ABSTRACT
SLAT (also known as DEF6) promotes T cell activation and differentiation by regulating NFAT-Ca2+ signaling. However, its role in TCR-mediated inside-out signaling, which induces integrin activation and T cell adhesion, a central process in T cell immunity and inflammation, has not been explored. Here, we show that SLAT is crucial for TCR-induced adhesion to ICAM-1 and affinity maturation of LFA-1 in CD4+ T cells. Mechanistic studies revealed that SLAT interacts, through its PH domain, with a key component of inside-out signaling, namely the active form of the small GTPase Rap1 (which has two isoforms, Rap1A and Rap1B). This interaction has been further shown to facilitate the interdependent recruitment of Rap1 and SLAT to the T cell immunological synapse upon TCR engagement. Furthermore, a SLAT mutant lacking its PH domain drastically inhibited LFA-1 activation and CD4+ T cell adhesion. Finally, we established that a constitutively active form of Rap1, which is present at the plasma membrane, rescues the defective LFA-1 activation and ICAM-1 adhesion in SLAT-deficient (Def6−/−) T cells. These findings ascribe a new function to SLAT, and identify Rap1 as a target of SLAT function in TCR-mediated inside-out signaling.
KEY WORDS: SLAT, Def6, Rap1, Integrin activation, T cell-adhesion, Inside-out signaling, Immunological synapse
Summary: This study reveals a new signaling pathway that mediates TCR-induced LFA-1 activation, involving SWAP-70-like adaptor of T cells (SLAT).
INTRODUCTION
An efficient adaptive immune response depends on the capacity of leukocytes, especially T cells, to quickly recirculate through blood and lymph, to extravasate into lymphoid organs and inflamed tissues through adhesion to endothelial cells and extracellular matrix, and to specifically respond to low amounts of peptide antigen following interaction with antigen-presenting cells (APC) (Ley and Kansas, 2004; Ley et al., 2007; Luster et al., 2005; von Andrian and Mackay, 2000). These adhesive interactions crucial for T cell trafficking and antigen recognition are facilitated by few specialized leukocyte integrins, such as β2 integrin leukocyte function-associated antigen-1 (LFA-1, also known as αLβ2 integrin) and the very late antigen 4 (VLA-4 or α4β1 integrin), which are structurally adapted to rapidly establish shear-resistant adhesions with their respective endothelial ligands, that is the intercellular adhesion molecules (ICAMs) and the vascular intercellular adhesion molecule (VCAM-1), respectively (Hogg et al., 2011). When expressed on the surface of resting lymphocytes, integrins are maintained in a bent and inactive (i.e. non-adhesive) conformation (Takagi et al., 2002), possessing low affinity for their ligands. In response to a number of physiological stimuli through chemokine receptors or T cell receptor (TCR), integrins become activated through the modulation of their conformation, leaving them competent for mediating high-affinity adhesion (Alon and Dustin, 2007; Dustin and Springer, 1989; Kim et al., 2003; Luo et al., 2007). In addition, upon TCR or chemokine receptor triggering, cell surface diffusion and clustering of integrins on the T cell surface enhances avidity for their ligand (Hogg et al., 2002). These two non-mutually exclusive, transient events (i.e. affinity and avidity modulations) that ultimately lead to integrin adhesiveness have collectively been termed ‘inside-out’ signaling (Abram and Lowell, 2009; Hogg et al., 2011; Kinashi, 2005), in contrast to ‘outside-in’ signaling, which occurs downstream of ligand binding to activated integrins, probably stabilizing adhesion and lowering the antigenic activation threshold for T cells (Carrasco et al., 2004).
Over the past decade, molecular events and selective signaling pathways leading to integrin activation upon TCR stimulation have been intensively investigated. Among the molecules that play crucial roles during TCR-mediated activation of β1 and β2 integrins (Dustin et al., 2004), the small GTPase Rap1 (which has two isoforms, Rap1A and Rap1B) has emerged as a major regulator of inside-out signaling in lymphocytes (Bos et al., 2001; Kinashi, 2005; Ménasché et al., 2007a; Mor et al., 2007; Sekine et al., 2009; Shimonaka et al., 2003). Indeed, Rap1A−/− T cells exhibit profound defects in cell adhesion, polarization, and migration to secondary lymphoid organs (Duchniewicz et al., 2006). Moreover, constitutively active Rap1 mutants (e.g. Rap1V12 or Rap1Q63E) potently increase the affinity (Katagiri et al., 2000; Reedquist et al., 2000) and avidity of LFA-1 in primary T cells (Sebzda et al., 2002), whereas a dominant-negative, nucleotide-free Rap1 (Rap1N17) mutant and Rap1-knockdown block TCR-induced integrin activation (Katagiri et al., 2000). Rap1 has also been shown to positively regulate T-cell–APC conjugates after TCR ligation (Katagiri et al., 2002). Several Rap1 effectors have been identified that bind active (i.e. GTP-bound) Rap1 and link Rap1 to integrins to promote the assembly of integrin-associated signaling complexes, such as Rap1 GTP interacting adapter molecule (RIAM; also known as APBB1IP), protein kinase D1 (PKD1; also known as PRKD1) and RapL (also known as RASSF5) (Katagiri et al., 2003; Kliche et al., 2006; Lee et al., 2009; Medeiros et al., 2005; Menasche et al., 2007b). Indeed, following TCR engagement, Rap1 relocalizes to the plasma membrane, where it can access integrins through adaptor functions of PKD1 and RIAM. In addition, RapL relocalization to the plasma membrane in response to TCR stimulation is needed for optimal binding to Rap1 and activation of LFA-1 (Raab et al., 2011).
SWAP-70-like adaptor of T cells (SLAT) (Tanaka et al., 2003), also known as DEF6 (Hotfilder et al., 1999) or IBP (Gupta et al., 2003b), is a guanine nucleotide exchange factor (GEF) for Cdc42 and Rac1 (Bécart et al., 2008; Gupta et al., 2003a), and is required for inflammatory responses mediated by Th1, Th2 and Th17 cells, reflecting its obligatory role in TCR-stimulated Ca2+ release from intracellular endoplasmic reticulum (ER) stores and, consequently in NFAT transcription factor activation (Bécart and Altman, 2009; Bécart et al., 2007; Canonigo-Balancio et al., 2009; Fos et al., 2014). Structurally, SLAT harbors, beginning at its N-terminus, a Ca2+-binding EF-hand domain and an immunoreceptor tyrosine-based activation motif (ITAM)-like sequence, a phosphatidylinositol 3,4,5-trisphosphate (PIP3)-binding pleckstrin homology (PH) domain, and a Dbl-homology (DH) domain exhibiting GEF activity (Gupta et al., 2003a; Oka et al., 2007). Previous structure-function analysis of SLAT has unveiled that: (1) Lck-dependent phosphorylation of two tyrosine residues in its ITAM-like sequence mediates SLAT translocation to the immunological synapse upon antigen stimulation and is essential for SLAT to exert its pivotal role in NFAT-dependent CD4+ T cell differentiation (Bécart et al., 2008), and (2) both the N-terminal EF-hand domain and the PH domain independently and directly interact with type 1 inositol 1,4,5-triphosphate receptor (IP3R1) to mediate TCR-induced Ca2+ signaling (Fos et al., 2014). Furthermore, the SLAT homologue SWAP-70 has been shown to control B cell homing to lymphoid organs in an inflammatory context by regulating integrin-mediated adhesion and cell polarization (Pearce et al., 2006), as well as being required for mast cell migration and adhesion to fibronectin (Sivalenka and Jessberger, 2004). These results prompted us to explore the potential function and mechanistic aspects of SLAT in the lymphocyte adhesion cascade, and more particularly in TCR-mediated integrin activation.
Here, we report that SLAT transduces TCR-mediated integrin inside-out signals in CD4+ T cells by directly interacting with activated (GTP-bound) Rap1 GTPase through its PH domain. This interaction is required for the interdependent and concomitant recruitment of Rap1 and SLAT to the plasma membrane and subsequently for integrin activation. These findings shed light on a new scaffold function of SLAT, mediated by its PH domain, required for promoting Rap1-dependent inside-out integrin signaling and thus modulating the T cell adhesion cascade.
