.25 sec accumulation time, from 410.0 to 550.0 precursor m/z range) followed by 70 sequential DIA windows of 3 Daltons each. Methods 7–11) these are the same as method 6 except that the TOF-MS scan range and 3 Dalton DIA isolation windows acquired data from 550.0 to 690.0 m/z, 690.0 to 830.0 m/z, 830.0 to 970.0 m/z, 970.0 to 1110.0 m/z, and 1110.0 to 1250.0 m/z, respectively. Method 12) the mass spectrometer method was set to go through 1742 cycles, where each cycle performed one TOF-MS scan type (0.10 sec accumulation time, across the 400.0 to 1200.0 precursor m/z range) followed by 32 sequential DIA windows of 26 Daltons each.
Table 1.
Multiple DIA method precursor range and mass-isolation window sizes evaluated.
| DIA method | Precursor range (m/z) | Number of sequential DIA windows | Isolation width (m/z) | Duty cycle (sec) |
|---|---|---|---|---|
| 1 | 410–690 | 56 | 6 | 3.1 |
| 2 | 550–830 | 56 | 6 | 3.1 |
| 3 | 690–970 | 56 | 6 | 3.1 |
| 4 | 970–1250 | 56 | 6 | 3.1 |
| 5 | 550–750 | 40 | 6 | 4.3 |
| 6 | 410–550 | 70 | 3 | 3.9 |
| 7 | 550–690 | 70 | 3 | 3.9 |
| 8 | 690–830 | 70 | 3 | 3.9 |
| 9 | 830–970 | 70 | 3 | 3.9 |
| 10 | 970–1110 | 70 | 3 | 3.9 |
| 11 | 1110–1250 | 70 | 3 | 3.9 |
| 12 | 400–1200 | 32 | 26 | 3.4 |
In shotgun mode the mass spectrometer was set to perform one TOF-MS scan type (0.25 sec accumulation time, in a 350 to 1600 m/z window) followed by 50 information dependent acquisition (IDA)-mode MS/MS-scans on the most intense candidate ions having a minimum intensity of 150 counts. Each MS/MS scan was operated under vender specified high-sensitivity mode with an accumulation time of 0.05 secs and a mass tolerance of 100 ppm. Precursor ions selected for MS/MS scans were excluded for 30 secs to reduce the occurrence of redundant peptide sequencing.
DIA data analysis parameters
Protalizer DIA software (Vulcan Analytical, Birmingham, AL) was used to analyze every DIA file with settings previously described for Sciex 5600 QTOFs.[22] The Swiss-Prot E. coli database downloaded March 17th 2015 was used as the reference proteome for all MS/MS searches. A precursor and fragment-ion tolerance for QTOF instrumentation was used for the Protalizer spectral-library free identification algorithm.[22] A multistage spectra deconvolution approach was applied as previously described, except only fragment-ions where the intensity was ≥ 70% of the scan containing the most intense value were retained to associate fragment-ions that are more likely derived from the same peptide based on the chromatographic elution profile. The ≥ 70% fragmention deconvolution setting was used because this allowed the most proteins to be quantified in three different DIA methods evaluated in Supplemental Figure 1. Potential modifications included in the searches were phosphorylation at S, T, and Y residues, N-terminal acetylation, N-terminal loss of ammonia at C residues, and pyroglutamic acid at N-terminal E and Q residues. Carbamidomethylation of C residues was searched as a fixed modification. The maximum valid protein and peptide expectation score from the X! Tandem search engine used for peptide and protein identification on reconstructed spectra was set to 0.005.
For DIA quantification by Protalizer the maximum number of b and y series fragment-ion transitions were set to nine excluding those with m/z values below 300 and not containing at least 10% of the relative intensity of the strongest fragment-ion assigned to a peptide. A minimum of five fragment-ions were required for a peptide to be quantified (except where indicated otherwise in Figure 7). In datasets where a minimum of seven consistent fragment-ions were not detected for the same peptide ion in each of the three files compared in a triplicate analysis, the algorithm identified the file with the largest sum fragment-ion AUC and extracted up to seven of these in the other files using normalized retention time coordinates based on peptides detected by the Protalizer algorithm in all the files in a dataset.
Figure 7.
