ABSTRACT
The scaffold protein Shoc2 accelerates activity of the ERK1 and ERK2 (ERK1/2, also known as MAPK3 and MAPK1) pathway. Mutations in Shoc2 result in Noonan-like RASopathy, a developmental disorder with a wide spectrum of symptoms. The amplitude of the ERK1/2 signals transduced through the complex is fine-tuned by the HUWE1-mediated ubiquitylation of Shoc2 and its signaling partner RAF-1. Here, we provide a mechanistic basis of how ubiquitylation of Shoc2 and RAF-1 is controlled. We demonstrate that the newly identified binding partner of Shoc2, the (AAA+) ATPase PSMC5, triggers translocation of Shoc2 to endosomes. At the endosomes, PSMC5 displaces the E3 ligase HUWE1 from the scaffolding complex to attenuate ubiquitylation of Shoc2 and RAF-1. We show that a RASopathy mutation that changes the subcellular distribution of Shoc2 leads to alterations in Shoc2 ubiquitylation due to the loss of accessibility to PSMC5. In summary, our results demonstrate that PSMC5 is a new and important player involved in regulating ERK1/2 signal transmission through the remodeling of Shoc2 scaffold complex in a spatially-defined manner.
KEY WORDS: Scaffold, ERK1/2, PSMC5, Remodeling, Shoc2 scaffold
Summary: Our studies describe a new, multi-level paradigm in which allosteric modifications alter the ability of the scaffold protein Shoc2 to actively accelerate transmission of ERK1/2 signals.
INTRODUCTION
Extensive studies of the signaling pathway mediated by extracellular signal-regulated kinases 1 and 2 (ERK1/2, also known as MAPK3 and MAPK1, respectively) have resulted in the identification of a large number of protein–protein interactions as well as multiple substrates, elucidating the complexity of the ERK1/2 pathway (Brown and Sacks, 2008; Chen et al., 2001; Katz et al., 2007). The branching of signals within the ERK1/2 pathway is regulated by scaffold proteins. Scaffolds assemble multi-component interacting modules and target these modules to specific cellular locations, thereby enhancing phosphorylation of specific substrates (Brown and Sacks, 2008, 2009; Pullikuth and Catling, 2007). Recent studies have uncovered surprising dynamics within these complexes (Garbett and Bretscher, 2014). However, the mechanisms by which scaffolds control the ERK1/2 pathway are poorly understood (Bhattacharyya et al., 2006; Brown and Sacks, 2009; Good et al., 2011).
The scaffold protein Shoc2 tethers Ras, RAF-1 and the catalytic subunit of protein phosphatase 1c (PP1c, also known as PPP1CA) to accelerate ERK1/2 signals (Rodriguez-Viciana et al., 2006). Silencing of Shoc2 expression leads to a dramatic decrease in ERK1/2 activity in a myriad of cells and in C. elegans (Rodriguez-Viciana et al., 2006; Sieburth et al., 1998; Yoshiki et al., 2010). The physiological significance of Shoc2 was underlined by studies showing that a mouse endothelial Shoc2 knockout leads to embryonic lethality (Yi et al., 2010). Mutations in Shoc2 affecting either Shoc2 localization (Cordeddu et al., 2009) or the assembly of the Shoc2 scaffold complex (Hannig et al., 2014) result in RASopathy – a congenital syndrome with a spectrum of overlapping symptoms, further emphasizing the importance of Shoc2. We have previously demonstrated that upon activation of the ERK1/2 pathway, Shoc2 translocates from the cytosol to late endosomes and/or multivesicular bodies (MVBs), possibly as part of the spatio-temporal regulation of signaling through the Ras–RAF-1 module (Galperin et al., 2012). We have also reported that the E3 ligase HUWE1 modulates ubiquitylation of Shoc2 and the ubiquitylation of Shoc2-associated RAF-1 (Jang et al., 2014). Our studies suggest that ubiquitylation is utilized as a negative-feedback mechanism modulating the ability of the non-catalytic scaffold Shoc2 to mediate the signaling activity of the ERK1/2 pathway (Jang et al., 2014). Deciphering the mechanisms by which Shoc2 regulates activity of the ERK1/2 pathway is necessary for understanding the physiological function of this essential scaffold.
In this study, we have identified the ATPase PSMC5 as a new component in the Shoc2–Ras–RAF-1 scaffold complex. PSMC5 (also called rtp6 or Sug1) belongs to a functionally diverse protein family of the AAA+ ATPases (for ‘ATPases associated with diverse cellular activities’) (Ferry et al., 2009; Su et al., 2000). A widespread mechanism underlying the functionally diverse AAA+ ATPases is the energy-dependent structural remodeling, unfolding and disassembly of macromolecules and protein complexes. Examples of active remodeling and destabilization catalyzed by AAA+ enzymes include protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction and the regulation of gene expression (Hanson and Whiteheart, 2005; Sauer and Baker, 2011). Although PSMC5 is mainly implicated in proteolysis as a part of a 19S regulatory complex of the 26S proteasome, degradation is not the only fate for a protein substrate that comes in contact with this ATP-dependent unfolding enzyme (Ferry et al., 2009). A growing body of evidence indicates that PSMC5 has a non-proteolytic function and, acting as part of the so-called ‘AAA Proteins Independent of 20S’ (APIS) complex, acts independently from other proteasome subunits (Gonzalez et al., 2002; Makino et al., 1999). For instance, several biochemical and genetic studies have indicated that PSMC5 plays a distinct proteasome-independent role in regulating transcription activation and elongation, DNA repair and chromatin remodeling (Ferdous et al., 2002; Ferry et al., 2009; Gonzalez et al., 2002; Sulahian et al., 2006; Swaffield et al., 1992). PSMC5 is also involved in facilitating misfolding and aggregation of proteins with a poly(Q) expansion in Huntington's disease (Rousseau et al., 2009). These activities rely on the non-proteolytic function of PSMC5 as a remodeling chaperone. The role of PSMC5 in regulating the ERK1/2 cascade has not been previously reported or explored.
Here, we show that PSMC5 modulates the ability of the E3 ligase HUWE1 to ubiquitylate Shoc2 and the Shoc2 signaling partner RAF-1. We also establish that PSMC5 mediates redistribution of the Shoc2 multi-protein module to late endosomes and/or MVBs where it sequesters HUWE1 from the complex. Such remodeling of the complex results in the attenuated ubiquitylation of Shoc2 and RAF-1 with corresponding changes in ERK1/2 activity. Our studies provide evidence that the mislocalized RASopathy mutant of Shoc2 (Ser2Gly) is hyper-ubiquitylated owing to the loss of accessibility to PSMC5. The ability of PSMC5 to control the assembly of Shoc2 complexes provides a new multi-layered paradigm for cross-talk between dynamics within the scaffold complex assembly as well as cellular distribution and the dynamics of ERK1/2 signaling.
RESULTS
Interaction of Shoc2 with PSMC5
Shoc2 interacts with its signaling partners, Ras and RAF-1 through the unstructured N-terminal domain and the leucine-rich curvature of Shoc2 is left available to interact with additional components of the scaffolding module (Jang et al., 2014; Jeoung et al., 2013). To further investigate the composition and dynamics of the Shoc2 scaffold complex, we performed a yeast two-hybrid screen in the human adult and fetal heart library using full-length Shoc2 as a bait as described previously (Jang et al., 2014). Six isolates of the evolutionarily conserved 45 kDa ATPase PSMC5 were identified. All of these isolates were mapped and contained nearly the entire C-terminal ATPase domain (amino acids 244–406) (Fig. 1A).
Fig. 1.
Shoc2 interacts with PSMC5. (A) The ‘bait’ and the ‘prey’ regions of Shoc2 and PSMC5 in the yeast two-hybrid screening. (B) HEK 293FT cells were co-transfected with HA–PSMC5 or HA–M-Ras, and GST–Shoc2. GST–Shoc2 was immunoprecipitated (IP) and analyzed by immunoblotting (IB) using anti-HA and -GST antibodies. (C) HEK 293FT cells were co-transfected with HA–PSMC5 or HA–M-Ras and Shoc2–tRFP. HA–PSMC5 and HA–M-Ras were immunoprecipitated and analyzed using anti-HA and -Shoc2 antibodies. (D) PSMC5 and Shoc2 antibodies were used to detect endogenous PSMC5 in Shoc2 immunoprecipitates of HEK 293FT cells. (E) Recombinant His–Shoc2 was mixed with GST–PSMC5 coupled to glutathione–Sepharose beads. Complexes were analyzed using anti-Shoc2 and -GST antibodies. (F) Cos-SR or -LV1 cells were co-transfected with HA–M-Ras and GST–PSMC5. GST–PSMC5 was immunoprecipitated and analyzed using anti-HA, -GST and -tRFP antibodies.
To validate the interaction of Shoc2 and PSMC5, we ectopically expressed a GST-fused Shoc2 (GST–Shoc2) with the HA-tagged PSMC5 (HA–PSMC5) in HEK 293FT cells. HA-tagged M-Ras was used as a positive control (Fig. 1B). HA–PSMC5 was readily immunoprecipitated with GST–Shoc2. To ensure that the interaction was not due to the GST tag, we also tested tRFP-fused Shoc2 (Shoc2–tRFP) with HA–PSMC5. We again found that Shoc2–tRFP co-precipitated with HA–PSMC5 (Fig. 1C). Using HEK 293FT cells we were able to show that endogenous Shoc2 co-precipitates with endogenous PSMC5, confirming this interaction under more physiological conditions (Fig. 1D). To determine whether Shoc2 and PSMC5 are direct binding partners or interact through a complex, we purified recombinant His-tagged full-length Shoc2 and GST-tagged PSMC5 (GST–PSMC5) that self-associates to form a functional dimer (Zhu et al., 2007). We found that Shoc2 and PSMC5 interact in vitro, demonstrating a direct binding between Shoc2 and PSMC5 (Fig. 1E).
