Version Changes
Revised. Amendments from Version 1
In version 2 of the manuscript, we have reworded and improved the clarity of the text in several places, to better convey the differences between the contrasted approaches and clarify the questions that are addressed by each analysis. We have also expanded the supplementary material with additional analyses of accuracy of abundance estimates among paralogous genes, and added more detailed descriptions of aspects such as the calculation of each abundance measure and the generation of the incomplete annotation files, information that was previously only available in the respective Datasets. The Discussion section has been extended, mainly to include a discussion of differential expression analysis methods incorporating variance estimates.
Abstract
High-throughput sequencing of cDNA (RNA-seq) is used extensively to characterize the transcriptome of cells. Many transcriptomic studies aim at comparing either abundance levels or the transcriptome composition between given conditions, and as a first step, the sequencing reads must be used as the basis for abundance quantification of transcriptomic features of interest, such as genes or transcripts. Various quantification approaches have been proposed, ranging from simple counting of reads that overlap given genomic regions to more complex estimation of underlying transcript abundances. In this paper, we show that gene-level abundance estimates and statistical inference offer advantages over transcript-level analyses, in terms of performance and interpretability. We also illustrate that the presence of differential isoform usage can lead to inflated false discovery rates in differential gene expression analyses on simple count matrices but that this can be addressed by incorporating offsets derived from transcript-level abundance estimates. We also show that the problem is relatively minor in several real data sets. Finally, we provide an R package ( tximport) to help users integrate transcript-level abundance estimates from common quantification pipelines into count-based statistical inference engines.
Keywords: RNA-seq, quantification, gene expression, transcriptomics
Introduction
Quantification and comparison of isoform- or gene-level expression based on high throughput sequencing reads from cDNA (RNA-seq) are arguably among the most common tasks in modern computational molecular biology. Currently, one of the most widely used approaches amounts to defining the genomic locations of a set of non-overlapping targets (typically, genes) and using the number of aligned reads overlapping a target as a measure of its abundance, or expression level. Several software packages have been developed for performing such “simple” counting (e.g., featureCounts 1 and HTSeq-count 2). More recently, the field has seen a surge in methods aimed at quantifying the abundances of individual transcripts (e.g., Cufflinks 3, RSEM 4, BitSeq 5, kallisto 6 and Salmon 7). These methods provide higher resolution than simple counting, and by circumventing the computationally costly read alignment step, some (notably, kallisto and Salmon) are also considerably faster. However, isoform quantification is more complex than the simple counting, due to the high degree of overlap among transcripts. Currently, there is no consensus regarding the optimal resolution or method for quantification and downstream analysis of transcriptomic output.
Another point of debate is the unit in which abundances are given. The traditional R/FPKM 8, 9 (reads/fragments per kilobase per million reads) have been largely superseded by the TPM 10 (transcripts per million), since the latter is more consistent across libraries. Regardless, all these units attempt to “correct for” sequencing depth and feature length and thus do not reflect the influence of these on quantification uncertainty. In order to account for these aspects, most statistical tools for analysis of RNA-seq data operate instead on the count scale. Most of these tools were designed to be applied to simple read counts, and the degree to which their performance is affected by using fractional estimated counts resulting from portioning reads aligning to multiple transcripts is still an open question. The fact that the most common sequencing protocols provide reads that are much shorter than the average transcript implies that the observed read counts depend on a transcript’s length as well as its abundance; thus, simple counts are arguably less accurate measures than TPMs of the true abundance of RNA molecules from given genes. The use of gene counts as input to statistical tools typically assumes that the length of the expressed part of a gene does not change across samples and thus its impact can be ignored for differential analysis.
In the analysis of transcriptomic data, as for any other application, it is of utmost importance that the question of interest is precisely defined before a computational approach is selected. Often, the interest lies in comparing the transcriptional output between different conditions, and most RNA-seq studies can be classified as either: 1) differential gene expression (DGE) studies, where the overall transcriptional output of each gene is compared between conditions; 2) differential transcript/exon usage (DTU/DEU) studies, where the composition of a gene’s isoform abundance spectrum is compared between conditions, or 3) differential transcript expression (DTE) studies, where the interest lies in whether individual transcripts show differential expression between conditions. DTE analysis results can be represented on the individual transcript level, or aggregated to the gene level, e.g., by evaluating whether at least one of the isoforms shows evidence of differential abundance.