RESULTS
SLAT is crucial for TCR-induced adhesion to ICAM-1 and LFA-1 affinity maturation in CD4+ T cells
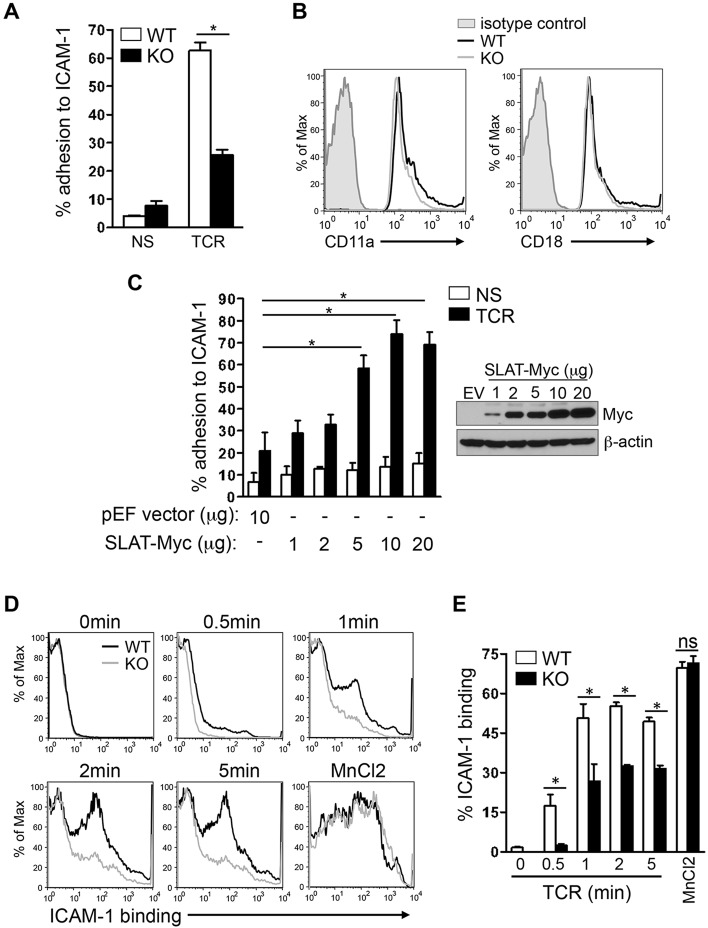
To define the role of SLAT in regulating LFA-1 function following TCR engagement, we first compared the ability of SLAT-deficient (Def6−/−) and wild-type (WT) CD4+ T cells to adhere to Fc-ICAM-1-coated plates following TCR stimulation. Def6−/− CD4+ T cells displayed a substantially impaired TCR-induced adhesion to ICAM-1 (Fig. 1A). This defect was not due to an altered expression of β2-integrin as flow cytometric analysis revealed comparable levels of CD11a (also known as ITGAL) and CD18 (β2-integrin, also known as ITGB2) (i.e. the two molecules comprising LFA-1) on the surface of Def6−/− and WT CD4+ T cells (Fig. 1B). We thus examined the effect of ectopic SLAT expression on the TCR-mediated adhesion to ICAM-1. SLAT-transfected Jurkat T cells showed a dose-dependent increase in TCR-induced adhesion to ICAM-1 relative to control transfectants (Fig. 1C) independently of any change in the surface expression level of CD18 integrin (Fig. S1A, two left panels), confirming the involvement of SLAT in TCR-mediated LFA-1-dependent adhesion to ICAM-1 in T cells. To further assess the role of SLAT in modulation of LFA-1 affinity, we analyzed the ability of soluble Fc-ICAM-1 to bind SLAT-sufficient and SLAT-deficient T cells after TCR stimulation by flow cytometry. Fig. 1D shows that loss of SLAT in CD4+ T cells significantly reduced binding of soluble Fc-ICAM-1 after TCR stimulation at all the time points tested, whereas pharmacological activation of LFA-1 by MnCl2, which bypasses TCR signals and directly induces the high-affinity conformation of LFA-1, completely bypassed the defective adhesion displayed by Def6−/− T cells (Fig. 1D,E).
Fig. 1.
SLAT is required for TCR-induced adhesion to ICAM-1 and LFA-1 affinity maturation in CD4+ T cells. (A) Purified splenic WT and Def6−/− (KO) CD4+ T cells were left untreated (NS) or stimulated with 10 µg/ml anti-CD3 mAb (TCR) and subsequently analyzed for their ability to bind plate-bound Fc-ICAM-1 (mean±s.d., n=4). (B) WT (black line) and KO (gray line) CD4+ T cells were analyzed for the surface expression of CD11a and CD18 by flow cytometry, with normal IgG used as an isotype control (shaded). (C) Jurkat T cells transfected with 10 µg empty pEF vector (EV) or with pEF vector encoding Myc-tagged SLAT (1–20 µg) were either left unstimulated (NS) or stimulated with 10 µg/ml anti-CD3 mAb OKT3 (TCR), and subsequently analyzed for adhesion to plate-bound Fc-ICAM-1. SLAT expression was analyzed by anti-Myc immunoblotting and β-actin expression served as a loading control. Adhesion data represents the means±s.d. of four independent experiments. (D,E) WT or KO CD4+ T cells were analyzed by flow cytometry for their ability to bind soluble Fc-ICAM-1 (as a measurement of LFA-1 affinity) in response to anti-CD3 (10 µg/ml) mAb stimulation for the indicated times or 1 mM MnCl2 (positive control) treatment for 5 min. Quantitative analysis of the results shown in D, representing the mean±s.d. percentage of ICAM-1 binding (determined in triplicates) is shown in E. One out of five representative experiments is shown. *P<0.01; ns, not significant (Student's t-test).
Interdependent recruitment of SLAT and Rap1 to the plasma membrane and immunological synapse upon TCR stimulation
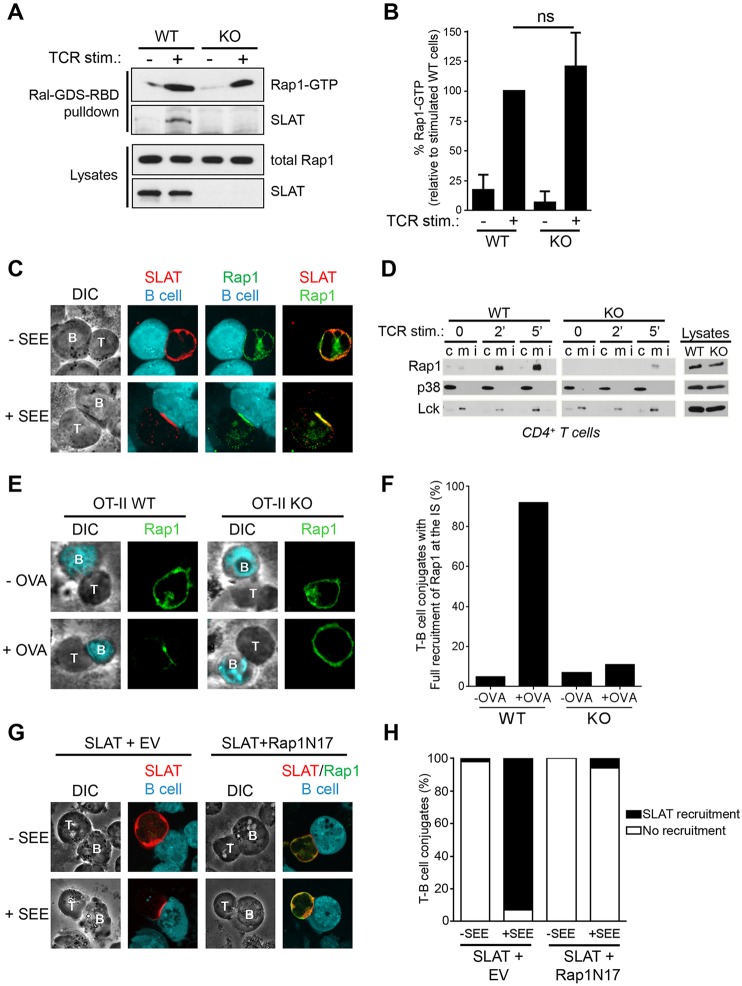
The small GTPase Rap1 has emerged as an important regulator of adhesiveness in lymphocytes (Kinashi, 2005). Both the activation of Rap1 and its transport to the plasma membrane are known to regulate LFA-1 activation (Bivona et al., 2004; Kliche et al., 2006; Menasche et al., 2007b). Switching between the inactive and active states of Rap1 is regulated by specific GEFs and GTPase-activating proteins (GAPs). Considering the previously reported GEF activity of SLAT towards Cdc42 and Rac1, we assessed whether SLAT was involved in regulating Rap1 activity through its GEF activity by using a pulldown assay that employs the GST-coupled Rap1-binding domain (RBD) of the Rap1 effector, RalGDS (RalGDS-RBD). We found that WT or Def6−/− T cells exhibited similar TCR-induced Rap1 activation following 2 min of stimulation and, interestingly, this analysis also unveiled an interaction between Rap1-GTP and SLAT (Fig. 2A,B). Furthermore, we assessed whether SLAT, previously found to translocate to the cell membrane and immunological synapse upon TCR stimulation (Bécart et al., 2008; Tanaka et al., 2003), might regulate Rap1 translocation to the membrane following TCR engagement. Therefore, we first investigated the localization of Rap1 with regard to the immunological synapse in staphylococcal enterotoxin E (SEE)-stimulated Jurkat T cells, with SLAT localization to the immunological synapse serving as an indicator of proper immunological synapse formation and TCR stimulation. T-cell-expressed Rap1 localized to the immunological synapse upon stimulation with SEE-pulsed Raji B cells (Fig. 2C). We further examined the subcellular localization of Rap1 in WT and Def6−/− T cells upon TCR stimulation. Strikingly, we observed that, in contrast to WT T cells, Rap1 was not enriched in the membrane fraction in Def6−/− T cells in response to TCR triggering (Fig. 2D). To substantiate the role of SLAT in regulating Rap1 relocalization to the immunological synapse in the context of an antigen-specific primary T cell response, we crossed Def6−/− mice with OT-II TCR-transgenic mice expressing an ovalbumin (Ova)-specific TCR, and evaluated the localization of Rap1. Consistent with our biochemical observations, Ova peptide stimulation induced translocation of Rap1 to the contact site only in conjugates formed between WT OT-II cells and dendritic cells, but not in Def6−/− OT-II T-cell–dendritic-cell conjugates (Fig. 2E,F). In addition, we examined the effect of a dominant-negative mutant of Rap1 (i.e. Rap1N17) on SLAT recruitment to the immunological synapse following TCR engagement. We observed that T-cell-expressed Rap1N17 drastically inhibited SLAT recruitment to the immunological synapse upon stimulation with SEE-pulsed Raji B cells (Fig. 2G,H). Taken together, these results reveal that SLAT and active Rap1 interact and are interdependently co-recruited to the plasma membrane and immunological synapse in an antigen-dependent manner.
Fig. 2.