Finally, to determine whether PSMC5 is part of the Shoc2–Ras–RAF-1 signaling complex, we expressed GST–PSMC5 and HA–M-Ras in stable cells either constitutively depleted of Shoc2 (LV1) or depleted and then reconstituted with the shRNA-insensitive Shoc2–tRFP (SR) (Jeoung and Galperin, 2014). As expected, PSMC5 was able to precipitate HA–M-Ras only from cells expressing Shoc2 (SR) (Fig. 1F). Cumulatively, these results indicate that PSMC5 is a previously undescribed interacting partner in the Shoc2–Ras–RAF-1 scaffold complex.
PSMC5 and Shoc2 colocalize on late endosomes and/or MVBs
Knowing that PSMC5 is an ATPase subunit of the 19S proteasome, we next set out to determine whether PSMC5 is involved in the turnover of Shoc2 or any of the Shoc2-interacting partners. Surprisingly, the depletion of PSMC5 in cells had no effect on the protein levels of Shoc2, RAF-1, HUWE1 or PP1c either under steady-state conditions or in cells treated with EGF, suggesting that PSMC5 is not involved in controlling the turnover of the proteins in the Shoc2 complex (Fig. S1). These results prompted us to investigate a possible proteasome-independent function of PSMC5 in the Shoc2 scaffold complex. Given that depletion of PSMC5 affects proteasome activity and thereby might alter functions and levels of various proteins, small interfering RNA (siRNA) depletion of PSMC5 is used conservatively in the following experiments (i.e. only to assess proteins in the Shoc2 complex).
We next compared the subcellular localization of PSMC5 and Shoc2 using live-cell fluorescence microscopy. For studies in living cells we utilized the tagRFP (tRFP)-fusion protein of Shoc2 described in our earlier studies (Galperin et al., 2012). Given that overexpression of PSMC5 does not impair the function of the proteasome (Rousseau et al., 2009), we utilized a previously characterized fluorescently tagged PSMC5 (CFP–PSMC5) (Laporte et al., 2008). Fluorescence microscopy revealed that Shoc2–tRFP and CFP–PSMC5 are distributed in the cytosol (Fig. 2A). Surprisingly, we also found a pool of CFP–PSMC5 and Shoc2–tRFP colocalized in intracellular vesicular structures. These findings were reminiscent of our earlier observations of Shoc2 translocating on endosomes (Fig. S2) (Galperin et al., 2012). Shoc2–tRFP- and CFP–PSMC5-containing vesicles were distributed throughout cells. The majority of these vesicles appeared as larger Shoc2–tRFP-decorated ‘donut-like’ profiles of similar intensity with easily recognized limiting membrane. Nearly all (98%) Shoc2–tRFP-positive vesicles colocalized with PSMC5. Occasionally, we observed PSMC5-positive vesicles that did not contain Shoc2 (Fig. 2A, arrow). Some vesicles showed rapid movement, but most were relatively static with movement over short distances (Movie 1).
Fig. 2.
PSMC5 and Shoc2 colocalize on late endosomes and/or MVBs. (A) Cos-SR cells transfected with CFP–PSMC5 were followed by live-cell fluorescence microscopy. The arrow points to a PSMC5-positive vesicle and does not contain Shoc2–tRFP. (B) Cos-SR cells expressing GST–PSMC5 and CFP–Rab7 were followed by live-cell fluorescence microscopy. (C) Cos-SR cells expressing GST–PSMC5 were fixed, immunostained for TSG101 and followed by immunofluorescence microscopy. Insets show high magnification images of the regions indicated by white rectangles. Scale bars: 10 µm. (D) Post-nuclear supernatants (PNS) from Cos1 cells were layered on 8–42% sucrose gradients and subjected to ultracentrifugation. The indicated proteins were identified by immunoblotting (IB) using specific antibodies in crude endosome (CE) or Golgi and endoplasmic reticulum (G/ER) fractions. (E) Shoc2 or HUWE1 were immunoprecipitated from the crude endosomal or Golgi and ER fractions and analyzed by using specific anti-HUWE1, -RAF-1 and -Shoc2 antibodies. (F) Cos-SR cells expressing control siRNA (siNT), GST–PSMC5 or siRNA targeting PSMC5 (siPSMC5) were followed by live-cell fluorescence microscopy. Insets show high magnification images of the regions indicated by white rectangles. Scale bars: 10 µm. (G) Crude endosomal and Golgi and ER subcellular fractions were prepared from Cos1 cells expressing GST–PSMC5. PSMC5 was immunoprecipitated and analyzed using anti-PSMC5 and -Shoc2 antibodies. (H) Crude endosomal subcellular fractions were prepared from Cos1 cells transiently transfected with non-targeting siRNA (siNT) or PSMC5 siRNA (siPSMC5). The expression of the indicated proteins was analyzed using specific antibodies.
To define the Shoc2- and PSMC5-containing vesicular compartments, Cos1 cells stably depleted of endogenous Shoc2 and expressing Shoc2–tRFP (SR) were transiently transfected with GST–PSMC5 and either CFP–Rab5 or CFP–Rab7, resident proteins of early and late endosomes, respectively. Shoc2 and PSMC5 were found on a population of CFP–Rab7-labeled late endosomes (Fig. 2B), but were not presented on early endosomes labeled with CFP–Rab5 (Fig. S2A). The Shoc2-positive vesicles did not colocalize with LAMP1, a marker of lysosomes (Fig. S2B), suggesting that Shoc2 complexes are not targeted to lysosomes. Interestingly, many of the Shoc2–tRFP- and GST–PSMC5-positive vesicles often contained multi-membrane intra-luminal branches characteristic of large MVBs and in some instances reached 4–5 µm in size (Fig. S2C,D). Therefore, we expressed TSG101–RFP, a marker for MVBs, in cells expressing Shoc2–GFP and GST–PSMC5. Partial colocalization of Shoc2–GFP with TSG101–RFP was observed in live cells (Fig. S2E). To confirm these findings, we examined localization of Shoc2–tRFP and TSG101 by immunofluorescence microscopy in cells expressing GST–PSMC5 (Fig. 2C). Similar to the experiments in living cells, the Shoc2–tRFP- and GST–PSMC5-positive vesicles colocalized with TSG101-positive immunostaining (Fig. 2C). These data indicate that PSMC5–Shoc2 complexes are targeted to a subset of late endosomes and/or MVBs. Our attempts to visualize endogenous Shoc2–PSMC5 complexes using immunofluorescence remained inconclusive due to the highly immunoreactive cytosolic pool of PSMC5 that obscured the endosome-localized pool of endogenous Shoc2–PSMC5 complexes.
To assess the localization and interaction of Shoc2 and PSMC5 on endosomes further, we isolated a crude endosomal (late and early endosomes) fraction of Cos1 cells using a discontinuous sucrose density gradient centrifugation (Fig. 2D) (de Araujo et al., 2008). In these experiments, the crude endosomal fraction was enriched at the 8%–35% interface of the sucrose gradient, whereas the 35%–42% interface contained Golgi and endoplasmic reticulum (ER). To demonstrate that the crude endosomal fraction and the Golgi and ER fraction were free of any significant amount of plasma membrane, samples of equal protein amounts from both interfaces and the post nuclear supernatant (PNS) were subjected to western blot analysis with antibody against Na+/K+ ATPase. We did not find Na+/K+ATPase in the crude endosomal or Golgi and ER fractions, suggesting that both fractions were largely free of plasma membrane (Fig. 2D). To determine the purity of the obtained fractions we examined the distribution of the key markers of early endosomes (EEA1 and Rab5) and lysosomes (LAMP1) as well as EGFR. Rab5 was found only in the crude endosomal fraction, whereas LAMP1 and EEA1 were also present in the Golgi and ER fraction. Given that LAMP1 and EEA1 participate in post-Golgi trafficking events, these observations were not entirely surprising (Caster and Kahn, 2013; Luo et al., 2006).
Next, we analyzed the distribution of Shoc2 and its interacting partners RAF-1 and HUWE1 on the gradient. Shoc2 and RAF-1 were enriched at the crude endosomal interface (Fig. 2D), whereas HUWE1 was present at the crude endosomal and Golgi and ER interfaces (Fig. 2E). The proteins recovered from either the crude endosomal or the Golgi and ER interfaces were then subjected to immunoprecipitation with anti-Shoc2 antibodies. The total amount of homogenate that was used in these experiments was comparable to the amount of total lysate used in immunoprecipitation experiments in Fig. 1. In this assay, Shoc2 precipitated both HUWE1 and RAF-1 from the crude endosomal fraction but not from the Golgi and ER interface (Fig. 2E). Owing to the limitations in the detection of PSMC5 on endosomes, a substantial amount of PSMC5 in Shoc2 precipitates from the crude endosomal fraction was not always detectable. Nevertheless, similar to the results of live-cell microscopy (Fig. 2F), Shoc2–PSMC5 complexes were easily found at the crude endosomal interface of cells expressing the GST-tagged version of PSMC5 (Fig. 2F) and siRNA-mediated knockdown of PSMC5 resulted in decreased levels of Shoc2 protein in the crude endosomal fraction (Fig. 2H). In addition, phosphorylated RAF-1 (pSer338) was also detectable on PSMC5–Shoc2 positive endosomes using immunofluorescence (Fig. S2F). These data confirm our observations in Fig. 2A that Shoc2 and PSMC5 are partially localized on endosomes and indicate that PSMC5 is redistributed to the late endosomes and/or MVBs together with Shoc2. Moreover, these results also raised the possibility that PSMC5 in the Shoc2 complex might modulate the distribution of the Shoc2 complexes to endosomes. In addition, these data suggest that oligomerization of PSMC5 is necessary for targeting Shoc2 to endosomes. This notion is addressed in detail below.