Shotgun identification and quantification via MS1 EICs
An unpublished version of the Protalizer tool for shotgun data analysis was applied in this study. Peptide and protein identifications were made using the X! Tandem Sledgehammer search engine (version 2013.09.01.1) against the forward and reverse E. coli Swiss-Prot database used in the DIA analyses with a 50 ppm precursor and fragment-ion mass tolerance. Potential and fixed modifications searched were the same as those described for the DIA data analysis. A maximum of two trypsin miscleavages were included in the analysis and protein and peptide maximum valid expectation scores were set to 0.005. Only ‘top hit’ peptides were further analyzed by the pipeline. Redundant peptide identifications with the same charge and modification state were eliminated for downstream analysis in each file except for one scan corresponding to the largest sum fragmention intensity according to X! Tandem.
MS1 peaks corresponding to peptides identified by the MS/MS search were extracted with the OpenMS feature finder centroid tool (version 1.11.1).[25] Default settings were used with the following exceptions: a mass trace m/z tolerance of 0.05, mass trace min spectra of 4, isotopic pattern charge low 2 and high 3, isotopic pattern m/z tolerance of 0.05, seed min score of 0.5, feature overall min score of 0.5, feature min isotope fit of 0.5, and feature min trace score of 0.5. Peptide ions identified were matched to MS survey scan peaks using a tolerance of +/− 0.025 m/z, identical charge, and retention time within +/− 15 seconds from the MS/MS scan the peptide was sequenced.
A similar procedure for data normalization with endogenous reference peptides was applied as previously described for the DIA tool.[22] However, instead of using fragment-ion AUC sum values corresponding to peptides with the most consistent abundance and similar elution time as normalization factors for each peptide quantified, MS1 EIC peptide intensity values were applied.
Retention time alignment was applied to enable feature detection of peptides not identified by MS/MS in every sample. Peptide elution times were normalized in each file based on the difference in elution time for each sample peptide to reference peptide(s) in files a peptide was identified by X! Tandem and matched to an MS1 EIC. This elution time difference was then used to predict the retention time range in which precursor-ions were extracted in files where a peptide was not initially identified. A retention time extraction window of +/− 15 seconds was used during this step to match precursor EICs across all files. Peptide and protein relative abundance were calculated as described previously for the DIA platform.[22]
Availability of raw data and software result files
All .wiff raw files and results generated by the Protalizer data processing software were deposited in the ProteomeXchange Consortium[26] via the PRIDE partner repository[27] with the dataset identifier PXD002688.
RESULTS AND DISCUSSION
Overview of DIA and shotgun proteomics
In DIA methods where sequential precursor m/z ranges are acquired,[13] the mass spectrometer is set to scan MS/MS fragment-ion spectra from multiple isolation windows that sequentially step through a precursor mass range in a duty cycle of several seconds (Figure 1A). In contrast, the shotgun mode collects data by determining precursors with the strongest intensities and charge states typical of peptides (2–3+) followed by fragmenting these precursors with small mass-isolation windows (Figure 1B). In the remaining sections below we characterize various settings including the size of mass-isolation windows and the precursor ranges analyzed that have important implications in generating DIA results and benchmark identification and quantification metrics to shotgun results.
Figure 1.
Evaluation of 3, 6, and 26 m/z mass-isolation windows
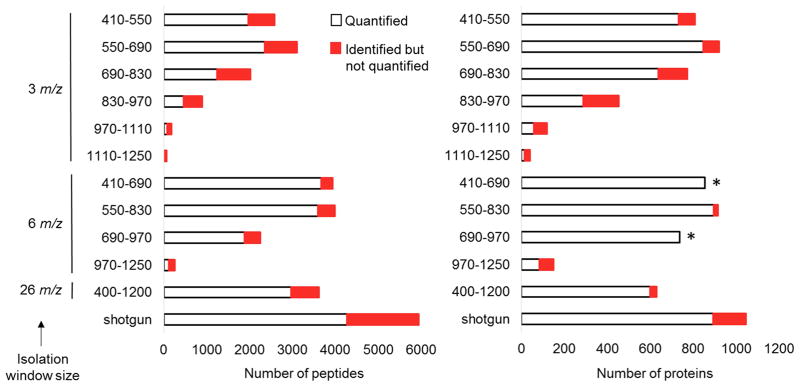
We designed multiple DIA methods shown in Table 1 to assess mass-isolation window size in a comprehensive manner across the entire tryptic peptide precursor m/z range. Each approach used an isolation window of 3, 6, or 26 m/z and had a duty cycle ranging from 3.4 to 4.3 seconds to provide well-defined peak shapes amenable for accurate quantification via fragment EICs. With this duty cycle, each 3 m/z mass-isolation window approach measured a 140 precursor m/z range and required 6 injections to analyze the mass range from 410–1250 m/z. Whereas, the 6 m/z isolation width methods analyzed a 280 m/z range and required 3 injections to assay the 410–1250 m/z precursor range. These 3 and 6 m/z isolation window approaches were benchmarked to 26 m/z SWATH windows initially applied by Gillet et al. that analyze an 800 m/z range in a single LC-DIA analysis.