Interdependent co-recruitment of SLAT and Rap1 to the plasma membrane and immunological synapse upon TCR stimulation. (A) WT or Def6−/− (KO) CD4+ T cells were either left untreated or stimulated with 10 µg/ml anti-CD3 mAb for 3 min. Active (GTP-loaded) Rap1 was precipitated using a GST–RalGDS-RBD fusion protein. Precipitates (Ral-GDS-RBD pulldown) and whole-cell lysates were analyzed by immunoblotting with the indicated antibodies. One out of four representative experiments is shown. (B) Bar graphs representing mean±s.d. densitometry data pooled from four independent pull-down experiments as shown in A. (C) Jurkat JA16 cells were co-transfected with Myc-tagged SLAT and Xpress-tagged Rap1. After 24 h, the cells were stimulated or not with SEE-pulsed CMAC-labeled Raji B cells and conjugates were bound to poly-L-lysine-coated coverslips, fixed, and stained with anti-Myc and anti-Xpress plus secondary Alexa-Fluor-555-coupled anti-rabbit-Ig or Alexa-Fluor-488-coupled anti-mouse-Ig antibodies, respectively. Individual and overlay of the green (SLAT), red (Rap1) and blue (B, B cell) images along with differential interference contrast (DIC) images are shown. T, T cell. Data shown are representative of three independent experiments. (D) Purified splenic CD4+ T cells from WT or Def6−/− (KO) mice were stimulated for the indicated times with an anti-CD3 mAb. Membrane (m), cytosolic (c) and detergent insoluble (i) fractions were prepared, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies (left panels). Expression of p38 MAPK (detected using an antibody recognizing all isoforms) and Lck in the cytosolic and membrane fractions, respectively, confirmed proper separation of the respective fractions. Analysis of protein expression in whole-cell lysates (right panels) revealed no difference of expression of Rap1, Lck or p38 MAPK in WT versus KO cells. (E) WT or Def6−/− OT-II CD4+ T cells were pulsed with OVA323–339 peptide (+OVA) or not pulsed (−OVA) dendritic cells for 20 min. T-cell–dendritic cell conjugates were then stained for Rap1 expression and localization. Corresponding DIC images are shown. (F) Quantitative analysis of the results shown in E. Rap1 and/or SLAT localization in the immunological synapse was analyzed in 100 T-dendritic cell conjugates. The graph represents the mean percentage of imaged cells scored in each group (recruitment versus no recruitment to the immunological synapse) and is representative of three experiments. (G) Jurkat JA16 cells were co-transfected with Xpress-tagged SLAT along with empty vector (EV) or Myc-tagged dominant-negative Rap1 (Rap1N17). After 24 h, the cells were stimulated or not with SEE-pulsed CMAC-labeled Raji B cells and conjugates were bound to poly-L-lysine-coated coverslips, fixed, and stained with anti-Xpress and anti-Myc plus secondary Alexa-Fluor-555-coupled anti-mouse-Ig or Alexa-Fluor-488-coupled anti-rabbit-Ig antibodies, respectively. Individual and overlay of the red (SLAT), green (Rap1N17) and blue (B cell) images along with DIC images are shown. (H) Quantitative analysis of the results shown in G, representing approximately 100 T-cell–B-cell conjugates analyzed.
The PH domain of SLAT is required for the interaction with active Rap1 and for the co-recruitment of SLAT and Rap1 to the immunological synapse
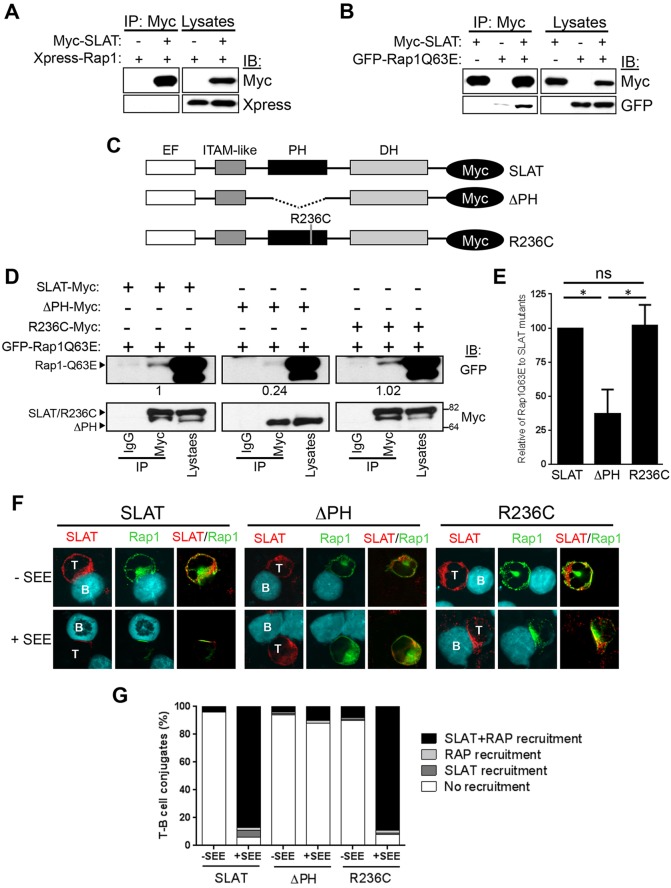
To gain more insights into the interplay between SLAT and Rap1, we first determined whether SLAT could directly associate with Rap1 by co-transfecting 293 T cells with Myc-tagged SLAT and either Xpress-tagged Rap1 (GDP-bound) or constitutively active forms of Rap1 (GFP-Rap1Q63E or Myc-tagged Rap1V12). Interestingly, WT Rap1 did not co-immunoprecipitate with SLAT (Fig. 3A), but constitutively active Rap1Q63E (Fig. 3B) or Rap1V12 (Fig. S2A) mutant proteins were detected in SLAT immunoprecipitates, suggesting that there is a specific and direct interaction of SLAT with active (GTP-bound) Rap1.
Fig. 3.
The PH domain of SLAT interacts with active Rap1 and is required to facilitate SLAT and Rap1 co-recruitment to the immunological synapse. (A,B) 293 T cells were co-transfected with Myc–SLAT and the indicated plasmids encoding Xpress–Rap1 (A) or GFP–Rap1Q63E (B). SLAT immunoprecipitation (IP) and cell lysates were analyzed by immunoblotting (IB). The results shown are representative of four independent experiments. (C) Schematic representation of Myc-tagged SLAT mutants. The EF hand, ITAM-like, PH and DH domains are shown. (D) 293 T cells were co-transfected with the indicated Myc-tagged SLAT plasmids together with GFP–Rap1Q63E. Cell lysates were immunoprecipitated with an anti-Myc antibody or normal IgG (as an immunoprecipitation control) and analyzed by immunoblotting with anti-Myc and anti-GFP antibodies. Cell lysates were also blotted with the same antibodies. Numbers under the Myc blots indicate the relative binding of Rap1Q63E to the SLAT mutants, as determined by densitometry. (E) Bar graphs representing the mean±s.d. relative binding of Rap1Q63E to SLAT mutants as determined by densitometry from three independent experiments as shown in D. (F) JA16 cells were co-transfected and stimulated as in Fig. 2C. Overlays of the red (SLAT) and green (Rap1) images are shown. B, B cell; T, T cell. The data are representative of three independent experiments. (G) Quantitative analysis of SLAT and Rap1 localization in the immunological synapse shown in F, representing approximately 100 T-cell–B-cell conjugates analyzed.
We further investigated the structural determinants of the SLAT–Rap1 interaction that mediates their relocalization to plasma membrane or immunological synapse following TCR ligation. RIAM and PKD1, two other Rap1-GTP-binding proteins that connect Rap1 activation to integrin activation, have been shown to associate, through their PH domain, with Src kinase-associated phosphoprotein of 55 kDa (SKAP55, also known as SKAP1) and Rap1, respectively (Lafuente et al., 2004; Medeiros et al., 2005; Menasche et al., 2007b). Considering that SLAT also harbors a PH domain, we assessed the effect of a SLAT mutant deleted of its PH domain (ΔPH) (Fig. 3C) on the interaction between SLAT and active Rap1 (Rap1Q63E) in transfected 293 T cells. We found that, compared to WT SLAT, the SLAT-ΔPH mutant displayed a substantially reduced interaction with active Rap1 (Fig. 3D,E). Notably, a similar effect was recapitulated in transfected Jurkat T cells (Fig. S2B). Moreover, a point mutation of R236 (R236C) in the SLAT PH domain, shown to be crucial for binding of the phosphoinositide 3-kinase (PI3K) product PIP3 to SLAT, did not interfere with the SLAT–Rap1 interaction (Fig. 3D,E). In parallel, we analyzed the immunological synapse localization of the SLAT mutants and Rap1 in conjugates formed between Jurkat T cells (co-transfected to express SLAT mutants and Rap1) and SEE-pulsed Raji B cells. The Rap1-interaction-deficient SLAT-ΔPH mutant was unable to translocate to the immunological synapse and concomitantly abolished Rap1 recruitment to the T-cell–APC interface upon SEE stimulation. However, the non-PIP3-binding R236C SLAT mutant that shows intact stimulus-induced membrane translocation (Bécart et al., 2008) did not abrogate Rap1 translocation to the immunological synapse (Fig. 3F,G). Taken together, these data indicate that the PH domain of SLAT is required for association with active Rap1 and for targeting both SLAT and Rap1 to the membrane and immunological synapse upon TCR stimulation and, furthermore, that SLAT fulfills these functions independently of its ability to bind lipids.