PSMC5 binds to leucine-rich repeat 21 of Shoc2
To better characterize the interaction between Shoc2 and PSMC5 and to determine the structural element(s) that mediate their interaction, a series of the previously validated Shoc2 deletion mutants was utilized (Fig. S3A) (Jeoung et al., 2013). We found that a fragment of Shoc2 corresponding to leucine-rich repeats (LRRs) 20 to 21 (ΔN-19) was sufficient for PSMC5 binding (Fig. S3B, lane 8). These data suggest that the minimal PSMC5 binding domain in Shoc2 is within LRRs 20 to 21, which does not overlap with the binding sites of other previously characterized Shoc2 partners.
When mapping the Shoc2–PSMC5 interaction, we found that the Shoc2–tRFP (ΔN-11) truncation mutant was localized to endosomes together with GST–PSMC5 as efficiently as full-length Shoc2–tRFP (Fig. 3A). Deletion of the C-terminus of Shoc2 (Δ21-C) abolished the distribution of Shoc2 to endosomes. Cumulatively, these results define a PSMC5 binding domain within the LRR21 of Shoc2 and provide evidence that the interaction of Shoc2 with PSMC5 is necessary for the localization of Shoc2 to endosomes.
Fig. 3.
The oligomerization and ATPase activity of PSMC5 are necessary for Shoc2–PSMC5 complex localization on late endosomes and/or MVBs. (A) Cos1 cells co-transfected with GST–PSMC5 and the Shoc2–tRFP truncated mutants depicted in Fig. S3A, were followed by live-cell fluorescence microscopy. (B) Cos-SR cells expressing the GST–PSMC5 mutants depicted in Fig. S3C were fixed, immunostained for GST and followed by immunofluorescence microscopy. (C) Cos-SR cells expressing the CFP–PSMC5 mutants depicted in Fig. S3C were followed by live-cell fluorescence microscopy. Insets show high magnification images of the regions of the cell indicated by white rectangles. Scale bars: 10 µm.
Recruitment of Shoc2–PSMC5 to late endosomes and/or MVBs requires PSMC5 oligomerization and its intact ATPase activity
Next, the functional features crucial for the ability of PSMC5 to facilitate recruitment of Shoc2 to endosomes were determined. We generated a PSMC5 deletion mutant lacking a coiled-coil domain, a region mediating the oligomerization of PSMC5, as well as truncated mutants lacking either the entire N-terminal part (amino acids 1–184) or the ATPase domain (amino acids 185–406) (Fig. S3C) (Bar-Nun and Glickman, 2012; Zhu et al., 2007). Consistent with the results of the yeast two-hybrid analysis (Fig. 1A), immunoprecipitation showed that PSMC5 recognizes Shoc2 through its ATPase domain and that the loss of the coil-coiled domain (ΔCC) in PSMC5 did not affect its interaction with Shoc2 (Fig. S3D). However, in cells expressing the PSMC5 ΔCC mutant we did not observe Shoc2 on endosomes (Fig. 3B). These results confirm that the binding of Shoc2 to PSMC5 is mediated by the region within the ATPase domain, but PSMC5 oligomerization through the coiled-coil domain is necessary for the endosomal localization of the PSMC5–Shoc2 complex.
The biochemistry of the AAA+ ATPases is well defined and the amino acid residues that are required for their ATPase activity in Walker A and Walker B motif of PSMC5 have been identified (Hanson and Whiteheart, 2005). To obtain further insights into the mechanisms of the PSMC5–Shoc2 interaction, we specifically disrupted the ATP binding and hydrolysis of PSMC5 by introducing point mutations changing the essential amino acid residue lysine 196 to methionine (K196M) in the Walker A motif, and glutamic acid 250 to glutamine (E250Q) in the Walker B motif (Koues et al., 2008; Zhu et al., 2007). The K196M mutation disrupts nucleotide binding in PSMC5, whereas the E250Q replacement perturbs the nucleotide hydrolysis of PSMC5 (Fig. S3C). Both the Walker A and Walker B mutants of PSMC5 retained the ability to bind Shoc2 (Fig. S3E). However, PSMC5 and Shoc2 were no longer localized to endosomes in the cells expressing these ATPase-deficient PSMC5 mutants (Fig. 3C). Our findings demonstrate that the PSMC5–Shoc2 interaction is mediated through the ATPase domain of PSMC5, but is independent of its ATPase activity. Taken together, our data indicate that the intact ATPase activity of PSMC5 and its ability to oligomerize are necessary for the effective recruitment of the PSMC5–Shoc2 complex to endosomes.
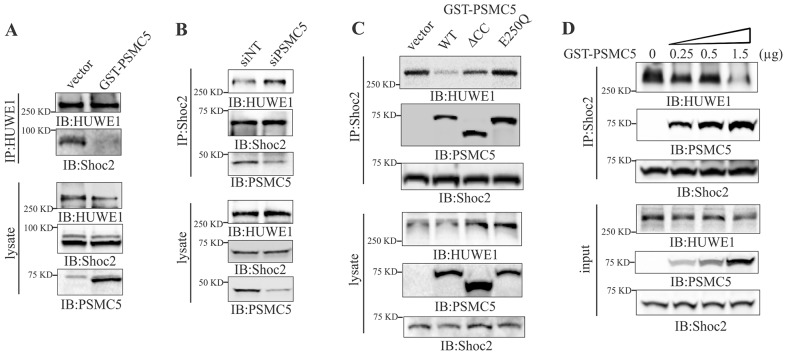
PSMC5 controls the composition of the Shoc2 scaffold complex and levels of ubiquitylation of the proteins in the complex
To perform their biological functions, multi-protein complexes undergo constant changes in their composition as well as in the conformation of the proteins in the complex. PSMC5 is known to play a critical role in remodeling of transcriptional complexes (Ferdous et al., 2002; Gonzalez et al., 2002; Swaffield et al., 1992). Thus, to explore whether PSMC5 regulates the composition of Shoc2 complexes, levels of oligomeric PSMC5 were increased by expressing GST–PSMC5 in cells. In cells with endogenous levels of PSMC5, HUWE1 readily precipitated Shoc2. However, in cells expressing oligomeric PSMC5, the interaction between Shoc2 and HUWE1 was clearly attenuated (Fig. 4A). Conversely, silencing of PSMC5 led to increased amounts of HUWE1 in complex with Shoc2 (Fig. 4B). Unlike the wild-type GST–PSMC5 (WT), expression of GST-tagged mutants of PSMC5 (E250Q and ΔCC) did not result in the loss of HUWE1 from the Shoc2 complex (Fig. 4C). We determined in our previous studies that Shoc2 or HUWE1 do not interact with GST (Jang et al., 2014). Taken together, these data support the hypothesis that PSMC5 plays a role in regulating the composition of the Shoc2 scaffold complexes.
Fig. 4.
PSMC5 modulates the interaction of Shoc2 with HUWE1. (A) Endogenous HUWE1 was immunoprecipitated (IP) from Cos1 cells transfected with GST–PSMC5 using an anti-HUWE1 antibody. The immunoprecipitates were analyzed by immunoblotting (IB) using anti-HUWE1 and -Shoc2 antibodies. (B) Endogenous Shoc2 was immunoprecipitated from Cos1 cells transiently transfected with non-targeting siRNA (siNT) or PSMC5 siRNA (siPSMC5) using anti-Shoc2 antibody. The immunoprecipitates were analyzed with anti-HUWE1, -PSMC5 and -Shoc2 antibodies. (C) Endogenous Shoc2 was immunoprecipitated from Cos1 cells transfected with full-length GST-PSMC5 (WT) or the GST–PSMC5 mutants (-ΔCC or -E250Q) using anti-Shoc2 antibody. The immunoprecipitates were analyzed with anti-HUWE1, -PSMC5 and -Shoc2 antibodies. (D) Lysates of Cos1 cells were mixed with purified in vitro GST–PSMC5 protein (0.25, 0.5 or 1.5 μg). Endogenous Shoc2 was immunoprecipitated using anti-Shoc2 antibody. The immunoprecipitates were analyzed with anti-HUWE1, -PSMC5 and -Shoc2 antibodies.
Next, we combined cellular lysates with GST–PSMC5 protein purified from E. coli and analyzed the HUWE1–Shoc2–PSMC5 complexes in vitro (Fig. 4D). Attenuated binding of Shoc2 and HUWE1 in the presence of increasing amounts of the purified GST–PSMC5 indicated that PSMC5 affects the stoichiometry in the Shoc2 complex and further confirmed that PSMC5 alters the composition of the proteins in the Shoc2 complex.
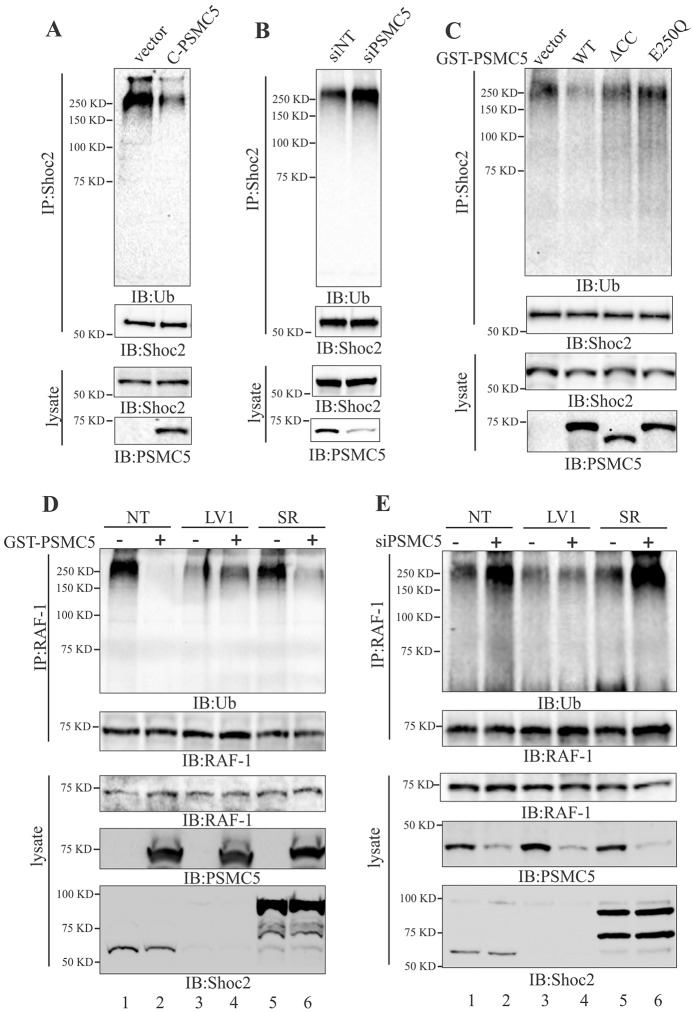
We reported that the ability of Shoc2 to accelerate ERK1/2 activity is regulated through HUWE1-mediated ubiquitylation of Shoc2 and RAF-1 (Jang et al., 2014). Therefore, we examined the effect of oligomeric PSMC5 on Shoc2 ubiquitylation. As shown in Fig. 5A, the ubiquitylation of endogenous Shoc2 was dramatically reduced in cells expressing PSMC5. By contrast, siRNA-mediated silencing of PSMC5 led to a considerable increase in Shoc2 ubiquitylation (Fig. 5B). This increase in Shoc2 ubiquitylation coincided with the increased binding of HUWE1 to Shoc2 (Fig. 4B). Expression of either the E250Q or ΔCC mutants of GST–PSMC5 did not affect the ubiquitylation of Shoc2 (Fig. 5C), further supporting the notion that PSMC5 changes the composition of the Shoc2 complex through sequestering HUWE1.