[13] All of the data collected were analyzed using Protalizer, a fully automated software platform that performs spectra deconvolution, MS/MS database searching for peptide identification, as well as label-free quantification of peptides and proteins across samples with fragment EICs.[22] Figure 2 shows the number of proteins and peptides identified and quantified with each of the DIA methods compared to the results from a shotgun analysis with the same LC-MS/MS system and tryptic digest. Although more proteins and peptides were identified by shotgun acquisition than any of the DIA methods tested (1,046 proteins versus 922), the number of quantifiable proteins using 6 m/z mass-isolation windows from 550–830 precursor m/z allowed the most proteins to be assayed in a single injection (897 compared to 894 proteins via shotgun). The lowered rate of converting identified peptides into those capable of being quantified with shotgun acquisition has been previously reported.[25] In this dataset, 62.7% of the shotgun sequenced peptides were matched to MS1 EICs. In contrast, 94.2% of peptides detected by the MS/MS database searches were able to be matched to a minimum of five fragment-ions in the most sensitive DIA method using 6 m/z isolation windows to analyze the 550–830 precursor m/z range.
Figure 2.
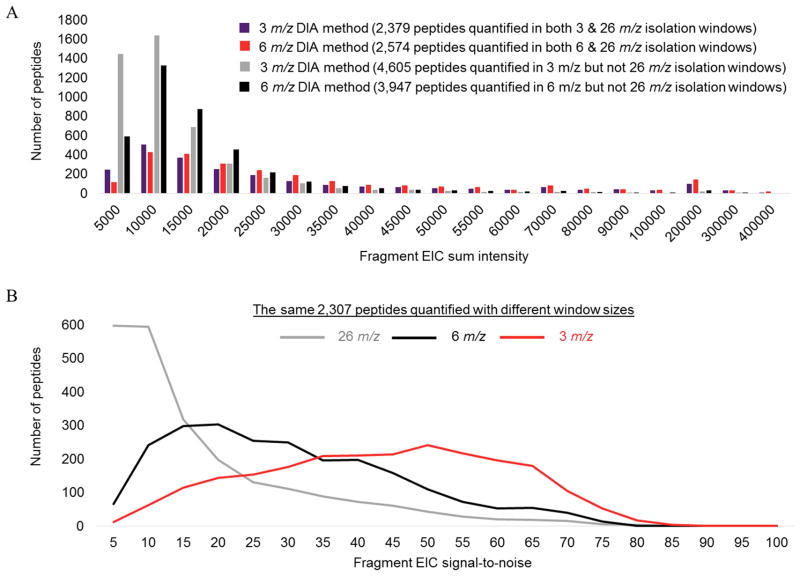
A noteworthy finding in our results is 26 m/z mass-isolation windows that analyze the largest precursor range in a single injection from 400–1200 m/z, identified only 630 proteins and quantified 600, whereas the DIA assay from 550–830 precursor m/z using 6 m/z windows both identified and quantified ~50% more proteins. We speculated the 26 m/z isolation window method effectively captures less useful information than methods that analyze only the very densely populated tryptic peptide range with smaller isolation windows because more specific isolation of peptides allows detection and quantification of lower abundance peptides. To determine if peptides detected with narrower isolation windows, but not with 26 m/z windows generally had smaller intensities, we compared the sum fragment EIC intensities of peptides that were not detected with the 26 m/z isolation window method but were with the two other approaches in Figure 3A. This data shows the peptides not detected with 26 m/z windows generally had small intensities. We then speculated that the reduced sensitivity observed with 26 m/z isolation windows may be due to an increase in co-eluting peptides in the same mass-isolation window causing a decrease in the signal-to-noise of fragment-ions and detection of peptides with small intensities. To test this, we determined the pool of peptides detected with each of the 3, 6, and 26 m/z isolation window DIA methods. A total of 2,307 of the same peptides detected with all three isolation windows and signal-to-noise histograms are shown in Figure 3B. Among the peptides detected with each method, the 3 m/z isolation windows had an average signal-to-noise of 42, the 6 m/z wide windows had a 28 average signal-to-noise, and the 26 m/z isolation windows had an average signal-to-noise of 16. Thus, indicating an inverse relationship between isolation window size and signal-to noise in DIA assays of complex peptide mixtures. We also evaluated another possible explanation for reduced sensitivity with 26 m/z windows - the detection of peptides near the upper and lower precursor m/z regions of each isolation window are not ideal due to quadrupole transmission imperfections or artifacts from the MS/MS database search. To investigate this, we compared the distribution of identified peptide precursor m/z values within two separate 26 m/z isolation windows to see if these were skewed to the center of windows, indicating a non-random distribution of peptide precursor values within each window. The 549–575 and 574–600 precursor m/z isolation windows are shown in Supplemental Figure 2 and indicated peptides were identified with a near random distribution, suggesting quadrupole transmission and the MS/MS database search were not factors causing the sensitivity reduction with 26 m/z isolation windows. In conclusion, our data indicate the decreased sensitivity with 26 m/z isolation windows is due to reduced signal-to-noise from co-eluting compounds that inhibit the detection of peptides with small intensities. A similar observation shown in Figure 3A comparing 3 and 6 m/z isolation windows was also apparent – 3 m/z isolation windows allows quantification of more low intensity peptides and has increased signal-to-noise across the same 2,307 peptides as shown in Figure 3B.