In this report, we make and give evidence for three claims: 1) gene-level estimation is considerably more accurate than transcript-level; 2) regardless of the level at which abundance estimation is done, inferences at the gene level are appealing in terms of robustness, statistical performance and interpretation; 3) taking advantage of transcript-level abundance estimates when defining or analyzing gene-level abundances leads to improved DGE results compared to simple counting for genes exhibiting DTU. The magnitude of the effect in a given data set thus depends on the extent of DTU, and the global impact is relatively small in several real data sets analyzed in this study.
To facilitate a broad range of analysis choices, depending on the biological question of interest, we provide an R/Bioconductor package, tximport, to import transcript lengths and abundance estimates from several popular quantification packages and export (estimated) count matrices and, optionally, average transcript length correction terms (i.e., offsets) that can be used as inputs to common statistical engines, such as DESeq2 11, edgeR 12 and limma 13.
Data and methods
Throughout this manuscript, we utilize two simulated data sets and four experimental data sets (Bottomly 14 [ Data set 3], GSE64570 15 [ Data set 4], GSE69244 16 [ Data set 5], GSE72165 17 [ Data set 6], see Supplementary File 1 for further details) for illustration. Details on the data generation and full records of the analyses are provided in the data sets and Supplementary File 1. The first simulated data set (sim1; Data set 1) is the synthetic human data set from Soneson et al. 18, comprising 20,410 genes and 145,342 transcripts and is available from ArrayExpress (accession E-MTAB-3766). This data set consists of three biological replicates from each of two simulated conditions, and differential isoform usage was introduced for 1,000 genes by swapping the relative expression levels of the two most dominant isoforms between conditions. For each gene in this data set, the total transcriptional output is the same in the two conditions (i.e., no overall DGE); it is worth noting that this is an extreme situation, but provides a useful test set for contrasting DGE, DTU and DTE. The second simulated data set (sim2; Data set 2) is a synthetic data set comprising the 3,858 genes and 15,677 transcripts from the human chromosome 1. It is available from ArrayExpress with accession E-MTAB-4119. Also here, we simulated two conditions with three biological replicates each. For this data set, we simulated both overall DGE, where all transcripts of the affected gene showed the same fold change between the conditions (420 genes), differential transcript usage (DTU), where the total transcriptional output was kept constant but the relative contribution from the transcripts changed (420 genes) and differential transcript expression (DTE), where the expression of 10% of the transcripts of each affected gene was modified (422 genes, 528 transcripts). The three sets of modified genes were disjoint. Again, this synthetic data set represents an extreme situation compared to most real data sets, but provides a useful test case to identify underlying causes of differences between results from various analysis pipelines.
In addition to Data set 1– Data set 6, which contain all code for reproducing our analyses, further method descriptions are given in Supplementary File 1.
http://dx.doi.org/10.5256/f1000research.7563.d114722
Dataset 1 (html) contains all the R code that was used to perform the analyses and generate the figures for the sim1 data set 30.
Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
http://dx.doi.org/10.5256/f1000research.7563.d114723
Data set 2 (html) contains all the R code that was used to perform the analyses and generate the figures for the sim2 data set 31.
Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
http://dx.doi.org/10.5256/f1000research.7563.d114724
Data set 3 (html) contains all the R code that was used to perform the analyses and generate the figures for the Bottomly data set 32.
Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
http://dx.doi.org/10.5256/f1000research.7563.d114725
Data set 4 (html) contains all the R code that was used to perform the analyses and generate the figures for the GSE64570 data set 33.
Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
http://dx.doi.org/10.5256/f1000research.7563.d114726
Data set 5 (html) contains all the R code that was used to perform the analyses and generate the figures for the GSE69244 data set 34.
Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
http://dx.doi.org/10.5256/f1000research.7563.d114730
Data set 6 (html) contain all the R code that was used to perform the analyses and generate the figures for the GSE72165 data set 35.
Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Gene abundance estimates are more accurate than transcript abundance estimates
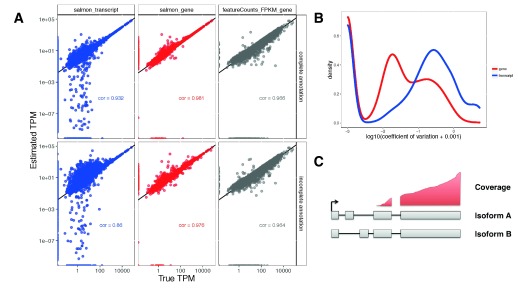
To evaluate the accuracy of abundance estimation with transcript and gene resolution, we used the quasi-mapping mode of Salmon 7 (v0.5.1) to estimate the TPM for each transcript in each of the data sets. Gene-level TPM estimates, representing the overall transcriptional output of each gene, were obtained by summing the corresponding transcript-level TPM estimates. For the two simulated data sets, the true underlying TPM of each feature was known and we could thus evaluate the accuracy of the estimates. Unsurprisingly, gene-level estimates were more accurate than transcript-level estimates ( Figure 1A, Supplementary Figures 4,5). We also derived TPM estimates from simple gene-level counts obtained from traditional alignment of the reads to the genome using STAR followed by counting with featureCounts, by dividing the read count for each gene with a reasonable measure of the length of the gene (the length of the union of its exons) and the total number of mapped reads, and scaling the estimates to sum to 1 million. The simple count estimates showed a lower correlation with the true TPMs than the Salmon estimates, in line with previous observations 19. It is worth noting that we are comparing entire (typical) workflows, and that differences may also occur if the set of reads that STAR is able to align to the genome is not identical to the set of reads that are contributing to the abundance estimation of Salmon. However, due to the large fraction of aligned reads and the high mapping rate with Salmon (both exceeding 99.8%, more than 95% of the reads were subsequently unambiguously assigned to genes by featureCounts), we do not expect this to have a major impact on the results shown in Figure 1A.
Figure 1 (sim2).
A: Accuracy of gene- and transcript-level TPM estimates from Salmon and scaled FPKM estimates derived from simple counts from featureCounts, in one of the simulated samples (sampleA1). Spearman correlations are indicated in the respective panels. Top row: using the complete annotation. Bottom row: using an incomplete annotation, with 20% of the transcripts randomly removed. Gene-level estimates are more accurate than transcript-level estimates. Gene-level estimates from Salmon are more accurate than those from featureCounts. B: Distribution of the coefficients of variation of gene- and transcript-level abundance estimates from Salmon, calculated across 30 bootstrap samples of one of the simulated samples (sampleA1). Gene-level estimates are less variable than transcript-level estimates. C: An example of unidentifiable transcript-level estimates, as uneven coverage does not cover the critical regions that would determine the amount that each transcript is expressed, while gene-level estimation is still possible.
Gene-level estimates derived from both simple counts and Salmon tended to show a high degree of robustness against incompleteness of the annotation catalog, as evidenced from estimation errors after first removing (at random) 20% of the transcripts ( Figure 1A, see also Supplementary File 1); in contrast, Salmon’s transcript estimate accuracies deteriorated. To further compare the merits of genome alignment-based vs alignment-free quantification, especially in their handling of multi-mapping reads, we investigated the accuracy of the abundance estimates within sets of paralogous genes ( Supplementary Figures 1–3). Also here, Salmon provided more consistently accurate estimates than STAR+ featureCounts. From the bootstrap estimates generated by Salmon, we also estimated the coefficient of variation of the abundance estimates. The gene-level estimates showed considerably lower variability than the transcript-level estimates in both simulated and experimental data ( Figure 1B, Supplementary Figures 6,7). Taken together, these observations suggest that the gene-level estimates are more accurate than transcript-level estimates and therefore potentially allow a more accurate and stable statistical analysis. A further argument in favor of gene-level analysis is the unidentifiability of transcript expression that can result from uneven coverage caused by underlying technical biases. While some extent of coverage variability might be alleviated by corrections for sequence- or position-specific biases 20, there remain cases where transcript expression cannot be inferred from data ( Figure 1C). Intermediate approaches, grouping together “indistinguishable” features are also conceiveable 21, but not yet standard practice.