Fig. 5.
PSMC5 controls levels of ubiquitylation of Shoc2 and RAF-1. (A–C) Shoc2 was immunoprecipitated (IP) from cells transfected with CFP–PSMC5 (A), PSMC5 siRNA (siPSMC5; B), or full-length GST-PSMC5 (WT) or the GST–PSMC5 mutants (ΔCC and E250Q) (C), and its ubiquitylation was detected by immunoblotting (IB) with anti-ubiquitin (Ub) antibody. (D,E) Cos-NT, -LV1 and -SR cells were transiently transfected with GST-PSMC5 (D) or PSMC5 siRNA (E). RAF-1 was precipitated and its ubiquitylation was detected with anti-ubiquitin antibody. The expression of PSMC5, Shoc2 and RAF-1 was analyzed using specific antibodies. siNT, non-targeting siRNA.
Given that Shoc2 is a scaffold that tethers HUWE1 to ubiquitylate RAF-1, we presumed that overexpression of PSMC5 would also affect HUWE1-mediated ubiquitylation of RAF-1. To determine whether ubiquitylation of RAF-1 is impacted, stable cells constitutively depleted of Shoc2 (LV1) and cells depleted and then reconstituted with the shRNA-insensitive Shoc2-tRFP (SR), as well as cells expressing non-targeting (NT) short hairpin RNA (shRNA) were utilized (Galperin et al., 2012). As predicted, PSMC5 overexpression induced a clear decrease in RAF-1 ubiquitylation in cells expressing Shoc2 (NT) (Fig. 5D, lanes 1 and 2), whereas in cells lacking Shoc2 (LV1), expression of PSMC5 had no effect on already reduced basal levels of RAF-1 ubiquitylation (Fig. 5D, lanes 3 and 4). Endogenous Shoc2 reconstituted with Shoc2–tRFP (SR) rescued the ability of PSMC5 to decrease RAF-1 ubiquitylation (Fig. 5D, lanes 5 and 6), demonstrating that the presence of Shoc2 is necessary for PSMC5-controlled RAF-1 ubiquitylation. Accordingly, we demonstrated that silencing of PSMC5 leads to increased ubiquitylation of RAF-1 only in cells expressing Shoc2 (Fig. 5E, lanes 1, 2 and 5, 6). In cells depleted of Shoc2, PSMC5 knockdown did not result in changes in ubiquitylation of RAF-1 (Fig. 5E, lanes 3 and 4). These data further support the notion that PSMC5 sequesters the E3 ligase HUWE1 from the Shoc2 complex, thereby reducing ubiquitylation of the proteins in the module.
Our previous studies have demonstrated that HUWE1-mediated Shoc2 and RAF-1 ubiquitylation attenuates the amplitude of the ERK1/2 phosphorylation (Jang et al., 2014). To assess how PSMC5 affects ERK1/2 activity, we examined ERK1/2 phosphorylation in cells expressing either full-length PSMC5 (WT) or the PSMC5 mutants (E250Q and ΔCC) (Fig. 6A,B). Whereas the full-length PSMC5 (WT) induced a twofold increase in ERK1/2 phosphorylation, with the maximum increase being observed 7 min after the stimulation of cells with EGF (Fig. 6A, lanes 4–6), expression of the ΔCC or E250Q mutants had no effect on ERK1/2 phosphorylation (Fig. 6A, lanes 7–12). Importantly, expression of PSMC5 (WT) in Shoc2-depleted cells (LV1) did not result in increased amplitude of the ERK1/2 phosphorylation (Fig. 6C).
Fig. 6.
PSMC5-modulated ubiquitylation is required to control the ERK1/2 pathway activity. (A,B) Cos1 cells were transfected with full-length GST–PSMC5 (WT) or the GST–PSMC5 mutants (ΔCC and E250Q). At 48 h post-transfection, the cells were serum-starved for 16 h and stimulated with EGF (0.2 ng/ml) for 7 and 15 min. The expression of the indicated proteins was analyzed by immunoblotting (IB) using specific antibodies. A representative blot from three experiments is shown in A. B shows the mean±s.d. (n=3) fold change for phosphorylated ERK1/2 (pERK1/2; 7 min of EGF treatment) normalized to the value for GAPDH in arbitrary units (phosphorylated ERK1/2/GAPDH) (a versus b, P<0.01, ANOVA followed by post hoc Tukey test) (B). (C) Cos-NT and -LV1 cells were transiently transfected with GST–PSMC5. At 48 h post-transfection, the cells were serum-starved for 16 h and stimulated with EGF (0.2 ng/ml) for 7 and 15 min. The expression of the indicated proteins was analyzed using specific antibodies. (D) Cos1 cells were transfected with GST–PSMC5. Cells were treated as described in A for indicated times. Shoc2 was precipitated and its ubiquitylation was detected with anti-ubiquitin (Ub) antibody. The results in each panel are representative of those from three independent experiments.
Next, we examined how GST–PSMC5 affects the ubiquitylation of Shoc2 in response to activation of the EGFR–ERK1/2 pathway. We found that expression of GST–PSMC5 abrogated the EGF-inducible ubiquitylation of Shoc2 (Fig. 6D, lanes 5–8) that is usually seen for Shoc2 (Fig. 6D, lanes 1–4) (Jang et al., 2014). GST–PSMC5 appears to abolish the ability of HUWE1 to catalyze the reversible ubiquitylation of Shoc2. Taken together, these results indicate that alterations in the mechanisms that control the assembly and/or disassembly of Shoc2–RAF-1–HUWE1 complexes impact on the dynamics of the Shoc2 ubiquitylation resulting in an altered amplitude of the ERK1/2 phosphorylation.
Ubiquitylation of Shoc2 is controlled by PSMC5 on endosomes
To gain further insights into the PSMC5-controlled ubiquitylation of Shoc2, we evaluated whether the Shoc2 localized to endosomes is ubiquitylated. As shown in Fig. 7A, Shoc2 precipitated from the enriched crude endosomal fractions was highly ubiquitylated. However, the ubiquitylation of Shoc2 was strikingly decreased in cells expressing GST–PSMC5. No effect on the overall ubiquitylation of the proteins in the crude endosomal fraction was detected (Fig. 7A, lysate).
Fig. 7.
PSMC5 controls ubiquitylation of Shoc2 on endosome. (A) A crude endosomal (CE) fraction was prepared from Cos1 cells expressing GST–PSMC5. Shoc2 was immunoprecipitated (IP) from the crude endosomal fraction and analyzed by immunoblotting (IB) using anti-ubiquitin (Ub) and -Shoc2 antibodies. (B) Shoc2 was immunoprecipitated from cells stably expressing either wild-type (WT) or the S2G mutant of Shoc2–tRFP using anti-RFP antibody. Immunoprecipitates were analyzed using anti-ubiquitin and -RFP antibodies. (C) Cos1 cells stably expressing the Shoc2-S2G mutant (Shoc2-S2G-tR) were transfected with GST–PSMC5. Shoc2 was immunoprecipitated using the anti-Shoc2 antibody. Shoc2 immunoprecipitates were analyzed using anti-ubiquitin and -Shoc2 antibodies. (D) Cos1 cells stably expressing the Shoc2-S2G mutant (Shoc2-S2G-tR) were depleted of HUWE1 by siRNA (siHUWE1). Shoc2 was immunoprecipitated using anti-RFP antibody. The immunoprecipitates were analyzed using anti-ubiquitin and -RFP antibodies. (E) Cos1 cells stably expressing either wild-type Shoc2–tRFP (WT) or the Shoc2-S2G mutant (Shoc2-S2G-tR) were transfected with GST–PSMC5. GST–PSMC5 was immunostained with GST antibody. Cells were fixed and followed by immunofluorescence microscopy. Insets show high magnification images of the regions of the cell indicated by white rectangles. Scale bars: 10 µm.