Figure 3.
To test if 50 ms dwell times had a negative impact on sensitivity, we included a longer dwell time method of 100 ms that analyzed peptides with precursor m/z values from 550–750. From comparing the number of peptides identified with this method, to a similar method acquiring peptides from 550–830 precursor m/z that also used 6 m/z isolation windows but with a 50 ms dwell time, each method had nearly an identical number of peptide identifications in the 550–750 precursor m/z range (data not shown). Thus, indicating the 50 ms dwell time used in this study did not compromise sensitivity with the 5600+ QTOF instrument.
Combining 3 and 6 m/z isolation window results across the 410–1250 precursor m/z range
For studies that have extensive access to instrumentation and sufficient starting material to permit multiple injections per sample analyzed, we determined the number of proteins and peptides that can be identified in any one of three technical replicates and quantified in the 410–1250 precursor m/z range in multiple injections. Using 3 m/z isolation windows in six separate injections allowed identification of 1,360 proteins and quantification of 1,133 proteins from the detection of 14,277 peptides and quantification of 7,004 peptides. In contrast, with 6 m/z isolation windows in three separate injections, we identified 1,049 proteins and quantified 1,017 proteins, whereas 9,277 peptides were identified and 6,537 were able to be quantified.
Spectral library-free DIA peptide identification by spectra deconvolution
Due to using larger mass-isolation windows in DIA mode compared to shotgun and SRM acquisition, highly complex MS/MS spectra are generated that contain fragment-ions from multiple precursors. Thus, making identification with MS/MS database search engines designed to match each fragment-ion spectrum to a single peptide-ion nontrivial. In order to reduce this issue in DIA studies, spectra deconvolution approaches are applied to simplify MS/MS spectra based on the principle that fragment-ions originating from the same precursor should have the same elution profile.[19–22] Additionally, the large precursor m/z range for each mass-isolation window in many DIA methods results in a vast number of potential peptide-ion matches in eukaryotic and prokaryotic proteomes that fall within the isolation window. In order to narrow the precursor m/z search space, several deconvolution methods allow replacing the large uncertainty in the precursor isolation window with a detected precursor that has similar elution characteristics to fragment-ions in the same isolation window.[20–22]
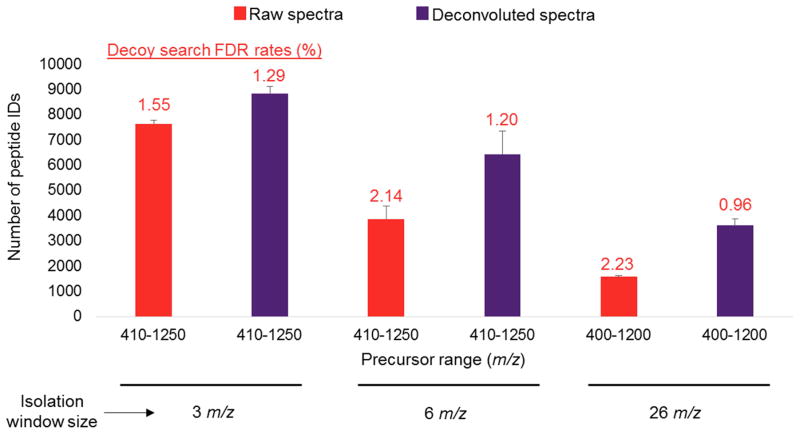