DTE is more powerful and easier to interpret on gene level than for individual transcripts
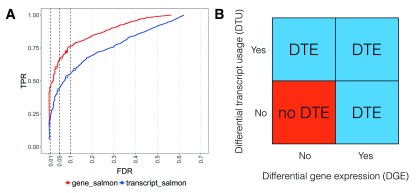
DTE is concerned with inference of changes in abundance at transcript resolution, and thus invokes a statistical test for each transcript. We argue that this can lead to several complications: the first is conceptual, since the rows (transcripts) in the result table will in many cases not be interpreted independently, since the researcher is often interested in comparing the results for transcripts from the same gene locus, and the second one is more technical, since the number of transcripts is considerably larger than the number of genes, which could lead to lower power due to the portioning of the total set of reads across a larger number of features and a potentially higher multiple testing penalty. We tested for DTE on the simulated data by applying edgeR 12 to the transcript counts obtained from Salmon (the application of count models to estimated counts is discussed in the next Section), and represented the results as transcript-level p-values or aggregated these to the gene level by using the perGeneQValue function from the DEXSeq 22 R package. Note that the transcript-level DTE test assesses the null hypothesis that individual transcripts do not change their expression, whereas the gene-level DTE test assesses the null hypothesis that all transcripts from a given gene exhibit no change in expression. Framing the DTE question at the gene level results in higher power, without sacrificing false discovery rate control ( Figure 2A). This is not surprising given the different null hypotheses, and, in fact, for many of the genes detected as true positives with the gene-level test, only a subset of the truly changing transcripts were detected ( Supplementary Figure 8). We note that this type of gene-level aggregation may favor genes in which one transcript shows strong changes, and that other approaches to increase power against specific alternatives are conceivable, e.g., capitalizing on the rich collection of methods for gene set analysis.
Figure 2 (sim2).
A: DTE detection performance on transcript- and gene-level, using edgeR applied to transcript-level estimated counts from Salmon. The statistical analysis was performed on transcript level and aggregated for each gene using the perGeneQValue function from the DEXSeq R package; aggregated results show higher detection power. The curves trace out the observed FDR and TPR for each significance cutoff value. The three circles mark the performance at adjusted p-value cutoffs of 0.01, 0.05 and 0.1. B: Schematic illustration of different ways in which differential transcript expression (DTE) can arise, in terms of absence or presence of differential gene expression (DGE) and differential transcript usage (DTU).
While DTE analysis is more suitable than DGE analysis for detecting genes with changes in absolute or relative isoform expression but no or only minor change in overall output ( Supplementary Figure 9), we argue that even gene-level DTE results may suffer from lack of interpretability. DTE can manifest in several different ways, as an overall differential expression of the gene or differential relative usage of its transcripts, or a combination of the two ( Figure 2B). We argue that the biological question of interest is in many cases more readily interpretable as a combination of DGE and DTU, rather than DTE. It has been our experience that results reported at the transcript level are still often cast to the gene level (i.e., given a differentially expressed transcript, researchers want to know whether also other isoforms of the gene are changing), suggesting that asking two specific gene-level questions (Is the overall abundance changing? Are the isoform abundances changing proportionally?) trumps the interpretability of one broad question addressing the transcript abundances (Are there changes in any of the isoform expression levels?), despite the increased need for multiple testing correction associated with performing two tests for each gene rather than one. There are of course also situations when a transcript-centric approach provides superior interpretability, for example in targeted experiments where specific isoforms are expected to change due to an administered treatment.
Incorporating transcript-level estimates leads to more accurate DGE results
DGE (i.e., testing for changes in the overall transcriptional output of a gene) is typically performed by applying a count-based inference method from statistical packages such as edgeR 12 or DESeq2 11 to gene counts obtained by read counting software such as featureCounts 1, HTSeq-count 2 or functions from the GenomicAlignments 23 R package. A lot has been written about how simple counting approaches are prone to give erroneous results for genes with changes in relative isoform usage, due to the direct dependence of the observed read count on the transcript length 24, and alternatives, such as Cuffdiff 24, which utilizes estimated transcript abundances, have been proposed. However, the extent of the problem in real data has not been thoroughly investigated. Here, we show that taking advantage of transcript-resolution estimates (e.g., obtained by Salmon) in count-based inference methods can lead to improved DGE results. We propose two alternative ways of integrating transcript abundance estimates into the DGE pipeline: to define an “artificial” count matrix, or to calculate offsets that can be used in the statistical modeling of the observed gene counts from, e.g., featureCounts. Both approaches are implemented in the accompanying tximport R package (available from http://bioconductor.org/packages/tximport).
For the DGE analyses, we defined three different gene-level count matrices for each data set (see also Supplementary File 1): 1) using featureCounts from the Rsubread 1 R package (denoted featureCounts below), 2) summing the estimated transcript counts from Salmon within genes ( simplesum), 3) summing the estimated transcript TPMs from Salmon within genes, and multiplying with the total library size in millions ( scaledTPM). We note that the scaledTPM values are artificial values, transforming underlying abundance measures to the “count scale” to incorporate the information provided by the sequencing depth. We further used the effective transcript lengths and estimated TPMs from Salmon to define average transcript lengths for each gene and each sample (normalization factors) as described in the Supplementary material, to be used as offsets for edgeR and DESeq2 when analyzing the featureCounts and simplesum count matrices ( featureCounts_avetxl and simplesum_avetxl).