LFA-1 activation and adhesion to ICAM-1 depend on the PH domain of SLAT
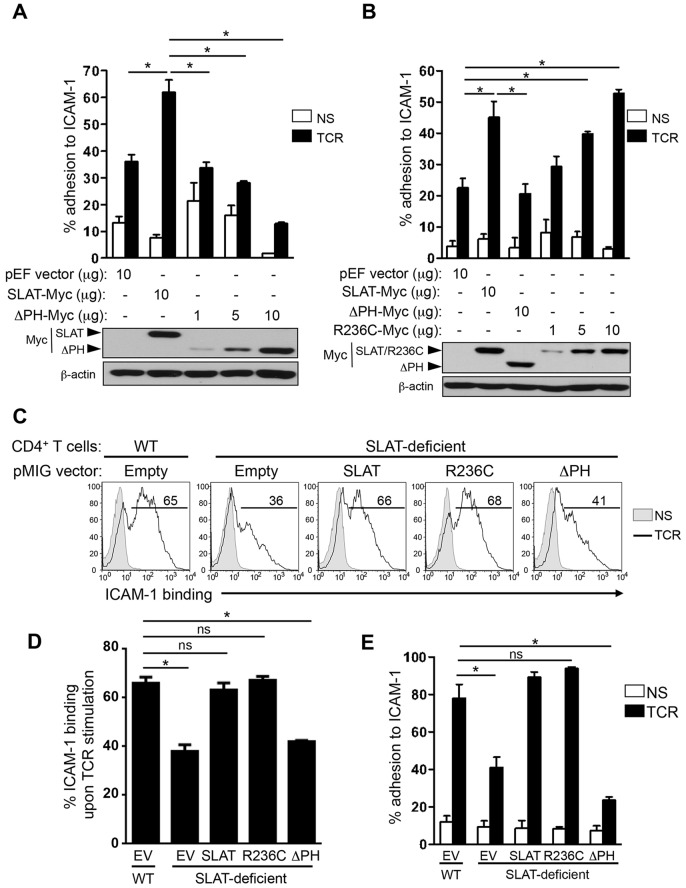
Next, to determine the functional relevance of the interaction between SLAT and Rap1, we ectopically expressed SLAT mutants in Jurkat T cells and assessed the adhesion of the transfectants to ICAM-1 upon TCR stimulation. In contrast to cells transfected with WT SLAT, which displayed enhanced adhesion, SLAT-ΔPH-transfected cells showed a dose-dependent decrease in their ability to adhere to ICAM-1 relative to control transfectants (Fig. 4A); conversely, R236C-transfected cells displayed a dose-dependent increase in adhesiveness to ICAM-1, which was similar to that displayed by WT SLAT transfectants (Fig. 4B). The apparent inability of SLAT-ΔPH to inhibit adhesion (Fig. 4B), in contrast to the inhibition observed with the same dose in Fig. 4A, likely reflects inter-experimental variation (e.g. different transfection efficiencies). We confirmed the physiological relevance of these results by infecting Def6−/− primary T cells with retroviruses expressing WT, ΔPH- or R236C-mutated SLAT and assessing, in parallel, the affinity maturation of LFA-1 and adhesion to ICAM-1 after TCR triggering. Consistently, R236C expression induced normal LFA-1 affinity and adhesion to ICAM-1 compared to cells expressing WT SLAT; in contrast, ΔPH-expressing cells exhibited reduced LFA-1 affinity maturation, similar to that displayed by empty-vector-transduced Def6−/− T cells (Fig. 4C,D), which paralleled the defective TCR-mediated adhesion to ICAM-1 (Fig. 4E). The ability of SLAT-ΔPH to inhibit LFA-1 adhesion was confirmed in transfected Jurkat T cells (Fig. S1B). As a control, we confirmed that the surface expression of CD11a and CD18 was similar in all infected T cells (Fig. S3A). Finally, the induction of high-affinity LFA-1 conformation by MnCl2 rescued the defective adhesion induced by the ΔPH mutant, indicating that the impaired affinity maturation accounts for the reduced adhesion observed with this ΔPH mutant (Fig. S3B). Collectively, these data indicate that interaction of the SLAT PH domain with active Rap1 is functionally crucial for optimal LFA-1 affinity maturation and subsequent adhesion in the context of TCR ligation.
Fig. 4.
TCR-induced LFA-1 dependent T cell adhesion depends on the SLAT PH domain. (A,B) Jurkat JA16T cells were transfected with empty pEF vector or pEF-Myc-SLAT (10 µg each) plus the indicated amounts of Myc-tagged SLAT-ΔPH mutant (A,B) or SLAT-R236C mutant (B). Cells were either left unstimulated (NS) or stimulated with OKT3 mAb (TCR) for 45 min, and subsequently analyzed for adhesion to plate-bound Fc-ICAM-1. Adhesion data represents the mean±s.d. of four independent experiments. Lower panels, expression of transfected proteins was detected by anti-Myc immunoblotting. An anti-β-actin immunoblot serves as a loading control. (C–E) Primary WT and Def6−/− (KO) CD4+ T cells were activated with anti-CD3 plus anti-CD28 mAbs and IL-2 and transduced with retroviral pMIG vectors expressing either GFP alone (Empty) or GFP plus the indicated SLAT cDNAs. Sorted GFP+ CD4+ T cells were left unstimulated (NS) or restimulated with anti-CD3 mAb (TCR) for 3 min (C,D) or 45 min (E), and analyzed either for their ability to bind soluble Fc-ICAM-1 by flow cytometry (C,D) or for their ability to adhere to plate-bound Fc-ICAM-1 (E). Numbers shown in FACS histograms (C) indicate the percentage of GFP+ CD4+ T cells able to bind soluble Fc-ICAM in a representative experiment. Data shown are representative of four independent experiments. (D,E) ICAM-1 binding and adhesion data represent the mean±s.d. of the percentage of four independent experiments. *P<0.01, WT versus Def6−/− cells; ns, not significant (two-tailed Student's t-test).
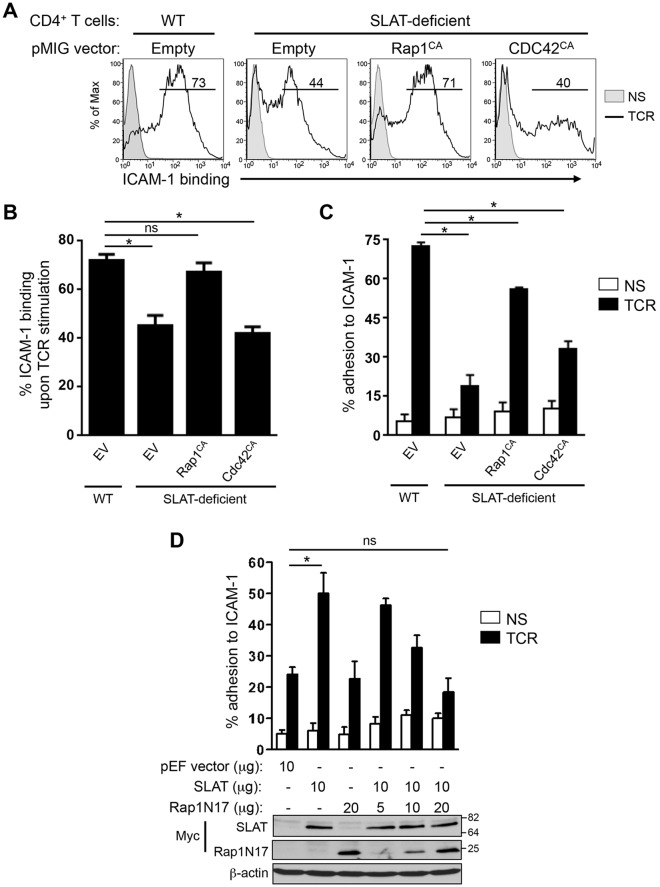
SLAT regulates TCR-mediated LFA-1 activation and T cell adhesion in a Rap1-dependent and Cdc42-independent manner
To further explore the functional relationship between Rap1 and SLAT, and to specifically assess whether Rap1 is a downstream partner of SLAT in TCR inside-out signaling, we examined the effect of constitutively active Rap1 (Rap1CA; Rap1V12) on TCR-mediated LFA-1 affinity maturation and adhesion in Def6−/− CD4+ T cells. Both LFA-1 affinity maturation (Fig. 5A,B) and adhesion to ICAM-1 (Fig. 5C) were largely restored when Def6−/− primary T cells were transduced with Rap1CA. Given this result, we also used the reverse approach by determining the effect of dominant-negative Rap1N17 on SLAT-mediated adhesion. Consistent with the results described above, we observed that Rap1N17 inhibited the SLAT-induced adhesion to ICAM-1 in a dose-dependent manner (Fig. 5D; Fig. S1B) independently of any change in the surface expression level of CD18 integrin (Fig. S1A). Finally, we have previously shown that SLAT plays a role in T cell activation and differentiation through its ability to activate the Cdc42 Rho GTPase through its GEF activity, which resides primarily in its DH domain (Bécart et al., 2008). Interestingly, in this study we show that the constitutively active Cdc42 (Cdc42CA) was largely incapable of rescuing defective TCR-mediated LFA-1 affinity maturation and T cell adhesion in Def6−/− CD4+ T cells (Fig. 5A–C). Taken together, these data indicate that Rap1CA, but not Cdc42CA, can rescue the TCR-mediated inside-out signaling in Def6−/− T cells, consistent with the association of active Rap1 with SLAT (Fig. 3) and with the interdependence of SLAT and active Rap1 in their co-recruitment to the immunological synapse (Fig. 2).
Fig. 5.
SLAT regulates TCR-mediated LFA-1 activation and T cell adhesion in a Rap1-dependent manner. (A–C) Primary WT and Def6−/− (KO) CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs plus IL-2, and transduced with retroviral pMIG vectors expressing either GFP alone (Empty) or GFP plus Rap1V12 (Rap1CA), or Cdc42Q61L (CDC42CA). Sorted GFP+ CD4+ T cells were left unstimulated (NS) or restimulated with anti-CD3 mAb (TCR) for 3 (A,B) or 45 (C) min, and analyzed for their ability to bind soluble Fc-ICAM-1 by flow cytometry (A,B) or adhere to plate-bound Fc-ICAM-1 (C). Numbers shown in FACS histograms (A) indicate the percentage of GFP+ CD4+ T cells that bound soluble Fc-ICAM-1 in a representative experiment, and the bar graphs shown in B and C represent the mean±s.d. of ICAM-1 binding of three independent experiments. (D) Jurkat JA16T cells were transfected with empty pEF vector or Myc–SLAT plasmids (10 µg each) along with dominant-negative Rap1 (Rap1N17) plasmid (5-20 µg). Cells were then either left unstimulated or stimulated for 45 min and subsequently analyzed for their ability to bind plate-bound Fc-ICAM-1. Lower panel, whole-cell lysates were immunoblotted with anti-Myc (SLAT and Rap1N17) and anti-β-actin antibodies to assess the expression of the transfected proteins and to control loading, respectively. Data are representative of three (A–C) and four (D) independent experiments. *P<0.01, WT versus Def6−/− cells; ns, not significant (two-tailed Student's t-test).