To assess the effect of DIA spectra deconvolution, we compared the number of peptide identifications using the Protalizer deconvolution approach[22] to unprocessed raw spectra. Figure 4 shows with 3 m/z isolation windows, an average of 16% more peptides were able to be identified in demultiplexed spectra from combining data from six precursor m/z ranges from 410–1250. Whereas deconvolution of data with 6 m/z isolation windows resulted in an average of 67% more peptides detected than searching raw spectra, and the difference was even more drastic for 26 m/z isolation windows where an increase of 129% was observed for deconvoluted versus raw DIA spectra. Supplemental Figure 3 shows the average number of peptide and protein identification with spectra deconvolution and searching raw spectra from each of the twelve DIA methods evaluated in the study and indicates spectra deconvolution increases the average number of proteins detected by 23%, 33%, and 45%, respectively, for the 3, 6, and 26 m/z isolation window methods. Taken together, our data indicate deconvoluting spectra is critical for maximizing peptide and protein detection sensitivity, especially for methods using large mass-isolation windows where there is a greater likelihood of multiple precursors eluting at the same time through an isolation window. In addition to increasing sensitivity, spectral deconvolution reduced false discovery rates (FDRs) in the 6 m/z mass-isolation window methods by an average of 44% and lowered the FDRs in the 26 m/z window method by 57%. Moreover, in the six separate precursor ranges analyzed with 3 m/z isolation windows, the overall FDR average was decreased by an average of 34%.
Figure 4.
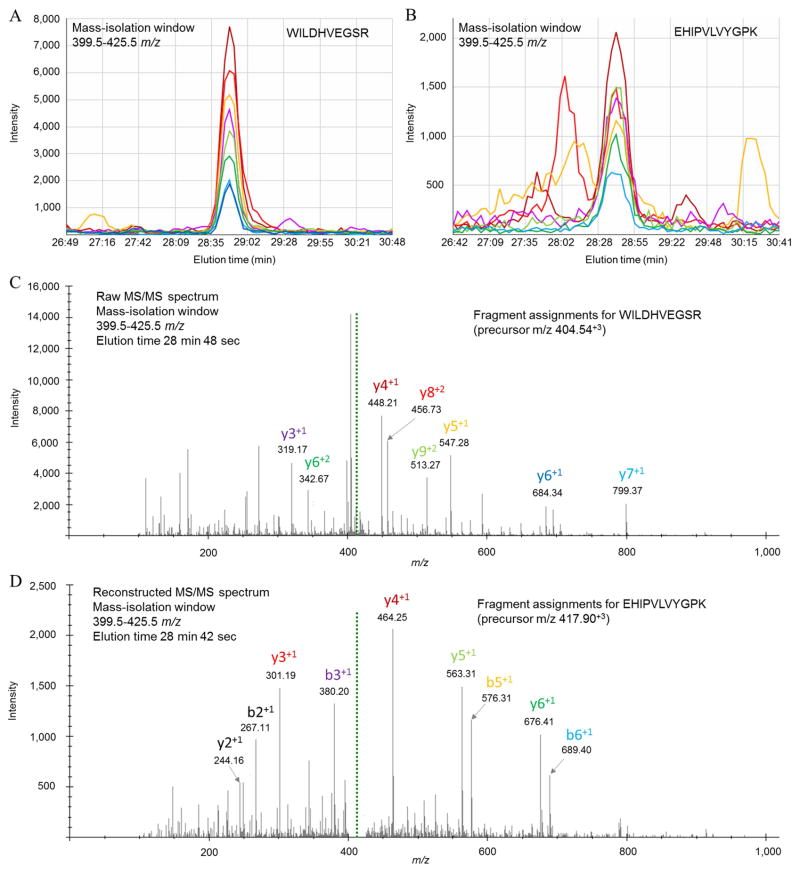
A representative example of two peptides that co-elute in the same 26 m/z mass-isolation window are presented in Figure 5. Fragment EICs of WILDHVEGSR and EHIPVLVYGPK are shown in Figure 5A–B, where the former peptide has a ~4-fold stronger intensity than the latter peptide with overlapping elution time. As a result, the EHIPVLVYGPK peptide is only able to be detected by an MS/MS database search engine with deconvolution of the raw tandem spectra, whereas the stronger WILDHVEGSR peptide is able to be identified with or without spectra deconvolution. The dominating signal strengths of WILDHVEGSR b and y fragment-ions are apparent in the raw MS/MS spectrum in Figure 5C that allow the spectrum to be matched specifically to the peptide sequence from the MS/MS database search. In contrast, fragment-ions belonging to the lower abundance EHIPVLVYGPK peptide are not readily apparent in the raw spectra shown in Figure 5C, but are in the deconvoluted spectra shown in Figure 5D where EHIPVLVYGPK is able to be confidently assigned to the reconstructed spectra.