Overall, the counts obtained by all methods were highly correlated ( Supplementary Figures 10–12), which is not surprising since any differences are likely to affect a relatively small subset of the genes. In general, the simplesum and featureCounts matrices led to similar conclusions in all considered data sets, even though there are differences between the two approaches in terms of how multi-mapping reads and reads partly overlapping intronic regions are handled 25. Previous studies have also shown that some loss of sensitivity for certain genes may be encountered from discarding multi-mapping fragments, which may be recovered through the use of transcript abundance estimators such as Salmon 21. The concordance between simplesum and featureCounts results also suggests that statistical methods based on the Negative Binomial assumption are applicable also to summarized, gene-level estimated counts, which is further supported by the similarity between the p-value histograms as well as the mean-variance relationships observed with the three types of count matrices ( Supplementary Figures 13–18).
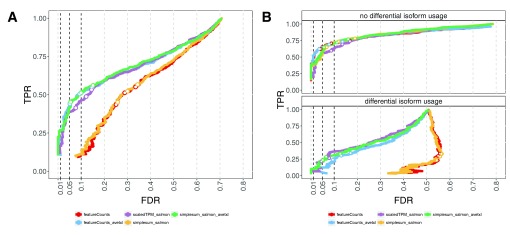
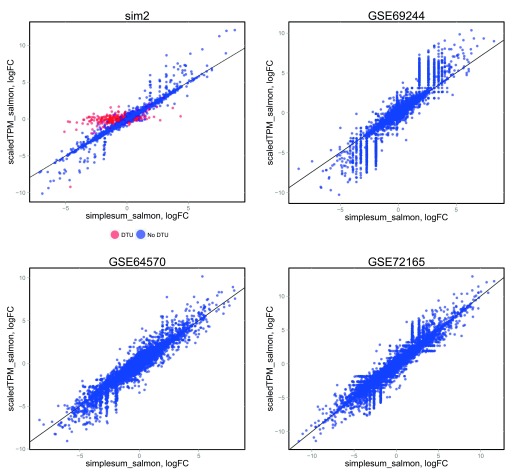
Accounting for the potentially varying average transcript length across samples when performing DGE, either in the definition of the count matrix (scaledTPM) or by defining offsets (featureCounts_avetxl, simplesum_avetxl), led to considerably improved false discovery rate (FDR) control compared to using the observed featureCounts or aggregated Salmon counts directly ( Figure 3A, Table 1). It is important to note that this improvement is entirely attributable to an improved handling of genes with changes in isoform composition between the conditions ( Figure 3B, Supplementary Figure 19), that we purposely introduced strong signals in the simulated data set in order to pinpoint these underlying causes, and that the overall effect in a real data set will depend on the extent to which considerable DTU is present. Experiments on various real data sets ( Supplementary Figure 20) show only small differences in the collections of significant genes found with the simplesum and simplesum_avetxl approaches, suggesting that the extent of the problem in many real data sets is limited, and that most findings obtained with simple counting are not induced by counting artifacts. Further support for this conclusion is shown in Figure 4 (see also Supplementary Figures 21–23 and Supplementary Table 1), where log-fold change estimates from edgeR, based on the simplesum and scaledTPM matrices, are contrasted. For the genes with induced DTU in the sim2 data set, log-fold changes based on the simplesum matrix are overestimated, as expected. However, this effect is almost absent in all the real data sets, again highlighting the extreme nature of our simulated data and suggesting that the effect of using different count matrices is considerably smaller for many real data sets. Table 1 further suggests that the lack of error control for simplesum and featureCounts matrices is more pronounced when there is a large difference in length between the differentially used isoforms. In the group with smallest length difference, where the longer differentially used isoform is less than 34% longer than the shorter one, all approaches controlled the type I error satisfactorily. It is worth noting that among all human transcript pairs in which both transcripts belong to the same gene, the median length ratio is 1.85, and for one third of such pairs the longer isoform is less than 38% longer than the shorter one (see Data set 1).
Figure 3 (sim2).
A: DGE detection performance of edgeR applied to three different count matrices (simplesum, scaledTPM, featureCounts), with or without including an offset representing the average transcript length (for simplesum and featureCounts, avetxl indicates that such offsets were used). Including the offset or using the scaledTPM count matrix leads to improved FDR control compared to using simplesum or featureCounts matrices without offset. The curves trace out the observed FDR and TPR for each significance cutoff value. The three circles mark the performance at adjusted p-value cutoffs of 0.01, 0.05 and 0.1. B: stratification of the results in A by the presence of differential isoform usage. The improvement in FDR control seen in A results from an improved treatment of genes with differential isoform usage, while all methods perform similarly for genes without differential isoform usage.