DISCUSSION
Integrin adhesion is essential for aspects of immune function such as antigen presentation and migration to lymph nodes and sites of inflammation. To bind its ligands, integrin needs to be converted into an active state in a highly regulated manner. TCR signals during initial contact between a T cell and an APC, inducing inside-out signaling resulting in integrin activation. In this study, we show that SLAT is involved in controlling TCR-mediated adhesion by interacting with Rap1, and facilitating its relocalization to the T cell membrane and immunological synapse. This new interplay between SLAT and Rap1 adds a piece to the molecular puzzle governing the TCR-mediated inside-out signaling and integrin activation in T cells.
Interestingly, we found that SLAT interacts with activated Rap1 through its PH domain. However, an R236C SLAT mutant, mutated in a residue crucial for PIP3 binding, retained its ability to bind Rap1, to be recruited to immunological synapse and to upregulate TCR-mediated LFA-1 affinity, suggesting that the SLAT PH domain might mediate these functions independently of phosphoinositide binding, and act as a scaffold or adaptor protein during TCR inside-out signaling. This finding is consistent with previous studies showing, for instance, that a majority of PH domains in yeast do not show specific binding to phospholipid head groups (Yu et al., 2004) nor do phosphoinositides alone define the location of those PH domains that are targeted to the membrane (Lemmon, 2004; Yu et al., 2004). Thus, PH-like domains are more versatile than initially thought. Our finding of a phosphoinositide-independent interaction between the SLAT PH domain and active Rap1 supports the emerging, although underappreciated, role of PH domains in protein–protein interactions (Balla, 2005; Kavran et al., 1998; Lemmon, 2004; Scheffzek and Welti, 2012), including our recent finding that SLAT regulates Ca2+ signaling through a direct interaction of its PH domain with the IP3R1 (also known as ITPR1) (Fos et al., 2014). Other examples include the interactions of PKD1 with active Rap1 (Medeiros et al., 2005), RIAM with SKAP-55 (Menasche et al., 2007b) and Itk kinase with calmodulin (Wang et al., 2014) through their respective PH domains.
It is important to note, however, that the adaptor function of the SLAT PH domain as a mediator of protein–protein interactions does not rule out another, indirect role of this domain in regulating the enzymatic GEF activity of SLAT. It has been shown in a number of DH-PH-module-containing ‘conventional’ GEFs that the PH domain contributes to the GEF activity of the bona fide catalytic DH domain (Kubiseski et al., 2003; Liu et al., 1998) by allosteric regulation of the catalytic module (Baumeister et al., 2003) or by stabilizing a catalytically competent conformation to promote nucleotide exchange (Yu et al., 2010). Therefore, deletion of the PH domain might indirectly affect SLAT GEF activity. Of note though, SLAT possesses an atypical arrangement of these domains with the PH domain being upstream of the DH domain, compared to conventional GEFs, in which the DH domain is N-terminal to the PH domain. Therefore, it is unclear whether the SLAT PH domain contributes to its enzymatic activity in the same way that PH domains contribute to the activity of conventional GEFs. Moreover, our finding that the constitutively active form of Cdc42, the small GTPase that is a target for the GEF activity of SLAT, cannot rescue the defective TCR-mediated LFA-1 activation in Def6−/− CD4+ T cells argues against a major role of the SLAT PH domain as a regulator of the catalytic activity, and suggests that the function of SLAT in integrin activation might be independent of its GEF activity for Cdc42. Nevertheless, definitive resolution of this question would require the future identification of key SLAT PH domain residues accounting for this interaction with Rap1, as well as the generation of minimally altered yet GEF-deficient SLAT DH domain mutants. Unfortunately, the primary sequence similarity between the SLAT DH domain and conventional DH domains in other GEFs, such as Vav proteins, including the DH domain of the closely related SWAP-70, is very low. This low similarity makes it extremely challenging to predict residues that are crucial for catalytic activity and, hence, to generate GEF-deficient SLAT point mutants. Hopefully, the future availability of such mutants will allow us to dissociate the ‘pure’ adaptor function of the SLAT PH domain from its potential contribution to the GEF activity of SLAT, as well as to precisely determine the importance of the catalytic activity of SLAT in its different functional effects.
Our results show that the co-recruitment of SLAT and Rap1 to the immunological synapse upon TCR ligation is interdependent because SLAT deletion prevented the localization of Rap1 at the immunological synapse and, conversely, a dominant-negative Rap1 mutant prevented the immunological synapse translocation of SLAT. However, the ability of constitutively active Rap1 to rescue the defective TCR-mediated inside-out signaling in Def6−/− T cells suggests that Rap1 is functionally a downstream target of SLAT. The co-recruitment of SLAT and Rap1 to the immunological synapse, mediated by the interaction of active Rap1 with the SLAT PH domain, would likely bring active Rap1 to the vicinity of plasma-membrane-localized integrins, thus facilitating TCR-mediated integrin activation.
This mode of regulation for Rap1 recruitment to the T cell plasma membrane (and immunological synapse) is reminiscent of that used by another PH-dependent Rap1-GTP interactor, namely, RIAM, which is part of a constitutive signaling module that includes adhesion and degranulation promoting adapter protein (ADAP, also known as FYB) and SKAP55, and acts as a scaffold to link Rap1 to talin and promote the formation of the ‘integrin activation complex’ at the plasma membrane (Kliche et al., 2006; Lee et al., 2009; Menasche et al., 2007b). Therefore, it would be interesting to determine if (and how) SLAT influences the interactions between the RIAM–ADAP–SKAP55 complex and Rap1-GTP, all the more so given that both SLAT and RIAM interact with active Rap1 through their respective PH domains (Menasche et al., 2007b). However, in our hands SLAT deficiency had no impact on the constitutive ADAP–SKAP55–RIAM module, nor did SLAT associate directly with RIAM (or ADAP) (Fig. S4), suggesting that there might be two distinct pools of active Rap1, one of which directly interacts with the SLAT PH domain while the other pool interacts with RIAM. Numerous Rap1 effector proteins have been described to regulate integrin activation in T cells, including RapL, which mediates Rap1-induced adhesion through spatial regulation of LFA-1 (Katagiri et al., 2006, 2003), and PKD1, which regulates the membrane localization of Rap1 (Medeiros et al., 2005). Additional studies will be required to determine how SLAT-mediated trafficking of active Rap1 relates to, intersects with or influences these other Rap1-regulating pathways.
SLAT promotes CD4+ T cell activation and differentiation by controlling NFAT activation through its GEF activity for Cdc42 (Bécart et al., 2008) and interaction with IP3R1 (Fos et al., 2014). However, its role in TCR-mediated inside-out signaling were largely independent of Cdc42 activation. These distinct functions of SLAT raise the possibility that different SLAT pools (or complexes) exist to mediate SLAT-dependent downstream signaling pathways, and future studies will determine whether these functions are linked or independent of each other. Furthermore, SLAT and RIAM have both been shown to regulate, beyond their role in T cell adhesion, NFAT activation, although through distinct mechanisms [SLAT acts by interacting with IP3R1 (Fos et al., 2014) whereas RIAM activates NFAT by controlling the activation of PLCγ1 (Patsoukis et al., 2009)]. Along the same line, ADAP has been shown to activate nuclear factor κB (NF-κB) signaling, an event not directly required for ADAP–SKAP55-dependent regulation of cell adhesion (Medeiros et al., 2007), emphasizing the fact that components of inside-out signaling pathways can influence distinct TCR signaling events beyond their adhesion function.
Taken together, our observations establish SLAT as a key regulator of T cell adhesion through its role in TCR-mediated inside-out signaling. Additional studies aiming at determining whether, similar to SLP-76 (also known as LCP2) (Horn et al., 2009), SLAT functions exclusively in TCR-induced inside-out signaling, or might also contribute to the chemokine-mediated inside-out signaling involved in T cell polarization, chemotaxis and intranodal migration, as does its related GEF, SWAP-70, in B cells (Pearce et al., 2006) and mast cells (Sivalenka and Jessberger, 2004), will help to further characterize the roles of SLAT in the CD4+ T cell response and inflammation. In this context, study of the molecular mechanisms underlying SLAT-mediated Rap1-dependent integrin activation could yield new targets for therapeutic intervention. This notion is further enhanced by the highly selective expression of SLAT in T cells (Bécart et al., 2007) and its well-established role in inflammatory diseases such as allergic lung inflammation and autoimmune disorders (Bécart et al., 2007; Canonigo-Balancio et al., 2009). Thus, SLAT might represent a promising drug target for treating T-cell-mediated inflammatory diseases, and an attractive alternative to integrin- and chemokine-directed therapies used preclinically (Abram and Lowell, 2009; Engelhardt and Kappos, 2008; Proudfoot, 2002).
MATERIALS AND METHODS
Antibodies and reagents
SLAT-specific antisera were prepared as described previously (Tanaka et al., 2003). Monoclonal antibodies (mAbs) specific for human CD3 (OKT3) were affinity purified from hybridoma-culture supernatants as described previously (Liu et al., 1997). Anti-p38 MAPK polyclonal antibody (clone 9212) and anti-Rap1 rabbit mAbs were obtained from Cell Signaling Technology, rabbit anti-RIAM mAb was from Abcam, anti-SKAP55 mouse antibody was from BD Biosciences and anti-ADAP rabbit polyclonal antibody was purchased from Upstate Biotechnology. The anti-Xpress mAb was from Invitrogen (for immunoblotting and immunofluorescence), the antibodies against c-Myc (9E10; for immunoblotting and immunoprecipitation), Lck, β-actin (C4), GFP (B-2) and His (H-3 or H15) were from Santa Cruz, the Flag mAb was from Sigma (clone M2). Normal mouse and rabbit Ig (used as immunoprecipitation controls) were purchased from R&D Systems.