Figure 5.
DIA quantification reproducibility
Although identifying peptides and proteins is a critical part of any DIA study, detecting meaningful differences across biological conditions compared requires reliable quantification.. Thus, we assessed whether peptides and proteins detectable by shotgun acquisition and quantified by MS1 EICs have similar quantitative precision to the most sensitive 6 m/z mass-isolation window DIA assay tested in this study from 550–830 precursor m/z. Our results comparing peptides in technical replicates shown in Figure 6A indicate DIA yielded more reproducible measurements than the shotgun MS1-intensity based peptide quantification. Specifically, the MS1 intensity based shotgun results had 465 of 4,092 total peptides quantified (11.3%) with a ≥ 30% fold-change difference across two technical replicates. In contrast, the DIA method applying 6 m/z isolation windows had 200 of 3,522 total peptides quantified (5.6%) with a ≥ 30% fold-change difference across two technical replicates – approximately half the number of peptides with poor reproducibility using a 30% fold-change cutoff. Moreover, histograms of the coefficient of variation at the protein level in Figure 6B indicate the DIA method has a 4-fold reduction in peptides quantified with a ≥ 20% or greater coefficient of variation (28 versus 119 proteins).
Figure 6.
Because our data indicate DIA fragment EIC quantification is more reproducible than shotgun-based MS1 quantification, and recent studies show SWATH fragment EICs have improved quantification in complex peptide digests,[21,28] we determined whether fragment EIC measurements in our dataset are inherently more precise than MS1 EICs. Figure 6C shows across 2,091 of the same peptide ions that could be matched to MS1 and fragment EICs, the variation in replicates quantified by MS1 EICs had more differences than fragment EICs. In addition, histograms of the coefficient of variation for precursor and fragment peptide EIC quantification is shown in Figure 6D, which indicated the fragment EIC approach had a 6.8% average coefficient of variation and the precursor EIC method had an 11.9% average coefficient of variation. Thus, not only does our data support the conclusion DIA is more reproducible than shotgun-MS1 EIC measurements, we also demonstrated that peptide quantification by fragment EICs is the reason for this improvement.
Impact of the number of fragment-ions and baseline subtraction on reproducibility
Limits on the number of fragment-ions used for deriving relative peptide quantification values across samples can be applied that emphasize sensitivity, measurement quality, or a balance between these metrics. To provide an example of this, Figure 7 shows technical replicates of peptides measured with the 550–830 precursor m/z DIA assay using 6 m/z mass-isolation windows with three fragment-ions compared to the same data analyzed with a minimum of five fragmentions. Also included in these comparisons were various amounts of baseline subtraction, although the amount of baseline noise that needs to be subtracted varies across different instrument platforms due to differences in the relative intensity scales and whether or not the on-board instrument software has already applied a baseline filter. Our data indicate using a moderate baseline subtraction to eliminate low signal-to-noise fragment-ions that balances the number of peptides that can be quantified versus assay reproducibility allows 3,522 peptides to be quantified with a minimum of five fragment-ions per peptide. In contrast, 3,919 peptides requiring a minimum of three fragment-ions detected were able to be quantified. However, the number of peptides quantified with a coefficient of variation above 20% were 167 when a minimum of five fragment-ions were used for peptide quantification and a moderate baseline subtraction compared to 281 peptides with three fragment-ions. Thus, our results indicate these data processing parameters should be carefully evaluated in DIA assays and more conservative settings that require a minimum of five fragment-ions for peptide quantification combined with a substantial baseline noise subtraction have the most favorable reproducibility.