Figure 4. Comparison of log-fold change estimates from edgeR, based on simplesum and scaledTPM count matrices, in four different data sets.
For the simulated data set ( sim2), where signals have been exaggerated to pinpoint underlying causes of various observations, genes with induced DTU (whose true overall log-fold change is 0) show a clear overestimation of log-fold changes when using simplesum counts. However, none of the real data sets contain a similar population of genes, suggesting that for many real data sets, simple gene counting leads to overall similar conclusions as accounting for underlying changes in transcript usage.
Table 1 (sim1). Observed false positive rates from a differential gene expression analysis using edgeR applied to various count matrices (with a nominal p-value cutoff at 0.05), limited to genes with true underlying differential isoform usage (recall that no genes are truly differentially expressed in this data set).
The results are stratified by “effect size” (the difference in relative abundance between the two differentially used isoforms) and the length ratio between the longer and the shorter of the differentially used isoforms. FPRs below the nominal p-value threshold (0.05) are marked in bold. For more details, see Data set 1.
| simplesum | featureCounts | simplesum_avetxl | featureCounts_avetxl | scaledTPM | |
|---|---|---|---|---|---|
| [0,0.33), [1,1.34) | 0.019 | 0.019 | 0.023 | 0.023 | 0.023 |
| [0.33,0.67), [1,1.34) | 0.059 | 0.059 | 0.059 | 0.059 | 0.059 |
| [0.67,1), [1,1.34) | 0.000 | 0.053 | 0.053 | 0.053 | 0.053 |
| [0,0.33), [1.34,2.57) | 0.075 | 0.070 | 0.070 | 0.065 | 0.065 |
| [0.33,0.67), [1.34,2.57) | 0.240 | 0.220 | 0.050 | 0.033 | 0.066 |
| [0.67,1), [1.34,2.57) | 0.420 | 0.540 | 0.038 | 0.077 | 0.038 |
| [0,0.33), [2.57,35.4] | 0.150 | 0.140 | 0.037 | 0.043 | 0.037 |
| [0.33,0.67), [2.57,35.4] | 0.650 | 0.650 | 0.060 | 0.060 | 0.034 |
| [0.67,1), [2.57,35.4] | 0.970 | 0.970 | 0.034 | 0.034 | 0.034 |
Discussion
In this article, we have contrasted transcript- and gene-resolution analyses in terms of both abundance estimation and statistical inference, and illustrated that gene-level results are often more accurate, powerful and interpretable than transcript-level results. Not surprisingly, however, accurate transcript-level estimation and inference play an important role in deriving appropriate gene-level results, and it is therefore imperative to continue improving abundance estimation and inference methods applicable to individual transcripts, since misestimation can propagate to the gene level. We have shown that when testing for changes in overall gene expression (DGE), traditional gene counting approaches may lead to an inflated false discovery rate compared to methods aggregating transcript-level TPM values or incorporating correction factors derived from these, for genes where the relative isoform usage differs between the compared conditions. These correction factors can be calculated from the output of transcript abundance programs, using e.g., the provided R package ( tximport). It is important to note that the average transcript length offsets must account for the differences in transcript usage between the samples and thus using (sample-independent) exon-union gene lengths will not improve performance.
On the six data sets studied here, simple counting with featureCounts led to very similar conclusions as estimated gene counts from Salmon, when combined with count-based statistical inference tools such as edgeR and DESeq2. Moreover, p-value distributions and mean-variance relationships were similar for actual and estimated counts. Taken together, this suggests that the negative binomial assumption made by the count-based tools is flexible enough to accommodate also estimated counts. All evaluated counting approaches, with and without the inclusion of average transcript length offsets, gave comparable DGE results for genes where DTU was not present. Thus, the extent of the FDR inflation in experimental data depends on the extent of DTU between the compared conditions; notably, our simulation introduced rather extreme levels of DTU, hence the inflated FDR, and the difference between the approaches was considerably smaller in real data sets. Recent studies have also shown that many genes express mainly one, dominant isoform 26 and for such genes, we expect that simple gene counting will work well.