For immunofluorescence, an anti-c-Myc rabbit antibody and an anti-Rap1 mAb were obtained from Santa Cruz or BD Biosciences, respectively. SEE was from Toxin Technology. Calcein-AM was from Invitrogen, human ICAM-1 Fc recombinant protein was from R&D Systems, goat anti-human IgG (Fc) was from Jackson ImmunoResearch, and allophycocyanin-labeled mouse anti-human IgG (Fc) was purchased from Southern Biotechnology. Phycoerythrin-conjugated anti-CD11a and -CD18 antibodies used for flow cytometry were purchased from Pharmingen. CellTracker Blue CMAC, Alexa-Fluor-488-conjugated goat anti-mouse-Ig antibody, and Alexa-Fluor-555-conjugated anti-rabbit-Ig antibody were from Life Technologies.
Plasmids
The previously described (Bécart et al., 2008; Fos et al., 2014) plasmids encoding Myc-tagged SLAT (full-length SLAT, SLAT-ΔPH and SLAT-R236C) were generated by PCR and subcloned into the pEF-Myc-His A/C mammalian vectors (Life Technologies) to encode an in-frame Myc tag epitope downstream of the insert. The R236C point mutation was introduced by use of a site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. The plasmids encoding His–ADAP, Flag–RIAM and GFP–Rap1Q63E were as previously reported (Menasche et al., 2007b) and were provided by Gary Koretzky (Weill Cornell Medical College, New York, NY). The Xpress-tagged WT mouse Rap1a and Myc-tagged human Rap1aV12 were subcloned into the pEF vector. Rap1aV12 was subcloned from the pEF-Myc expression vector into the BglII and EcoRI sites of the pMIG retroviral vector. A dominant-negative form of human Rap1 (PRK5myc Rap1a N17) was kindly provided by Monika Raab (Goethe University, Frankfurt, Germany). The retroviral vector of constitutively active Cdc42 (pMX GFP Cdc42Q61L) was purchased from Addgene.
Mice, cell culture, purification and transfection
Mice were maintained under specific pathogen-free conditions and used in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International. Def6−/− mice on a C57BL/6 background have been previously described (Bécart et al., 2007). T cells from OT-II TCR-transgenic mice were used as a source of Vβ5-Vα2 CD4+ T cells that specifically recognised amino acid residues 323–339 of Ova (OVA323–339). Six- to 10-week-old mice were used in all experiments. After erythrocyte lysis, lymph node and spleen CD4+ T cells were isolated by positive selection with anti-CD4 (L3T4) mAb-coated microbeads (MACS). T cells were cultured in RPMI-1640 medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 1 mM MEM nonessential amino acid solution and 100 U/ml each of penicillin G and streptomycin (Life Technologies). The Jurkat cell line JA16 was a kind gift of Jacques Nunes (INSERM, France) and was cultured in RPMI-1640 with 10% FBS. Raji B cells were obtained from the ATCC and grown in the same medium. Jurkat JA16 cells in logarithmic growth phase were transfected with plasmid DNAs by electroporation (Villalba et al., 2001). After 24 h, transfected cells were washed with serum-free RPMI-1640 medium, and used either in immunoprecipitation, adhesion or immunofluorescence microscopy experiments. 293 T cells were grown in DMEM medium plus 10% FBS and transfected using the TransIT®-LT1 transfection reagent (Mirus). After 24 h, transfected cells were harvested for immunoprecipitation experiments.
Retroviral transduction of primary CD4+ T cells
Platinum-E packaging cells (Morita et al., 2000) (0.5×106 cells) were plated in a six-well plate in 2 ml DMEM plus 10% FBS. After overnight incubation, the cells were transfected with 3 µg retroviral plasmid DNA with TransIT®-LT1 transfection reagent. After 24 h, the medium was replaced with RPMI plus 10% FBS. Cultures were maintained for 24 h, and the retroviral supernatant was harvested, filtered (0.45 µm), supplemented with 5 µg/ml polybrene and 100 U/ml IL-2, and then used to infect CD4+ T cells that had been preactivated for 24 h with anti-CD3 plus -CD28 mAbs plus recombinant IL-2 (100 U/ml). Plates were centrifuged for 1 h at 800 g and incubated for 8 h at 32°C and for 16 h at 37°C, followed by two additional retroviral infections at daily intervals.
Immunoprecipitation and immunoblot analysis
Myc–SLAT, His–ADAP, Flag–RIAM, Xpress–Rap1, and GFP–Rap1Q63E, R236C or ΔPH SLAT mutants were co-expressed in 293 T cells. At 1 day after transfection, 293 T cells were resuspended in lysis buffer (20 mM Tris-HCl pH 7.5, 137 mM NaCl, 2 mM EDTA, 10% glycerol and 0.2% NP-40), and the lysates were collected after centrifugation at 13,000 g for 20 min. Immunoprecipitation was performed by adding the indicated antibodies plus 30 μl of protein A/G PLUS-agarose (Santa Cruz Biotechnology) and incubating the lysates for 2 h at 4°C with gentle shaking. Samples were washed four times with lysis buffer, and the immunoprecipitates were dissolved in 1× Laemmli buffer, subjected to SDS-PAGE, transferred onto nitrocellulose membrane and immunoblotted with the indicated antibodies. For immunoprecipitation of endogenous proteins in CD4+ T cells, murine CD4+ T cells (107) were resuspended in serum-free medium and left unstimulated or incubated for 30 min on ice with 20 μg/ml of anti-mouse CD3ε mAb (145-2C11; BioLegend), followed by crosslinking with 20 μg/ml goat anti-hamster-IgG antibody (Pierce) for the indicated times at 37°C with gentle shaking. Cells were lysed in lysis buffer as indicated above, the lysates were centrifuged and supernatants were rotated for 2 h at 4°C with an anti-RIAM antibody (Abcam). Immune complexes were analyzed as described above using the indicated specific antibodies for immunoblot analysis.
Subcellular fractionation
Primary CD4+ T cells, isolated as described above, were activated for 48 h in plates coated with anti-CD3 mAb (5 μg/ml) plus soluble anti-CD28 mAb (2.5 μg/ml). The activated T cells were then cultured for 24 h without anti-CD3 or -CD28 mAbs but with recombinant human IL-2 (20 units/ml), washed, and restimulated with anti-CD3 mAb (10 μg/ml) for the indicated times. The cells were resuspended in ice-cold hypotonic lysis buffer and incubated on ice for 15 min. The cells were transferred to a 1-ml syringe and sheared by passing them five times through a 27-gauge needle. The lysates were centrifuged at 200 g for 10 min to remove nuclei and cell debris. The supernatant was transferred to ultracentrifuge Eppendorf tubes and centrifuged at 45,000 g for 30 min at 4°C. The supernatant (cytosol) was collected, and the pellet was resuspended in lysis buffer [20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, pH 8.0, 5 mM NaPiP, 1 mM sodium orthovanadate (Na3VO4), 20 mM NaPO4, pH 7.6, 3 mM β-glycerophosphate, 10 mM NaF, 1% Triton X-100, and 10 μg/ml each aprotinin and leupeptin], vortexed for 20 min at 4°C, and centrifuged again at 45,000 g for 30 min. The supernatant representing the membrane fraction was saved, and the detergent insoluble fraction resuspended in 1% SDS in water. Protein concentrations in the cytosolic and the membrane fraction were measured by the Bradford method, and each fraction was then diluted with Laemmli buffer to yield equal protein concentrations. Identical amounts of protein were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted with the indicated antibodies.
Immunofluorescence microscopy
Conjugation of transfected Jurkat JA16T cells and Raji B cells, staining, and preparation of samples for microscopy were performed as described previously (Charvet et al., 2005). Raji B cells or dendritic cells were prelabeled with CellTracker Blue CMAC. For antigen-specific stimulation, CD4+ T cells from Def6−/− or WT OT-II TCR-transgenic mice were incubated with dendritic cells pulsed with 10 μg/ml OVA323–339 peptide or non-pulsed dendritic cells. Cells were briefly centrifuged to promote conjugate formation and incubated at 37°C for 20 min in serum-free medium on poly-L-lysine-coated slides, then fixed with 4% paraformaldehyde, and permeabilized with 0.1% saponin in PBS with 1% BSA. The conjugates were stained with the primary antibodies rabbit anti-Myc, mouse anti-Xpress, mouse anti-Rap1 or SLAT antisera, followed by the secondary antibodies Alexa-Fluor-555-conjugated goat anti-rabbit-IgG, or Alexa-Fluor-488-conjugated goat anti-mouse-IgG. Immunofluorescence images were recorded on a Olympus Fluoview FV10i confocal microscope. A minimum of 100 randomly selected conjugates per slide were examined in each experiment.
Flow cytometry
Jurkat T cells or primary mouse CD4+ T cells were labeled with the following fluorochrome-conjugated antibodies (Pharmingen): anti-CD11a (phycoerythrin), or anti-CD18 (phycoerythrin) or isotype control (phycoerythrin-conjugated). Acquisition was performed on a FACSCalibur flow cytometer (BD) and analyzed with FlowJo software.
Rap1 activity assay
Purified primary WT or Def6−/− CD4+ T cells were blasted with plate-coated anti-CD3 (5 µg/ml) plus soluble anti-CD28 (2.5 µg/ml) mAbs and IL-2 (100 U/ml) for 2 days, and rested for 24 h. 25×106 cells/condition were either left untreated or incubated with anti-CD3 mAb 145-2C11 (BioLegend) and cross-linked with goat anti-hamster-IgG antibody (Pierce) for 3 min at 37°C. Subsequently, cells were resuspended in lysis buffer (25 mM Tris-HCl pH7.5, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 5% glycerol) for 5 min on ice, and active Rap1 was isolated using a GST-RalGDS–Rap1-binding domain (RBD) fusion protein (Millipore). The amount of bound Rap1 was quantified by western blotting using an anti-Rap1 antibody (Cell Signaling Technology).