One of the known Shoc2 mutations found in RASopathy patients leads to protein that is targeted to plasma membrane due to aberrant N-terminal myristoylation (Cordeddu et al., 2009). This S2G mutation precludes Shoc2 from being distributed to the late endosome and/or MVB compartment (Galperin et al., 2012). To examine ubiquitylation of this plasma-membrane-targeted Shoc2 mutant in the background of cells lacking endogenous Shoc2, we generated cells stably expressing the Shoc2–tRFP (S2G) mutant and depleted of endogenous Shoc2 (Shoc2-S2G-tR). Surprisingly, Shoc2-S2G ubiquitylation was noticeably higher than that of WT Shoc2 (Fig. 7B) and the expression of GST–PSMC5 had no effect on the ubiquitylation of the Shoc2-S2G mutant (Fig. 7C). Interestingly, the Shoc2-S2G mutant that had an intact PSMC5-binding motif was not able to precipitate GST–PSMC5 as effectively as WT Shoc2 (Fig. S4A). It is therefore likely that PSMC5 cannot modulate ubiquitylation of the Shoc2-S2G mutant due to its aberrant cellular distribution. To confirm this hypothesis, we examined the ubiquitylation of the Shoc2-S2G mutant in cells depleted of HUWE1. HUWE1 depletion resulted in decreased ubiquitylation of the Shoc2-S2G mutant (Fig. 7D), indicating that HUWE1 is the E3 ligase that ubiquitylates the plasma-membrane-targeted Shoc2. Immunofluorescence analysis of cells expressing either Shoc2-WT or the Shoc2-S2G mutant together with GST–PSMC5 further confirmed our findings as shown in Fig. 7A–D. As seen above, Shoc2-WT was readily recruited to the PSMC5-positive endosomes (Fig. 7E). However, we did not observe colocalization of the Shoc2-S2G mutant and PSMC5 on intracellular compartments, suggesting that PSMC5 controls HUWE1-mediated ubiquitylation of Shoc2 when proteins in the complex are targeted to late endosomes and/or MVBs.
PSMC5 regulates EGFR/RAF-1-induced cell growth and motility
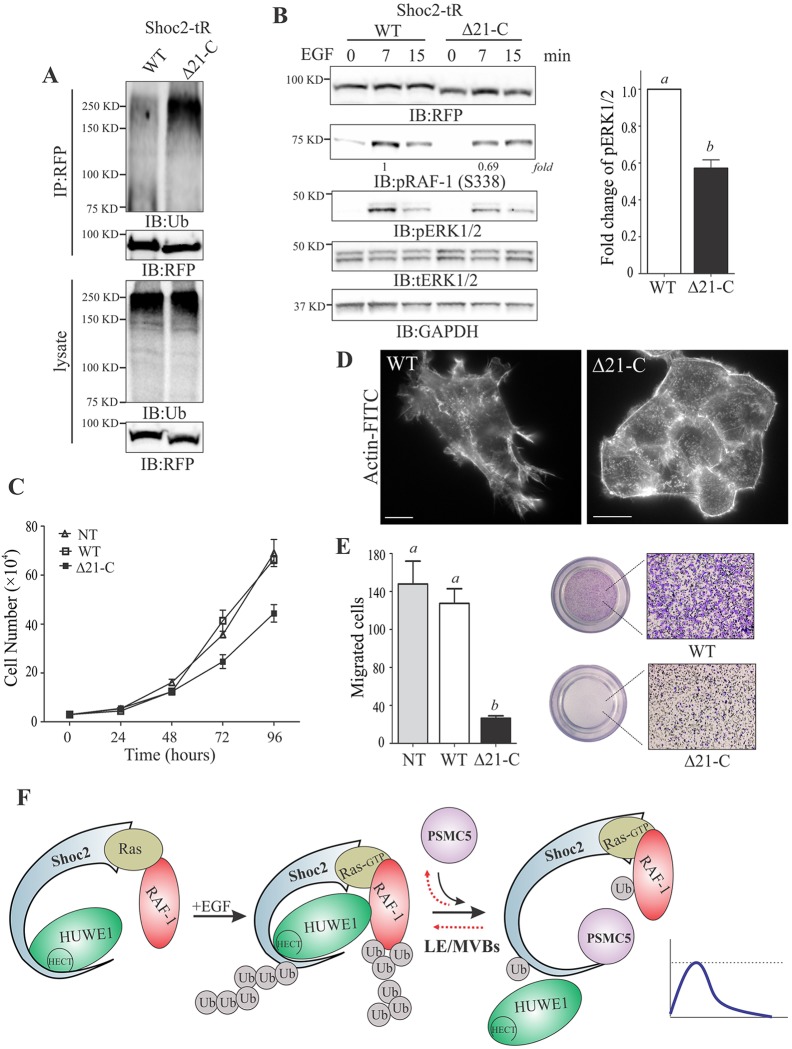
To assess the involvement of PSMC5 in the control of Shoc2-transduced ERK1/2 signals for cellular functions further, we have utilized a knockdown and rescue (KDAR) approach (Jeoung and Galperin, 2014). Here, we used the KDAR approach to constitutively deplete endogenous Shoc2 in HeLa and Cos1 cells and then rescue this with expression of the Shoc2–tRFP mutant lacking the PSMC5-binding domain (Δ21-C) (Figs 3 and 8; Fig. S4B,C). HeLa and Cos1 cells depleted of endogenous Shoc2 and then rescued with expression of the full-length Shoc2–tRFP, as well as cells expressing non-targeting shRNA (NT), were established in our previous studies (Jang et al., 2014; Jeoung and Galperin, 2014). All other counterparts in the Shoc2 complex, including PSMC5, were not affected or manipulated in any way offering a clear interpretation of how the loss of PSMC5 in the Shoc2 complex affects the cellular functions regulated by Shoc2-mediated ERK1/2 signals. The stability of the Shoc2-Δ21-C mutant and its ability to bind Ras and RAF-1 were assessed in our previous studies and found to be comparable to that of the full-length Shoc2 (Jeoung et al., 2013). In order to prevent clonal variations due to the different sites of viral genome incorporation, we utilized pooled populations of cells in the following experiments. First, we examined how the loss of PSMC5 binding affects the ubiquitylation of the Shoc2-Δ21-C mutant. We found that the levels of ubiquitylation of the Shoc2-Δ21-C mutant were dramatically higher than that of full-length Shoc2 (Fig. 8A). We then examined how increased ubiquitylation of the Shoc2-Δ21-C mutant affects its ability to accelerate ERK1/2 signals. The amplitude of RAF-1 phosphorylation at S338 and the phosphorylation of ERK1/2 were significantly lower in cells expressing the Shoc2-Δ21-C mutant than in cells expressing full-length Shoc2 (Fig. 8B). Similar results were observed with HeLa cells expressing either WT or the Shoc2-Δ21-C mutant (Fig. S4B). These findings indicate that loss of the PSMC5 binding leads to augmented ubiquitylation of Shoc2 followed by a decreased amplitude of ERK1/2 phosphorylation. Further analysis revealed that the growth rates of cells expressing the PSMC5-binding-deficient Δ21-C mutant of Shoc2 were markedly decreased as compared to the growth rates of cells expressing WT Shoc2 (Fig. 8C; Fig. S4C). Additionally, we observed that cells expressing the Shoc2-Δ21-C mutant showed an altered morphology and presented with a rounded epithelia-like shape. To assess whether the changes in cell morphology were caused by alterations in formations of lamellipodia and filopodia, we stained for filamentous actin (Fig. 8D). Cells expressing full-length Shoc2 had easily detectable lamellipodia, filopodia and actin stress fibers. However, cells expressing the Δ21-C mutant of Shoc2 lacked protrusions and exhibited a profound cortical actin staining. We then monitored whether changes in the morphology of the cells expressing the Shoc2-Δ21-C mutant led to their altered motility. To measure migration of cells expressing the Shoc2-Δ21-C mutant we used a Transwell assay (Fig. 8E). We found that cells expressing the Shoc2 mutant lacking the PSMC5-binding domain exhibited a markedly decreased directed migration.
Fig. 8.
PSMC5 modulates Shoc2-transduced cell growth and motility. (A) Shoc2 was immunoprecipitated (IP) from Cos1 cells stably expressing either full-length Shoc2-tRFP (WT) or the mutant of Shoc2 (Δ21-C). Shoc2 ubiquitylation was detected with anti-ubiquitin (Ub) antibody. (B) Cos1 cells stably expressing Shoc2-WT–tRFP or the Shoc2-Δ21-C–tRFP mutant were serum-starved and treated with 0.2 ng/ml of EGF for 7 and 15 min. The cell lysates were probed for RFP, phosphorylated RAF-1 (S338) (pRAF-1), phosphorylated ERK1/2 (pERK1/2), total ERK1/2 (tERK) and GAPDH. Blots from the multiple experiments were analyzed. Bars represent the mean±s.d. (n=3) for pERK1/2 (7 min of EGF treatment) normalized to the value for GAPDH in arbitrary units (a versus b, P<0.01, by Student's t-test). (C) Equal numbers of Cos1 cells constitutively depleted of endogenous Shoc2 or expressing either Shoc2-WT–tRFP or the Shoc2-Δ21-C–tRFP mutant as well as control cells (Cos-NT) were plated onto 24-well dishes. The cell numbers were counted 24, 48, 72 and 96 h after seeding. The graph depicts the mean±s.d. number from triplicate experiments. (D) Cos1 cells constitutively expressing either Shoc2–tRFP (WT) or the Shoc2-Δ21-C–tRFP mutant were fixed and labeled with phalloidin–FITC. Scale bars: 10 µm. (E) Cos1 cells constitutively depleted of endogenous Shoc2 and expressing Shoc2-WT–tRFP or the Shoc2-Δ21-C–tRFP mutant as well as control cells (Cos-NT) were allowed to migrate in a Transwell migration chamber coated with collagen for 4 h. The migrated cells were stained with Crystal Violet and counted in three random fields per membrane from three independent experiments. The graph depicts the mean±s.e.m. number from triplicate experiments (a versus b, P<0.01, ANOVA followed by post hoc Tukey test). The results in each panel are representative of those from three independent experiments. Images from the representative experiment are shown. (F) Schematic model of PSMC5 function in Shoc2-mediated ERK1/2 signals. Shoc2 incorporates the E3 ligase HUWE1 and AAA+ ATPase PSMC5 into the Ras and RAF-1 signaling complex. HUWE1 mediates ubiquitylation of Shoc2 and RAF-1 to fine-tune the RAF-1 activity. PSMC5 triggers redistribution of the Shoc2–HUWE1–Ras–RAF-1 complex to late endosomes and/or multivesicular bodies (LE/MVBs) to remodel and reactivate the signaling complex. Steps affected by loss of PSMC5 binding are shown by red dotted arrows. Impaired binding of PSMC5 and endosomal localization of Shoc2 affects mechanisms regulating assembly of the Shoc2 complexes, ultimately changing amplitude of the ERK1/2 activity. The blue line in the graph shown to the right indicates the threshold of ERK1/2 signaling amplitude controlled by Shoc2.