All evaluations in this study were performed using well-established count-based differential analysis tools. These methods take as input a matrix of counts, which is assumed to correctly represent the origin of each read in a particular set of libraries. However, due to sequence similarities among transcripts or genes, there is often a hidden uncertainty in the feature abundance estimates, even when the set of input reads is fixed. With the development of fast, alignment-free abundance estimation methods, this uncertainty can now be estimated rapidly using bootstrap approaches (see e.g. Figure 1b). Method development is currently underway in the field to account for this uncertainty in the differential expression analysis (e.g., MetaDiff 27, sleuth 6), which has the potential to improve performance of both DTE and DGE analyses. If such methods are based on (potentially transformed) aggregated transcript counts as gene-level abundance measures, DGE analysis will still be affected by the presence of DTU, and thus could benefit from the inclusion of average transcript length offsets, or by instead using the sum of transcript TPMs as gene abundance measures.
Our results highlight the importance of carefully specifying the question of interest before selecting a statistical approach. Summarization of abundance estimates at the gene level before performing the statistical testing should be the method of choice if the interest is in finding changes in the overall transcriptional output of a gene. However, it is suboptimal if the goal is to identify genes for which at least one of the transcripts show differences in transcriptional output, since it may miss genes where two transcripts change in opposite directions, or where a lowly expressed transcript changes. For gene-level detection of DTE (that is, whether any transcript showed a change in expression between the conditions), statistical testing applied to aggregated gene counts led to reduced power and slightly inflated FDR compared to performing the statistical test on the transcript level and aggregating results within genes ( Supplementary Figure 9). Statistical inference on aggregated transcript TPMs (scaledTPM) showed low power for detecting changes that did not affect the overall transcriptional output of the gene, as expected. An alternative to DTE analysis, for potential improved interpretability, is to perform a combination of DGE and DTU analyses, both resulting in gene-level inferences. Table 2 summarizes our results and give suggested workflows for the different types of analyses we have considered.
Table 2. Summary of suitable analysis approaches for the three types of comparative analyses discussed in the manuscript (DGE, DTE and DTU).
| Task | Input data | Software (examples) | Post-processing |
|---|---|---|---|
| DGE | Aggregated transcript counts +
average transcript length offsets, or simple counts + average transcript length offsets |
Salmon, kallisto, BitSeq, RSEM | |
| tximport | |||
| DESeq2, edgeR, voom/limma | |||
| DTE | Transcript counts | Salmon, kallisto, BitSeq, RSEM | Optional gene-level
aggregation |
| tximport | |||
| DESeq2, edgeR, sleuth, voom/limma | |||
| DTU/DEU | Transcript counts or bin counts,
depending on interpretation potential 18 |
Salmon, kallisto, BitSeq, RSEM | Optional gene-level
aggregation |
| DEXSeq |
Finally, we note that abundance estimation at the gene level can reduce the impact of technical biases on expression levels, which have been shown to lead to estimation errors, such as expression being attributed to the wrong isoform 28. Non-uniform coverage from amplification bias or from position bias (3’ coverage bias from poly-(A) selection) can result in unidentifiable transcript-level estimation. While correction of technical artifacts in coverage can be attempted computationally, through estimation of sequence- and position-specific biases 20, we note that such errors and estimation problems are also minimized when summarizing expression to the gene level. This being said, there may of course be situations where a direct transcript-level analysis is appropriate. For example, in a cancer setting where a specific deleterious splice variant is of interest (e.g., AR-V7 in prostate cancer 29), inferences directly at the transcript level may be preferred. However, while this may be preferred for individual known transcripts, transcriptome-wide differential expression analyses may not be warranted, given the associated multiple testing cost.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Soneson C et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
F1000Research: Dataset 1. Data set 1, 10.5256/f1000research.7563.d114722
F1000Research: Dataset 2. Data set 2, 10.5256/f1000research.7563.d114723
F1000Research: Dataset 3. Data set 3, 10.5256/f1000research.7563.d114724
F1000Research: Dataset 4. Data set 4, 10.5256/f1000research.7563.d114725
F1000Research: Dataset 5. Data set 5, 10.5256/f1000research.7563.d114726
F1000Research: Dataset 6. Data set 6, 10.5256/f1000research.7563.d114730
Software availability
Software access
Source code as at the time of publication
Archived source code as at the time of publication
Software license
tximport is released under a GNU Public License (GPL).
Acknowledgments
The authors would like to thank Magnus Rattray, Alexander Kanitz, Hubert Rehrauer, Xiaobei Zhou and Rafael Irizarry for helpful comments on earlier versions of this manuscript. The authors would also like to thank the reviewers, Rob Patro and Stephen Floor, for their excellent suggestions that have helped to improve this manuscript.