Adhesion assay
Flat-bottom 96-well plates (Maxisorb, Nunc) were coated with 5 µg/ml goat anti-human-IgG (Fc) antibody (Jackson ImmunoResearch) in PBS for 2 h at 37°C. After blocking the wells with 1% BSA for 1 h at 37°C, wells were coated overnight at 4°C with a fusion protein (1 µg/ml) of recombinant mouse ICAM-1 and human IgG1 Fc fragment (Fc-ICAM-1; R&D Systems). Cells were labeled in serum-free, Phenol-Red-free HBSS (containing CaCl2 and MgCl2) with 2.5 µM Calcein-AM (Invitrogen) for 30 min at 37°C, washed twice in HBSS medium, and incubated (3×105 cells/well) on ICAM-1-coated plates for 45 min at 37°C, in the presence or absence of 10 µg/ml anti-CD3 mAbs (2C11 for primary murine CD4+ T cells or OKT3 for human Jurkat T cells) or 1 mM MnCl2. Non-adherent cells were carefully removed by three consecutive washes with pre-warmed (37°C) washing buffer (HBSS+0.5% BSA). Input (time 0) and output (from adhering cells after 45 min incubation) fluorescence was read in the 96-well plate at the 490-nm excitation wavelength and 520-nm emission wavelength. Specific adhesion was obtained by subtracting background adhesion (mean reading for wells uncoated with ICAM-1-Fc) from the reading from each well. Specific adhesion was expressed as a percentage of output fluorescence divided by input fluorescence in triplicate wells.
Soluble ICAM-1-binding assay
Primary T cells or transfected Jurkat T cells were resuspended in Hanks Balanced Salt Solution (HBSS) containing 1 mM CaCl2 and 1 mM MgCl2 (Invitrogen). Cells were stimulated with 10 µg/ml anti-CD3 mAb (145-2C11 antibody for murine CD4+ T cells, or OKT3 antibody for Jurkat T cells) or an equal volume of PBS, in the presence of mICAM-1-Fc (20 μg/ml; R&D Systems) and allophycocyanin-conjugated anti-human-IgG1 antibody (Fc-specific; Southern Biotechnology) for 3 min at 37°C. Cells were fixed in 7.4% PFA on ice for 10 min, and then washed in HBSS. ICAM-1 binding was evaluated by flow cytometry, as a measurement of the high-affinity state of LFA-1.
Statistics
Data were analyzed using Prism software (GraphPad Software). Statistical significance was analyzed by two-tailed Student's t-tests. Unless otherwise indicated, data represent the mean±s.d., with P<0.01 considered statistically significant.
Acknowledgements
We thank Gary Koretzky (Univ. Pennsylvania), Chris Rudd (Cambridge University, UK) and Monika Raab (Goethe University, Frankfurt) for the ADAP, RIAM, and Rap1 plasmids, and all the members of the Cell Biology Division for helpful comments, and Cheryl Kim, Kurt Van Gunst and Anthony Jose for assistance with flow cytometry and cell sorting. This is manuscript number 1796 from La Jolla Institute for Allergy & Immunology.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.B. conceived and designed the study. S.B. and M.C. executed, analyzed and interpreted the data being published. S.B. and A.A. wrote the manuscript with input from all the authors. C.F. and A.C.B. generated mammalian and retroviral vectors used in the study and A.C.B. was in charge of the mouse colony involved in this study. K.L. provided advice in integrin signaling. All authors read and approved the final manuscript. S.B. and A.A. share senior authorship.
Funding
The work was supported by National Institutes of Health [grant number AI068320 to A.A.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.172742/-/DC1
References
- Abram C. L. and Lowell C. A. (2009). The ins and outs of leukocyte integrin signaling. Annu. Rev. Immunol. 27, 339-362. 10.1146/annurev.immunol.021908.132554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R. and Dustin M. L. (2007). Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity 26, 17-27. 10.1016/j.immuni.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Balla T. (2005). Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 118, 2093-2104. 10.1242/jcs.02387 [DOI] [PubMed] [Google Scholar]
- Baumeister M. A., Martinu L., Rossman K. L., Sondek J., Lemmon M. A. and Chou M. M. (2003). Loss of phosphatidylinositol 3-phosphate binding by the C-terminal Tiam-1 pleckstrin homology domain prevents in vivo Rac1 activation without affecting membrane targeting. J. Biol. Chem. 278, 11457-11464. 10.1074/jbc.M211901200 [DOI] [PubMed] [Google Scholar]
- Bécart S. and Altman A. (2009). SWAP-70-like adapter of T cells: a novel Lck-regulated guanine nucleotide exchange factor coordinating actin cytoskeleton reorganization and Ca2+ signaling in T cells. Immunol. Rev. 232, 319-333. 10.1111/j.1600-065X.2009.00839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécart S., Charvet C., Canonigo Balancio A. J., De Trez C., Tanaka Y., Duan W., Ware C., Croft M. and Altman A. (2007). SLAT regulates Th1 and Th2 inflammatory responses by controlling Ca2+/NFAT signaling. J. Clin. Invest. 117, 2164-2175. 10.1172/JCI31640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécart S., Balancio A. J. C., Charvet C., Feau S., Sedwick C. E. and Altman A. (2008). Tyrosine-phosphorylation-dependent translocation of the SLAT protein to the immunological synapse is required for NFAT transcription factor activation. Immunity 29, 704-719. 10.1016/j.immuni.2008.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona T. G., Wiener H. H., Ahearn I. M., Silletti J., Chiu V. K. and Philips M. R. (2004). Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164, 461-470. 10.1083/jcb.200311093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., de Rooij J. and Reedquist K. A. (2001). Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2, 369-377. 10.1038/35073073 [DOI] [PubMed] [Google Scholar]
- Canonigo-Balancio A. J., Fos C., Prod'homme T., Becart S. and Altman A. (2009). SLAT/Def6 plays a critical role in the development of Th17 cell-mediated experimental autoimmune encephalomyelitis. J. Immunol. 183, 7259-7267. 10.4049/jimmunol.0902573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Y. R., Fleire S. J., Cameron T., Dustin M. L. and Batista F. D. (2004). LFA-1/ICAM-1 interaction lowers the threshold of B cell activation by facilitating B cell adhesion and synapse formation. Immunity 20, 589-599. 10.1016/S1074-7613(04)00105-0 [DOI] [PubMed] [Google Scholar]
- Charvet C., Canonigo A. J., Billadeau D. D. and Altman A. (2005). Membrane localization and function of Vav3 in T cells depend on its association with the adapter SLP-76. J. Biol. Chem. 280, 15289-15299. 10.1074/jbc.M500275200 [DOI] [PubMed] [Google Scholar]
- Duchniewicz M., Zemojtel T., Kolanczyk M., Grossmann S., Scheele J. S. and Zwartkruis F. J. T. (2006). Rap1A-deficient T and B cells show impaired integrin-mediated cell adhesion. Mol. Cell. Biol. 26, 643-653. 10.1128/MCB.26.2.643-653.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M. L. and Springer T. A. (1989). T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature 341, 619-624. 10.1038/341619a0 [DOI] [PubMed] [Google Scholar]
- Dustin M. L., Bivona T. G. and Philips M. R. (2004). Membranes as messengers in T cell adhesion signaling. Nat. Immunol. 5, 363-372. 10.1038/ni1057 [DOI] [PubMed] [Google Scholar]
- Engelhardt B. and Kappos L. (2008). Natalizumab: targeting alpha4-integrins in multiple sclerosis. Neurodegener. Dis. 5, 16-22. 10.1159/000109933 [DOI] [PubMed] [Google Scholar]
- Fos C., Becart S., Canonigo Balancio A. J., Boehning D. and Altman A. (2014). Association of the EF-hand and PH domains of the guanine nucleotide exchange factor SLAT with IP(3) receptor 1 promotes Ca(2)(+) signaling in T cells. Sci. Signal. 7, ra93 10.1126/scisignal.2005565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Fanzo J. C., Hu C., Cox D., Jang S. Y., Lee A. E., Greenberg S. and Pernis A. B. (2003a). T cell receptor engagement leads to the recruitment of IBP, a novel guanine nucleotide exchange factor, to the immunological synapse. J. Biol. Chem. 278, 43541-43549. 10.1074/jbc.M308960200 [DOI] [PubMed] [Google Scholar]
- Gupta S., Lee A., Hu C., Fanzo J., Goldberg I., Cattoretti G. and Pernis A. B. (2003b). Molecular cloning of IBP, a SWAP-70 homologous GEF, which is highly expressed in the immune system. Hum. Immunol. 64, 389-401. 10.1016/S0198-8859(03)00024-7 [DOI] [PubMed] [Google Scholar]
- Hogg N., Henderson R., Leitinger B., McDowall A., Porter J. and Stanley P. (2002). Mechanisms contributing to the activity of integrins on leukocytes. Immunol. Rev. 186, 164-171. 10.1034/j.1600-065X.2002.18614.x [DOI] [PubMed] [Google Scholar]
- Hogg N., Patzak I. and Willenbrock F. (2011). The insider's guide to leukocyte integrin signalling and function. Nat. Rev. Immunol. 11, 416-426. 10.1038/nri2986 [DOI] [PubMed] [Google Scholar]
- Horn J., Wang X., Reichardt P., Stradal T. E., Warnecke N., Simeoni L., Gunzer M., Yablonski D., Schraven B. and Kliche S. (2009). Src homology 2-domain containing leukocyte-specific phosphoprotein of 76 kDa is mandatory for TCR-mediated inside-out signaling, but dispensable for CXCR4-mediated LFA-1 activation, adhesion, and migration of T cells. J. Immunol. 183, 5756-5767. 10.4049/jimmunol.0900649 [DOI] [PubMed] [Google Scholar]
- Hotfilder M., Baxendale S., Cross M. A. and Sablitzky F. (1999). Def-2, -3, -6 and -8, novel mouse genes differentially expressed in the haemopoietic system. Br. J. Haematol. 106, 335-344. 10.1046/j.1365-2141.1999.01551.x [DOI] [PubMed] [Google Scholar]