Taken together, the results strongly suggest that PSMC5 plays a key role in the mechanisms by which Shoc2 accelerates ERK1/2 signals. Furthermore, these results suggest that the ability of Shoc2 to cycle between its ubiquitylated and non-ubiquitylated forms is important for its function in transducing ERK1/2 signals for cell migration and growth.
DISCUSSION
The concept of scaffolding complexes being dynamic entities rather than stable complexes has recently garnered increased attention (Langeberg and Scott, 2015; Nussinov et al., 2013). However, very little is known about the dynamics within these scaffold complexes, and how these dynamics might relate to their physiological roles. In our earlier studies, we demonstrated that the E3 ligase HUWE1 provides a negative-feedback mechanism controlling the amplitude of the Shoc2-transduced ERK1/2 activity. These results suggested that ubiquitin modification of a non-catalytic Shoc2 scaffold plays an active role in the ability of Shoc2 to accelerate ERK1/2 activity (Jang et al., 2014). Here, the AAA+ ATPase PSMC5 is shown to be a newly described component in the Shoc2 scaffolding complex that facilitates control over the dynamics of the ERK1/2 signals transmitted through the Shoc2 complex. Our findings demonstrate that, as a part of a feedback-loop mechanism, PSMC5 (1) is recruited to endosomes together with Shoc2, (2) remodels Shoc2 complexes by sequestering the E3 ligase HUWE1 from the Shoc2 complex, and (3) modulates ubiquitylation of Shoc2 and RAF-1 to control the amplitude of the ERK1/2 signals in a spatially defined manner.
PSMC5 is a part of the Shoc2–Ras–RAF-1 signaling complex
PSMC5 is well known for its function as an unfoldase in the 19S regulatory particle of the proteasome (Forster et al., 2010). Furthermore, PSMC5 unfolds proteins in a proteasome-independent manner [e.g. transcription complexes or poly(Q) protein aggregates of Huntingtin] (Rousseau et al., 2009; Sun et al., 2002). Here, we provide compelling evidence that PSMC5 is an integral partner in the Shoc2 scaffolding complex. We show that a pool of endogenous Shoc2 is in the same molecular complex with PSMC5 and M-Ras. We demonstrated that PSMC5 interacts with Shoc2 in vitro, in the absence of other components of the complex, and determined that the Shoc2 C-terminal LRR (LRR 21-C) domain is sufficient for PSMC5 binding to Shoc2 (Figs 1 and 3). We also established that PSMC5 is a part of the Shoc2 signaling complex localized to endosomes (Fig. 2; Fig. S2). Interestingly, another subunit of the proteasome XAPC7 (also known as PSMA7, RC6-1 and HSPC in mammals) has been identified as specifically interacting with the late endosomal protein Rab7 and to be recruited to late endosomes and/or MVBs through this interaction (Dong et al., 2004). XAPC7 has been reported to interfere with the transport of membrane proteins from early to late endosomes without affecting the metabolism of ubiquitylated proteins (Dong et al., 2004).
PSMC5 remodels Shoc2 complexes on endosomes
Our results show that, in the context of the Shoc2 complex, PSMC5 ATPase activity leads to the loss of HUWE1 from the Shoc2 complex. Changes in the stoichiometry of the HUWE1–PSMC5–Shoc2 scaffolding module cause a dramatic shift in the dynamics of Shoc2 ubiquitylation (Figs 5–7) without affecting the stability of the proteins in the complex (Fig. S1). Our data establish that Shoc2 provides a binding surface for PSMC5 and is the core of the HUWE1–PSMC5 feedback loop. Deficiencies in the ATPase activity or the ability of PSMC5 to oligomerize did not affect the Shoc2–PSMC5 interaction (Fig. S3C–E). However, silencing of Shoc2 abrogates the effect of PSMC5 on ubiquitylation of RAF-1, supporting the notion that Shoc2 is the platform that holds multi-protein complexes and allows for the dynamics of the feedback mechanisms regulating ERK1/2 signals.
One of the most intriguing observations of this study is the finding that PSMC5 controls ubiquitylation of Shoc2 in a spatially-restricted manner when the Shoc2–PSMC5 complexes are localized on late endosomes and/or MVBs. The ubiquitylation of the plasma-membrane-localized Shoc2 is not controlled by PSMC5 (Fig. 7). Our results also demonstrate that PSMC5 uses its ATP-driven force to recruit Shoc2 to endosomes. The ATP-hydrolysis- and the oligomerization-deficient mutants of PSMC5 lost their ability to recruit Shoc2 to late endosomes and/or MVBs (Fig. 3B,C). These findings are consistent with our observations that PSMC5 oligomerization and intact ATP hydrolysis are essential to control HUWE1-mediated ubiquitylation of Shoc2 and RAF-1 (Figs 4 and 5), and suggest that endosomes provide the specific microenvironmental requirement for signaling. Moreover, our finding of enlarged PSMC5-positive MVBs indicates that proteins of endocytic machinery regulating membrane fusion are involved, and future studies are needed to fully elucidate the mechanisms that govern Shoc2 signaling complexes. Although late endosomes and MVBs are traditionally regarded as the place of signal attenuation, signaling complexes that promote signal ERK1/2 activation were also found on these compartments (Disanza et al., 2009). For instance, in the case of the adaptor complex p14–mp1 (also known as LAMTOR2–LAMTOR3) (Teis et al., 2002) or neutrophillin-mediated activation of the late-endosomal TrkA receptor (Hisata et al., 2007), late endocytic compartments provide spatially confined intracellular platforms controlling both signal intensity and specificity (Sorkin and Goh, 2009). It is plausible that Shoc2- and PSMC5-containing endosomes are an example of signal compartmentalization to control the intensity of this signal. This notion would be consistent with our previous hypothesis that Shoc2-positive late endosomes and/or MVBs are not associated with the classical ‘signaling’ endosomes but likely represent a specialized population of late endosomes (Galperin et al., 2012).
Functional implications of PSMC5-modulated ubiquitylation of Shoc2
We found that the loss of PSMC5 binding by Shoc2 alters the amplitude of RAF-1 and ERK1/2 phosphorylation (Fig. 8). Moreover, cells expressing the PSMC5-binding-deficient mutant of Shoc2 (Δ21-C) exhibit lower rates of growth and decreased motility, likely due to the aberrations in the Shoc2-mediated ERK1/2 activity. These data indicated that the Shoc2 module incorporates the AAA+ PSMC5 unfoldase to provide an additional layer of control over the amplitude of the ERK1/2 signals transmitted through the module. HUWE1-mediated ubiquitylation of Shoc2 and RAF-1 allows for the dynamic range of RAF-1 activity to be fine-tuned, whereas PSMC5 controls the ability of HUWE1 to modify the noncatalytic scaffold Shoc2 thereby actively monitoring assembly and, possibly, stability of molecules in the complex. We have yet to understand whether PSMC5-induced changes in the composition of the Shoc2 complex ultimately lead to changes in Shoc2 conformation and dissociation of HUWE1 from the complex or, alternatively, in the active ATP-dependent unfolding of HUWE1 and its dissociation from the Shoc2 complex. It is tempting to speculate that loss of HUWE1 from the complex and de-ubiquitylation of Shoc2 elicits a conformational change in the scaffold that ‘reactivates’ the ability of Shoc2 to positively regulate ERK1/2 cascade signal transmission. Given that PSMC5 was found to be associated with HUWE1 (Besche et al., 2009) as well as being able to interact directly with another HECT E3-ligase, Ufd4p (Xie and Varshavsky, 2000), unfolding of HUWE1 by PSMC5 is also plausible. Future studies exploring the direct interaction between HUWE1 and PSMC5 in the context of the Shoc2 complex will resolve these questions and provide important mechanistic details on active remodeling within the Shoc2 complex.
In this study, we provide new evidence that ubiquitin modification of a non-catalytic Shoc2 scaffold plays an active role in controlling Shoc2 complex assembly and ERK1/2 activity. We also demonstrate that, by coupling Ras–RAF-1 phosphorylation signals with Shoc2 ubiquitylation, the ubiquitin machinery governs assembly of the Shoc2 complex. Therefore, we propose a model that is based on what is currently understood for the mechanisms modulating the Shoc2-mediated ERK1/2 signals (Fig. 8F). This model posits that, in the context of a Shoc2 scaffolding platform, activation of the ERK1/2 pathway induces HUWE1-mediated ubiquitylation of Shoc2 and the subsequent ubiquitylation of RAF-1. These modifications of Shoc2 and RAF-1 lead to changes in the amplitude of ERK1/2 signaling. The mechanical ATP-fueled force of PSMC5 then triggers sequestration of HUWE1 from the complex. Our current model also suggests that remodeling of the Shoc2 complex occurs upon targeting the complex to late endosomes and/or MVBs. Given that changes in Shoc2 ubiquitylation represent a very dynamic process, it will be important to identify the deubiquitinating enzyme assisting in the ‘reactivation’ of the Shoc2 module. It is conceivable that additional Shoc2 partners contribute to the process of the complex remodeling. Additionally, it is possible that PSMC5-containing Shoc2 complexes are pre-assembled in cells and growth factor stimuli trigger activation of PSMC5 and complex remodeling. Future studies of Shoc2 complexes will resolve the basis for this hypothesis and identify additional proteins regulating dynamics within the scaffold complex as well as the full extent of the mechanisms by which signals transmitted through the complex are regulated.