Prototype for the most sensitive single-injection DIA method
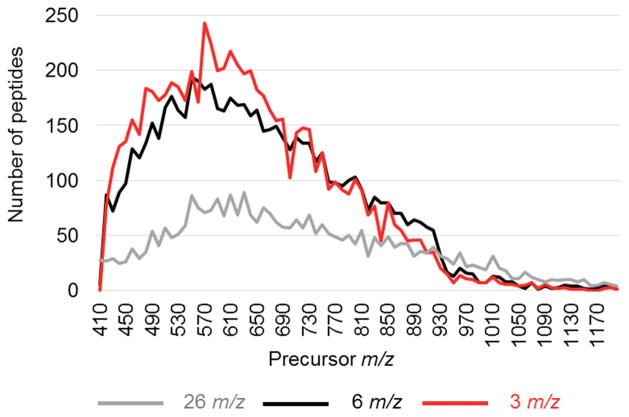
A comprehensive analysis of tryptic peptides across the entire mass range shown in Figure 8 indicates the most information-rich precursor region in tryptic peptide digests is from approximately 450–730 m/z. Of the total 6,537 peptides quantified by fragment EICs in a minimum of two out of three replicates with 6 m/z isolation windows ranging from 410–1250 precursor m/z, 4,465 (68%) were in the 450–730 precursor m/z range. Although this exact 280 m/z range was not analyzed in this study, the 6 m/z isolation windows used to analyze the 410–690 and 690–830 precursor m/z ranges shown in Table 1 provide proof-of-principle for this optimized assay for future studies.
Figure 8.
The data shown in Figure 8 also indicates the less-specific, 26 m/z mass-isolation windows actually have enhanced sensitivity relative to the more narrow 3 and 6 m/z isolation window methods above 920 m/z. This finding suggests the decrease in precursor-ion density in this range,[23] coupled with the substantial carbon isotope envelopes in these large peptides, allows 26 m/z windows to quantify more peptides because the 3 and 6 m/z isolation widths disperse these large peptides into multiple isolation windows causing a reduction in fragment-ion intensity.
CONCLUSIONS AND OUTLOOK
We have described an improved single injection LC-DIA method that differs from the initial SWATH approach [13] by targeting the most dense peptide precursor m/z range in complex tryptic peptide digests in order to obtain the maximum amount of information possible from a single injection DIA approach. Our findings show that applying 6 m/z isolation windows to assay the 450–730 precursor m/z range, rather than 26 m/z windows across the 400–1200 precursor m/z range, increases the signal-to-noise of peptide fragment EICs by an average of 37% and number of peptides quantified by 35%. Although we emphasized the most effective isolation windows and mass ranges for the most sensitive single injection LC-DIA analysis in order to minimize the amount of sample and instrumentation required for practical reasons, our data also suggests a rational basis for DIA experimental designs when samples can be reinjected multiple times. Specifically, our data showed a substantial signal-to-noise difference between 3, 6, and 26 m/z window methods indicating an inverse relationship between isolation window size and signal-to-noise consistent with the premise underlying the high specificity and quantitative accuracy of targeted SRM.[7–8] An interesting exception to the signal-to-noise and window size relationship was observed for the small population of peptides with precursor values greater than 920 m/z. These large peptides have substantial 13C isotope distributions causing them to have a diminished signal that is distributed over as much as a 3 m/z range for peptides in a 2+ charge state. Thus, causing the peptide to fall into multiple DIA isolation windows when using only 3 and 6 m/z isolation widths. Not only does this suggest DIA assays that analyze peptides with precursor m/z values greater than 920 should use isolation windows greater than 6 m/z to maximize sensitivity, it also indicates targeted SRM assays would benefit by considering this effect to reduce false negative peptide detection rates.
Because multiple DIA methods have been described that apply MS1 EICs for relative peptide quantification,[12,15–16] we sought to ascertain if these DIA approaches could be expected to have better quantitative performance than shotgun-based proteomics. Comparisons of the peptide quantification reproducibility across technical replicates in the same DIA analyses and group of 2,091 peptides indicated the fragment-based EIC approach had approximately half the amount of variation compared to MS1 EIC quantification, suggesting the improved precision of DIA is a product of fragment EICs. This has implications for DIA approaches that apply MS1 EIC-based quantification, since our data suggests DIA MS1 EICs have similar quantitative reproducibility to findings obtained with MS1 EICs in shotgun analyses. Furthermore, this finding is also relevant for recently described DIA methods that utilize varying isolation window widths based on precursor-ion densities that are not held constant across specific precursor m/z ranges and samples analyzed - because such assays rely on MS1 EICs for relative peptide quantification.[29] While the quantitative comparison of MS1 EICs compared to fragment EICs and QTOF instrument parameters in this study provides a framework for implementing DIA, future studies on other instruments such as quadrupole-Orbitraps are needed to ensure the findings are relevant across different mass spectrometers.
Altogether, our findings indicate the LC-DIA methods described offer better reproducibility than shotgun based proteomics and allow more peptides and proteins to be quantified with the same instrument and LC conditions. This practical resource for DIA assay settings described in this study, combined with the recent availability of DIA software,[21–22] should facilitate access to DIA workflows for proteomics laboratories seeking more reliable quantitative findings in future proteomic studies.