- Katagiri K., Hattori M., Minato N., Irie S.-k., Takatsu K. and Kinashi T. (2000). Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol. Cell. Biol. 20, 1956-1969. 10.1128/MCB.20.6.1956-1969.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K., Hattori M., Minato N. and Kinashi T. (2002). Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol. Cell. Biol. 22, 1001-1015. 10.1128/MCB.22.4.1001-1015.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K., Maeda A., Shimonaka M. and Kinashi T. (2003). RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 4, 741-748. 10.1038/ni950 [DOI] [PubMed] [Google Scholar]
- Katagiri K., Imamura M. and Kinashi T. (2006). Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol. 7, 919-928. 10.1038/ni1374 [DOI] [PubMed] [Google Scholar]
- Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y. and Lemmon M. A. (1998). Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273, 30497-30508. 10.1074/jbc.273.46.30497 [DOI] [PubMed] [Google Scholar]
- Kim M., Carman C. V. and Springer T. A. (2003). Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301, 1720-1725. 10.1126/science.1084174 [DOI] [PubMed] [Google Scholar]
- Kinashi T. (2005). Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5, 546-559. 10.1038/nri1646 [DOI] [PubMed] [Google Scholar]
- Kliche S., Breitling D., Togni M., Pusch R., Heuer K., Wang X., Freund C., Kasirer-Friede A., Menasche G., Koretzky G. A. et al. (2006). The ADAP/SKAP55 signaling module regulates T-cell receptor-mediated integrin activation through plasma membrane targeting of Rap1. Mol. Cell. Biol. 26, 7130-7144. 10.1128/MCB.00331-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiseski T. J., Culotti J. and Pawson T. (2003). Functional analysis of the Caenorhabditis elegans UNC-73B PH domain demonstrates a role in activation of the Rac GTPase in vitro and axon guidance in vivo. Mol. Cell. Biol. 23, 6823-6835. 10.1128/MCB.23.19.6823-6835.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E. M., van Puijenbroek A. A. F. L., Krause M., Carman C. V., Freeman G. J., Berezovskaya A., Constantine E., Springer T. A., Gertler F. B. and Boussiotis V. A. (2004). RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell 7, 585-595. 10.1016/j.devcel.2004.07.021 [DOI] [PubMed] [Google Scholar]
- Lee H.-S., Lim C. J., Puzon-McLaughlin W., Shattil S. J. and Ginsberg M. H. (2009). RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284, 5119-5127. 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A. (2004). Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 32, 707-711. 10.1042/BST0320707 [DOI] [PubMed] [Google Scholar]
- Ley K. and Kansas G. S. (2004). Selectins in T-cell recruitment to non-lymphoid tissues and sites of inflammation. Nat. Rev. Immunol. 4, 325-336. 10.1038/nri1351 [DOI] [PubMed] [Google Scholar]
- Ley K., Laudanna C., Cybulsky M. I. and Nourshargh S. (2007). Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678-689. 10.1038/nri2156 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wenger R. H., Zhao M. and Nielsen P. J. (1997). Distinct costimulatory molecules are required for the induction of effector and memory cytotoxic T lymphocytes. J. Exp. Med. 185, 251-262. 10.1084/jem.185.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang H., Eberstadt M., Schnuchel A., Olejniczak E. T., Meadows R. P., Schkeryantz J. M., Janowick D. A., Harlan J. E., Harris E. A. S. et al. (1998). NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell 95, 269-277. 10.1016/S0092-8674(00)81757-2 [DOI] [PubMed] [Google Scholar]
- Luo B.-H., Carman C. V. and Springer T. A. (2007). Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619-647. 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster A. D., Alon R. and von Andrian U. H. (2005). Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 6, 1182-1190. 10.1038/ni1275 [DOI] [PubMed] [Google Scholar]
- Medeiros R. B., Dickey D. M., Chung H., Quale A. C., Nagarajan L. R., Billadeau D. D. and Shimizu Y. (2005). Protein kinase D1 and the beta 1 integrin cytoplasmic domain control beta 1 integrin function via regulation of Rap1 activation. Immunity 23, 213-226. 10.1016/j.immuni.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Medeiros R. B., Burbach B. J., Mueller K. L., Srivastava R., Moon J. J., Highfill S., Peterson E. J. and Shimizu Y. (2007). Regulation of NF-kappaB activation in T cells via association of the adapter proteins ADAP and CARMA1. Science 316, 754-758. 10.1126/science.1137895 [DOI] [PubMed] [Google Scholar]
- Ménasché G., Kliche S., Bezman N. and Schraven B. (2007a). Regulation of T-cell antigen receptor-mediated inside-out signaling by cytosolic adapter proteins and Rap1 effector molecules. Immunol. Rev. 218, 82-91. 10.1111/j.1600-065X.2007.00543.x [DOI] [PubMed] [Google Scholar]
- Menasche G., Kliche S., Chen E. J. H., Stradal T. E. B., Schraven B. and Koretzky G. (2007b). RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation. Mol. Cell. Biol. 27, 4070-4081. 10.1128/MCB.02011-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., Dustin M. L. and Philips M. R. (2007). Small GTPases and LFA-1 reciprocally modulate adhesion and signaling. Immunol. Rev. 218, 114-125. 10.1111/j.1600-065X.2007.00538.x [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T. and Kitamura T. (2000). Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063-1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Oka T., Ihara S. and Fukui Y. (2007). Cooperation of DEF6 with activated Rac in regulating cell morphology. J. Biol. Chem. 282, 2011-2018. 10.1074/jbc.M605153200 [DOI] [PubMed] [Google Scholar]
- Patsoukis N., Lafuente E. M., Meraner P., Kim J. s., Dombkowski D., Li L. and Boussiotis V. A. (2009). RIAM regulates the cytoskeletal distribution and activation of PLC-gamma1 in T cells. Sci. Signal. 2, ra79 10.1126/scisignal.2000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Angeli V., Randolph G. J., Junt T., von Andrian U., Schnittler H.-J. and Jessberger R. (2006). Signaling protein SWAP-70 is required for efficient B cell homing to lymphoid organs. Nat. Immunol. 7, 827-834. 10.1038/ni1365 [DOI] [PubMed] [Google Scholar]
- Proudfoot A. E. I. (2002). Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106-115. 10.1038/nri722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Smith X., Matthess Y., Strebhardt K. and Rudd C. E. (2011). SKAP1 protein PH domain determines RapL membrane localization and Rap1 protein complex formation for T cell receptor (TCR) activation of LFA-1. J. Biol. Chem. 286, 29663-29670. 10.1074/jbc.M111.222661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedquist K. A., Ross E., Koop E. A., Wolthuis R. M., Zwartkruis F. J., van Kooyk Y., Salmon M., Buckley C. D. and Bos J. L. (2000). The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 148, 1151-1158. 10.1083/jcb.148.6.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K. and Welti S. (2012). Pleckstrin homology (PH) like domains – versatile modules in protein–protein interaction platforms. FEBS Lett. 586, 2662-2673. 10.1016/j.febslet.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Sebzda E., Bracke M., Tugal T., Hogg N. and Cantrell D. A. (2002). Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 3, 251-258. 10.1038/ni765 [DOI] [PubMed] [Google Scholar]
- Sekine Y., Ikeda O., Tsuji S., Yamamoto C., Muromoto R., Nanbo A., Oritani K., Yoshimura A. and Matsuda T. (2009). Signal-transducing adaptor protein-2 regulates stromal cell-derived factor-1 alpha-induced chemotaxis in T cells. J. Immunol. 183, 7966-7974. 10.4049/jimmunol.0902096 [DOI] [PubMed] [Google Scholar]
- Shimonaka M., Katagiri K., Nakayama T., Fujita N., Tsuruo T., Yoshie O. and Kinashi T. (2003). Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J. Cell Biol. 161, 417-427. 10.1083/jcb.200301133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalenka R. R. and Jessberger R. (2004). SWAP-70 regulates c-kit-induced mast cell activation, cell-cell adhesion, and migration. Mol. Cell. Biol. 24, 10277-10288. 10.1128/MCB.24.23.10277-10288.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J., Petre B. M., Walz T. and Springer T. A. (2002). Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599-611. 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Bi K., Kitamura R., Hong S., Altman Y., Matsumoto A., Tabata H., Lebedeva S., Bushway P. J. and Altman A. (2003). SWAP-70-like adapter of T cells, an adapter protein that regulates early TCR-initiated signaling in Th2 lineage cells. Immunity 18, 403-414. 10.1016/S1074-7613(03)00054-2 [DOI] [PubMed] [Google Scholar]
- Villalba M., Bi K., Rodriguez F., Tanaka Y., Schoenberger S. and Altman A. (2001). Vav1/Rac-dependent actin cytoskeleton reorganization is required for lipid raft clustering in T cells. J. Cell Biol. 155, 331-338. 10.1083/jcb.200107080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Andrian U. H. and Mackay C. R. (2000). T-cell function and migration — Two sides of the same coin. N. Engl. J. Med. 343, 1020-1034. 10.1056/NEJM200010053431407 [DOI] [PubMed] [Google Scholar]
- Wang X., Boyken S. E., Hu J., Xu X., Rimer R. P., Shea M. A., Shaw A. S., Andreotti A. H. and Huang Y. H. (2014). Calmodulin and PI(3,4,5)P(3) cooperatively bind to the Itk pleckstrin homology domain to promote efficient calcium signaling and IL-17A production. Sci. Signal. 7, ra74 10.1126/scisignal.2005147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D. and Lemmon M. A. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13, 677-688. 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Yu B., Martins I. R. S., Li P., Amarasinghe G. K., Umetani J., Fernandez-Zapico M. E., Billadeau D. D., Machius M., Tomchick D. R. and Rosen M. K. (2010). Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 140, 246-256. 10.1016/j.cell.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]