It is clear, however, that uncontrolled ERK1/2 activity transduced through the Shoc2 complex leads to changes in cellular behavior (i.e. altered cell growth and motility) (Fig. 8) and dramatic consequences during embryonic development (e.g. Noonan-like RASopathy). Given that developmental processes crucially rely on tightly controlled signaling activities, aberrations in the mechanisms that control the timing and amplitude of ERK1/2 signals will have important implications in development. Not surprisingly, alterations in this intricate mechanism due to the Shoc2-S2G mutation cause RASopathy with pathological conditions ranging from distinctive craniofacial dysmorphisms and a wide spectrum of congenital heart defects to variable neurocognitive impairments, brain anomalies and Moyamoya syndrome (Capalbo et al., 2012; Choi et al., 2015; Gripp et al., 2013; Hoban et al., 2012).
In summary, these studies are the first to identify the AAA+ ATPase PSMC5 as a part of ERK1/2 signaling and provide insights into the new mechanism modulating ubiquitylation of Shoc2 and RAF-1, and ERK1/2 activity, by controlling the assembly of signaling scaffold complexes. Ours is also the first report demonstrating a role for the PSMC5 ATPase in remodeling signaling complexes in a mammalian system and suggesting that this role in controlling post-translational modification is evolutionarily conserved.
MATERIALS AND METHODS
Antibodies and other reagents
EGF was obtained from BD Biosciences. Antibodies against the following proteins were used: GST, RAF-1, HA, PSMC5, GAPDH, phosphorylated ERK1/2 (pERK1/2) and EGFR (Santa Cruz Biotechnology); Shoc2 (Proteintech); EEA1 and phosphorylated RAF-1 (pRAF-1; Cell Signaling); Rab5 (BD Biosciences); RFP and Na+/K+ ATPase (Thermo Scientific); FLAG (SydLabs); HUWE1 (Bethyl); ubiquitin (Covance); PP1c (Millipore); Rpt1 (Enzo); LAMP1 (DSHB); TSG101 (GeneTex); anti-Cyclin B1 antibody was provided by Tianyan Gao (University of Kentucky, Lexington, KY).
Yeast two-hybrid screening assays
Full-length human Shoc2 was cloned into the lexA vector pB27 as an N-LexA-Shoc2-C fusion and screened against a human embryo ventricle and heart prey cDNA library. Yeast two-hybrid screens were performed by Hybrigenics SA (http://www.hybrigenics-services.com).
Cell culture, constructs and transfections
HEK 293FT cells (Invitrogen), HeLa (ATCC), Cos1 (ATCC), and stable cell lines (NT, LV1, SR, S2G, Δ21-C) (derivative of Cos1 cells) were grown in DMEM (Sigma) containing 10% FBS. Shoc2–tRFP and its mutants were described previously (Galperin et al., 2012; Jeoung et al., 2013). The plasmid carrying full-length PSMC5 was obtained from Open Biosystems (Thermo Scientific). The transfections of DNA constructs were performed using TransIT® (Mirus Bio LLC) reagents. siRNA transfections were performed as described previously (Jang et al., 2014). The siRNA sequences used to target the PSMC5 and HUWE1 transcripts were described previously (Jang et al., 2014; Koues et al., 2008).
Sucrose gradient subcellular fractionation
Sucrose gradient subcellular fractions were prepared as described (de Araujo et al., 2008). Briefly, Cos1 cells were grown on 15-cm dishes, washed and scraped with a rubber policeman in cold PBS. The cells were then pelleted, resuspended in homogenization buffer (250 mM sucrose, 3 mM imidazole pH 7.4 containing protease and phosphatase inhibitors) and homogenized. Homogenization was carried out until ∼90% of cells were broken without major breakage of the nucleus, as monitored by microscopy. The samples were centrifuged for 10 min at 2000 g at 4°C and the resulting supernatant was designated as the post nuclear supernatant (PNS). The PNS were adjusted to 40.6% sucrose concentration and then overlaid with 1.5 volumes of 35% sucrose and the rest of the tube was filled with 8.6% sucrose. Sucrose gradients were centrifuged for 6 h at 100,000 g at 4°C and the crude endosomal fraction and Golgi and ER membrane fraction were collected.
Denaturing immunoprecipitation for in vivo ubiquitylation assay, immunoprecipitation and western blot analysis
Denaturing immunoprecipitation, immunoprecipitation and western blot analysis were performed as described previously (Jang et al., 2014).
Interaction of recombinant proteins
GST- and His-tagged proteins were affinity purified and stored in PBS and 10% glycerol. Equal aliquots of PSMC5 coupled to glutathione–Sepharose beads were then incubated with recombinant His–Shoc2 at 4°C for 2 h. Beads were washed four times with cell lysis buffer and eluted with 2× Laemmli sample buffer.
Immunofluorescence staining and analysis
Cell immunostaining, image acquisition and analysis were described previously (Galperin et al., 2012). For immunostaining with anti-TSG101, anti-GST and anti-pRAF-1 antibodies cells were permeabilized with 0.05% saponin prior to cell fixation with 3% paraformaldehyde. For actin staining, cells were fixed with 4% paraformaldehyde, permeabilized using 0.1% Triton for 1 min and labeled with phalloidin–FITC for 20 min at room temperature.
Migration assay
Migration assays were performed using 8.0-μm pore 24-well TC inserts (Greiner Bio-one, Monroe, NC). Filters were coated with 15 μg/ml collagen at 37°C for 30 min. Cells were trypsinized and collected with serum-free medium containing soybean trypsin inhibitor (1 mg/ml). Cells were centrifuged (500 g for 5 min), washed and resuspended in serum-free medium. Cells (5×104) were then added to the upper chamber and the lower chamber was filled with complete medium with 10% serum. Cells were allowed to migrate at 37°C for 4 h. After removing non-migrated cells, membranes were fixed in methanol and stained with 1% Crystal Violet. Migrated cells were counted in three random fields per membrane under the microscope at ×20. Each assay was repeated more than three times.
Cell growth
Equal number of cells were seeded onto 24-well plates at a density of 3×104 cells per well in the appropriate culture medium with supplements. Cells were trypsinized and counted at 24, 48, 72 and 96 h after seeding using a cell counter (TC10™ Automated Cell Counter, Bio-Rad, Inc.).
Statistical analysis
Results are expressed as means±s.d. The statistical significance of the differences between groups was determined using either Student's t-test or one-way ANOVA (followed by the Tukey's test). P<0.05 was considered statistically significant. All statistical analyses were carried out using GraphPad Prism (GraphPad Software).
Acknowledgements
We thank Drs Matthew Gentry, Tianyan Gao, Louis Hersh, Charles Waechter, Shoshana Bar-Nun and Stacy Smith for providing reagents and critical reading of the manuscript; the Genetic Technologies Core at the Department of Molecular and Cellular Biochemistry (University of Kentucky) for assistance with the production of lentiviruses; the Protein Core at the Department of Molecular and Cellular Biochemistry (University of Kentucky) for assistance with protein purification; the UK Flow Cytometry & Cell Sorting core facility for assistance in cell sorting.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
E.G. and E.R.J. planned the experiments, analyzed and interpreted data. E.R.J., H.I.J. and P.S. performed all biochemical assays. E.G. collected and analyzed fluorescence microscopy data. G.P. purified lentiviruses to support biochemical assays. M.J. performed transwell assays. E.G. and E.R.J. drafted the manuscript.