Supplementary Material
Acknowledgments
Funding to support the Sciex 5600+ mass spectrometry system was acquired through the National Center for Research Resources S10 Program (RR027015) for shared instrumentation to K.D.G. Research support for R.E.M. was from the National Institutes of Health R01 (MH094445) and the Lindsay Brinkmeyer Schizophrenia Research Fund. Additional funding for both R.E.M. and A.J.F. were from the L.I.F.E. Foundation Award and a U.N.C.I. pilot award.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Stahl DC, Swiderek KM, Davis MT, Lee TD. Data-controlled automation of liquid chromatography/tandem mass spectrometry analysis of peptide mixtures. J Am Soc Mass Spectrom. 1996;7:532. doi: 10.1016/1044-0305(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 3.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 5.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74:4741. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 7.Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol Cell Proteomics. 2005;4:1134. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9:761. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, Rinner O. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat Methods. 2004;1:39. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 12.Plumb R, Johnson K, Rainville P, Smith B, Wilson I, Castro-Perez J, Nicholson J. UPLC MSE; a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun Mass Spectrom. 2006;20:1989. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 13.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:1. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panchaud A, Scherl A, Shaffer SA, von Haller D, Kulasekara D, Miller S, Goodlett D. PAcIFIC how to dig deeper into the proteomics ocean. Anal Chem. 2009;81:6481. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger T, Cox J, Mann M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol and Cell Proteomics. 2010;9:2252. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. Drift time- specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014;11:167. doi: 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- 17.Tate S, Larsen B, Bonner R, Gingras AC. Label-free quantitative proteomics trends for protein-protein interactions. J Proteomics. 2013;81:91. doi: 10.1016/j.jprot.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Carvalho PC, Han X, Xu T, Cociorva D, da Carvalho MG, Barbosa VC, Yates JR. XDIA: improving on the label-free data-independent analysis. Bioinformatics. 2010;26:847. doi: 10.1093/bioinformatics/btq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bern M, Finney G, Hoopmann M, Merrihew G, Toth M, MacCoss MJ. Deconvolution of mixture spectra from ion-trap data-independent- acquisition tandem mass spectrometry. Anal Chem. 2010;82:833. doi: 10.1021/ac901801b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong J, Schwahn A, Downard K. ETISEQ – an algorithm for automated elution time ion sequencing of concurrently fragmented peptides for mass spectrometry-based proteomics. BMC Bioinformatics. 2009;10:244. doi: 10.1186/1471-2105-10-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsou CC, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras AC, Nesvizhskii AI. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heaven MR, Cobbs AL, Gunawardena HP, Funk AJ, Pacheco NL, Olsen ML, Ford MJ, Shaffer SA, Norris JL. A data-Independent acquisition tool for peptide identification, label-free quantification, reporting of differentially regulated proteins, and archiving chromatograms. Submitted for publication in. Mol Cell Proteomics. 2015 [Google Scholar]
- 23.Scherl A, Shaffer SA, Taylor GK, Kulasekara HD, Miller SI, Goodlett D. Genome-specific gas-phase fractionation strategy for improved shotgun proteomic profiling of proteotypic peptides. Anal Chem. 2008;80:1182. doi: 10.1021/ac701680f. [DOI] [PubMed] [Google Scholar]
- 24.Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, Edwards MJ, Greis KD, Shertzer HG, Fisher AB, Lentsch AB. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisser H, Nahnsen S, Grossmann J, Nilse L, Quandt A, Brauer H, Sturm M, Kenar E, Kohlbacher O, Aebersold R, Malmström L. An automated pipeline for high-throughput label-free quantitative proteomics. J Proteome Res. 2013;12:1628. doi: 10.1021/pr300992u. [DOI] [PubMed] [Google Scholar]
- 26.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkey RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vizcaíno JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Perez-Riverol Y, Reisinger F, Rios D, Wang R, Hermjakob H. The proteomics identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rardin MJ, Schilling B, Cheng LY, MacLean BX, Sorensen DJ, Sahu AK, MacCoss MJ, Vitek O, Gibson BW. MS1 peptide ion intensity chromatograms in MS2 (SWATH) data independent acquisitions. Improving post acquisition analysis of proteomic experiments. Mol Cell Proteomics. 2015;14:2405. doi: 10.1074/mcp.O115.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruderer R, Bernhardt OM, Gandhi T, Miladinović SM, Cheng LY, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C, Vitek O, Rinner O, Reiter L. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14:1400. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.