Funding Statement
MDR and CS acknowledge support from the “RNA & Disease” National Center of Competence in Research, an SNSF project grant (143883) and from the European Commission through the 7th Framework Collaborative Project RADIANT (Grant Agreement Number: 305626). MIL was supported by NIH grant 5T32CA009337-35.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved]
Supplementary material
Supplementary file 1.
Supplementary File 1 (pdf) contains more detailed information about the data sets, supplementary methods and supplementary figures referred to in the text.
.
References
- 1. Liao Y, Smyth GK, Shi W: featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 2. Anders S, Pyl PT, Huber W: HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trapnell C, Roberts A, Goff L, et al. : Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li B, Dewey CN: RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glaus P, Honkela A, Rattray M: Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012;28(13):1721–1728. 10.1093/bioinformatics/bts260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bray N, Pimentel H, Melsted P, et al. : Near-optimal RNA-Seq quantification. arXiv:1505.02710. 2015. Reference Source [Google Scholar]
- 7. Patro R, Duggal G, Kingsford C: Accurate, fast, and model-aware transcript expression quantification with Salmon. bioRxiv. 2015. 10.1101/021592 [DOI] [Google Scholar]
- 8. Mortazavi A, Williams BA, McCue K, et al. : Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 9. Trapnell C, Williams BA, Pertea G, et al. : Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wagner GP, Kin K, Lynch VJ: Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131(4):281–285. 10.1007/s12064-012-0162-3 [DOI] [PubMed] [Google Scholar]
- 11. Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robinson MD, McCarthy DJ, Smyth GK: edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ritchie ME, Phipson B, Wu D, et al. : limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bottomly D, Walter NA, Hunter JE, et al. : Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One. 2011;6(3):e17820. 10.1371/journal.pone.0017820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang S, Marín-Juez R, Meijer AH, et al. : Common and specific downstream signaling targets controlled by Tlr2 and Tlr5 innate immune signaling in zebrafish. BMC Genomics. 2015;16(1):547. 10.1186/s12864-015-1740-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Currais A, Goldberg J, Farrokhi C, et al. : A comprehensive multiomics approach toward understanding the relationship between aging and dementia. Aging (Albany NY). 2015;7(11):937–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang AJ, Ortega FE, Riegler J, et al. : Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature. 2015;527(7577):240–244. 10.1038/nature15721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soneson C, Matthes KL, Nowicka M, et al. : Differential transcript usage from RNA-seq data: isoform pre-filtering improves performance of count-based methods. bioRxiv. 2015. 10.1101/025387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanitz A, Gypas F, Gruber AJ, et al. : Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome Biol. 2015;16(1):150. 10.1186/s13059-015-0702-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roberts A, Trapnell C, Donaghey J, et al. : Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robert C, Watson M: Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol. 2015;16(1):177. 10.1186/s13059-015-0734-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anders S, Reyes A, Huber W: Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–17. 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lawrence M, Huber W, Pagès H, et al. : Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trapnell C, Hendrickson DG, Sauvageau M, et al. : Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao S, Xi L, Zhang B: Union Exon Based Approach for RNA-Seq Gene Quantification: To Be or Not to Be? PLoS One. 2015;10(11):e0141910. 10.1371/journal.pone.0141910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzàlez-Porta M, Frankish A, Rung J, et al. : Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14(7):R70. 10.1186/gb-2013-14-7-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jia C, Guan W, Yang A, et al. : MetaDiff: differential isoform expression analysis using random-effects meta-regression. BMC Bioinformatics. 2015;16(1):208. 10.1186/s12859-015-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Love MI, Hogenesch JB, Irizarry RA: Modeling of RNA-seq fragment sequence bias reduces systematic errors in transcript abundance estimation. bioRxiv. 2015. 10.1101/025767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Antonarakis ES, Lu C, Wang H, et al. : AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–38. 10.1056/NEJMoa1315815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soneson C, Love MI, Robinson MD: Data set 1 in: Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]
- 31. Soneson C, Love MI, Robinson MD: Data set 2 in: Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]
- 32. Soneson C, Love MI, Robinson MD: Data set 3 in: Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]
- 33. Soneson C, Love MI, Robinson MD: Data set 4 in: Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]
- 34. Soneson C, Love MI, Robinson MD: Data set 5 in: Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]
- 35. Soneson C, Love MI, Robinson MD: Data set 6 in: Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]