Funding
This project was supported by grants from the National Cancer Institute [grant number R00CA126161 to E.G.]; the National Institute of General Medical Sciences (NIGMS) [grant number P20GM103486] (formally supported by the Center for Research Resources); the National Institute of General Medical Sciences [grant number R01GM113087 to E.G.]; the American Cancer Society [grant number RSG-14-172-01-CSM to E.G.]; and from American Heart Association [grant number 15PRE25090207 to H.J.]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the NIGMS. The Genetic Technologies and Protein cores mentioned above are supported in part by a grant from the National Institute of General Medical Sciences [grant number P20GM103486]. The UK Flow Cytometry & Cell Sorting core facility is supported in part by the Office of the Vice President for Research, the Markey Cancer Center and an NCI Center Core Support Grant [grant number P30CA177558 to the University of Kentucky Markey Cancer Center]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.177543/-/DC1
References
- Bar-Nun S. and Glickman M. H. (2012). Proteasomal AAA-ATPases: structure and function. Biochim. Biophys. Acta 1823, 67-82. 10.1016/j.bbamcr.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Besche H. C., Haas W., Gygi S. P. and Goldberg A. L. (2009). Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48, 2538-2549. 10.1021/bi802198q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R. P., Reményi A., Good M. C., Bashor C. J., Falick A. M. and Lim W. A. (2006). The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311, 822-826. 10.1126/science.1120941 [DOI] [PubMed] [Google Scholar]
- Brown M. D. and Sacks D. B. (2008). Compartmentalised MAPK pathways. Handb. Exp. Pharmacol. 186, 205-235. 10.1007/978-3-540-72843-6_9 [DOI] [PubMed] [Google Scholar]
- Brown M. D. and Sacks D. B. (2009). Protein scaffolds in MAP kinase signalling. Cell Signal. 21, 462-469. 10.1016/j.cellsig.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo D., Scala M. G., Melis D., Minopoli G., Improda N., Palamaro L., Pignata C. and Salerno M. (2012). Clinical Heterogeneity in two patients with Noonan-like Syndrome associated with the same SHOC2 mutation. Ital. J. Pediatr. 38, 48 10.1186/1824-7288-38-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caster A. H. and Kahn R. A. (2013). Recruitment of the Mint3 adaptor is necessary for export of the amyloid precursor protein (APP) from the Golgi complex. J. Biol. Chem. 288, 28567-28580. 10.1074/jbc.M113.481101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C. and Cobb M. H. (2001). MAP kinases. Chem. Rev. 101, 2449-2476. 10.1021/cr000241p [DOI] [PubMed] [Google Scholar]
- Choi J.-H., Oh M.-Y., Yum M.-S., Lee B. H., Kim G.-H. and Yoo H.-W. (2015). Moyamoya syndrome in a patient with noonan-like syndrome with loose anagen hair. Pediatr. Neurol. 52, 352-355. 10.1016/j.pediatrneurol.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Cordeddu V., Di Schiavi E., Pennacchio L. A., Ma'ayan A., Sarkozy A., Fodale V., Cecchetti S., Cardinale A., Martin J., Schackwitz W. et al. (2009). Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 41, 1022-1026. 10.1038/ng.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo M. E., Huber L. A. and Stasyk T. (2008). Isolation of endocitic organelles by density gradient centrifugation. Methods Mol. Biol. 424, 317-331. 10.1007/978-1-60327-064-9_25 [DOI] [PubMed] [Google Scholar]
- Disanza A., Frittoli E., Palamidessi A. and Scita G. (2009). Endocytosis and spatial restriction of cell signaling. Mol. Oncol. 3, 280-296. 10.1016/j.molonc.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Chen W., Welford A. and Wandinger-Ness A. (2004). The proteasome alpha-subunit XAPC7 interacts specifically with Rab7 and late endosomes. J. Biol. Chem. 279, 21334-21342. 10.1074/jbc.M401022200 [DOI] [PubMed] [Google Scholar]
- Ferdous A., Kodadek T. and Johnston S. A. (2002). A nonproteolytic function of the 19S regulatory subunit of the 26S proteasome is required for efficient activated transcription by human RNA polymerase II. Biochemistry 41, 12798-12805. 10.1021/bi020425t [DOI] [PubMed] [Google Scholar]
- Ferry C., Gianni M., Lalevee S., Bruck N., Plassat J.-L., Raska I. Jr, Garattini E. and Rochette-Egly C. (2009). SUG-1 plays proteolytic and non-proteolytic roles in the control of retinoic acid target genes via its interaction with SRC-3. J. Biol. Chem. 284, 8127-8135. 10.1074/jbc.M808815200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F., Lasker K., Nickell S., Sali A. and Baumeister W. (2010). Toward an integrated structural model of the 26S proteasome. Mol. Cell. Proteomics 9, 1666-1677. 10.1074/mcp.R000002-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin E., Abdelmoti L. and Sorkin A. (2012). Shoc2 is targeted to late endosomes and required for Erk1/2 activation in EGF-stimulated cells. PLoS ONE 7, e36469 10.1371/journal.pone.0036469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D. and Bretscher A. (2014). The surprising dynamics of scaffolding proteins. Mol. Biol. Cell 25, 2315-2319. 10.1091/mbc.E14-04-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Delahodde A., Kodadek T. and Johnston S. A. (2002). Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296, 548-550. 10.1126/science.1069490 [DOI] [PubMed] [Google Scholar]
- Good M. C., Zalatan J. G. and Lim W. A. (2011). Scaffold proteins: hubs for controlling the flow of cellular information. Science 332, 680-686. 10.1126/science.1198701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp K. W., Zand D. J., Demmer L., Anderson C. E., Dobyns W. B., Zackai E. H., Denenberg E., Jenny K., Stabley D. L. and Sol-Church K. (2013). Expanding the SHOC2 mutation associated phenotype of Noonan syndrome with loose anagen hair: structural brain anomalies and myelofibrosis. Am. J. Med. Genet. A 161, 2420-2430. 10.1002/ajmg.a.36098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig V., Jeoung M., Jang E. R., Phillips J. A. III and Galperin E. (2014). A Novel SHOC2 variant in rasopathy. Hum. Mutat. 35, 1290-1294. 10.1002/humu.22634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I. and Whiteheart S. W. (2005). AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519-529. 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- Hisata S., Sakisaka T., Baba T., Yamada T., Aoki K., Matsuda M. and Takai Y. (2007). Rap1-PDZ-GEF1 interacts with a neurotrophin receptor at late endosomes, leading to sustained activation of Rap1 and ERK and neurite outgrowth. J. Cell Biol. 178, 843-860. 10.1083/jcb.200610073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban R., Roberts A. E., Demmer L., Jethva R. and Shephard B. (2012). Noonan syndrome due to a SHOC2 mutation presenting with fetal distress and fatal hypertrophic cardiomyopathy in a premature infant. Am. J. Med. Genet. A 158A, 1411-1413. 10.1002/ajmg.a.35318 [DOI] [PubMed] [Google Scholar]
- Jang E. R., Shi P., Bryant J., Chen J., Dukhante V., Gentry M. S., Jang H., Jeoung M. and Galperin E. (2014). HUWE1 is a molecular link controlling RAF-1 activity supported by the Shoc2 scaffold. Mol. Cell. Biol. 34, 3579-3593. 10.1128/mcb.00811-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung M. and Galperin E. (2014). Visualizing of signaling proteins on endosomes utilizing knockdown and reconstitution approach. Methods Enzymol. 534, 47-63. 10.1016/B978-0-12-397926-1.00003-2 [DOI] [PubMed] [Google Scholar]
- Jeoung M., Abdelmoti L., Jang E. R., Vander Kooi C. W. and Galperin E. (2013). Functional integration of the conserved domains of Shoc2 scaffold. PLoS ONE 8, e66067 10.1371/journal.pone.0066067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M., Amit I. and Yarden Y. (2007). Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta 1773, 1161-1176. 10.1016/j.bbamcr.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koues O. I., Dudley R. K., Truax A. D., Gerhardt D., Bhat K. P., McNeal S. and Greer S. F. (2008). Regulation of acetylation at the major histocompatibility complex class II proximal promoter by the 19S proteasomal ATPase Sug1. Mol. Cell. Biol. 28, 5837-5850. 10.1128/MCB.00535-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeberg L. K. and Scott J. D. (2015). Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 16, 232-244. 10.1038/nrm3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Salin B., Daignan-Fornier B. and Sagot I. (2008). Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181, 737-745. 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Nakatsu F., Furuno A., Kato H., Yamamoto A. and Ohno H. (2006). Visualization of the post-Golgi trafficking of multiphoton photoactivated transferrin receptors. Cell Struct. Funct. 31, 63-75. 10.1247/csf.31.63 [DOI] [PubMed] [Google Scholar]
- Makino Y., Yoshida T., Yogosawa S., Tanaka K., Muramatsu M. and Tamura T.-a. (1999). Multiple mammalian proteasomal ATPases, but not proteasome itself, are associated with TATA-binding protein and a novel transcriptional activator, TIP120. Genes Cells 4, 529-539. 10.1046/j.1365-2443.1999.00277.x [DOI] [PubMed] [Google Scholar]
- Nussinov R., Ma B. and Tsai C.-J. (2013). A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochim. Biophys. Acta 1834, 820-829. 10.1016/j.bbapap.2012.12.014 [DOI] [PubMed] [Google Scholar]
- Pullikuth A. K. and Catling A. D. (2007). Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell Signal. 19, 1621-1632. 10.1016/j.cellsig.2007.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Oses-Prieto J., Burlingame A., Fried M. and McCormick F. (2006). A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol. Cell 22, 217-230. 10.1016/j.molcel.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Rousseau E., Kojima R., Hoffner G., Djian P. and Bertolotti A. (2009). Misfolding of proteins with a polyglutamine expansion is facilitated by proteasomal chaperones. J. Biol. Chem. 284, 1917-1929. 10.1074/jbc.M806256200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T. and Baker T. A. (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587-612. 10.1146/annurev-biochem-060408-172623 [DOI] [PubMed] [Google Scholar]
- Sieburth D. S., Sun Q. and Han M. (1998). SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell 94, 119-130. 10.1016/S0092-8674(00)81227-1 [DOI] [PubMed] [Google Scholar]
- Sorkin A. and Goh L. K. (2009). Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 315, 683-696. 10.1016/j.yexcr.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Su K., Yang X., Roos M. D., Paterson A. J. and Kudlow J. E. (2000). Human Sug1/p45 is involved in the proteasome-dependent degradation of Sp1. Biochem. J. 348, 281-289. 10.1042/bj3480281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulahian R., Sikder D., Johnston S. A. and Kodadek T. (2006). The proteasomal ATPase complex is required for stress-induced transcription in yeast. Nucleic Acids Res. 34, 1351-1357. 10.1093/nar/gkl012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Johnston S. A. and Kodadek T. (2002). Physical association of the APIS complex and general transcription factors. Biochem. Biophys. Res. Commun. 296, 991-999. 10.1016/S0006-291X(02)02026-0 [DOI] [PubMed] [Google Scholar]
- Swaffield J. C., Bromberg J. F. and Johnston S. A. (1992). Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activation domain in GAL4. Nature 357, 698-700. 10.1038/357698a0 [DOI] [PubMed] [Google Scholar]
- Teis D., Wunderlich W. and Huber L. A. (2002). Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3, 803-814. 10.1016/S1534-5807(02)00364-7 [DOI] [PubMed] [Google Scholar]
- Xie Y. and Varshavsky A. (2000). Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. USA 97, 2497-2502. 10.1073/pnas.060025497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Chen M., Wu X., Yang X., Xu T., Zhuang Y., Han M. and Xu R. (2010). Endothelial SUR-8 acts in an ERK-independent pathway during atrioventricular cushion development. Dev. Dyn. 239, 2005-2013. 10.1002/dvdy.22343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiki S., Matsunaga-Udagawa R., Aoki K., Kamioka Y., Kiyokawa E. and Matsuda M. (2010). Ras and calcium signaling pathways converge at Raf1 via the Shoc2 scaffold protein. Mol. Biol. Cell 21, 1088-1096. 10.1091/mbc.E09-06-0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Wani G., Yao J., Patnaik S., Wang Q.-E., El-Mahdy M. A., Praetorius-Ibba M. and Wani A. A. (2007). The ubiquitin–proteasome system regulates p53-mediated transcription at p21waf1 promoter. Oncogene 26, 4199-4208. 10.1038/sj.onc.1210191 [DOI] [PubMed] [Google Scholar]