Highlights
-
•
Loline and ergot alkaloids are fundamental for Clavicipitaceae.
-
•
These secondary metabolism clusters are found in other, far related, species.
-
•
Phylogenetics establish a close relationship between clusters of different species.
-
•
Detection of large amounts of gene loss supports the HGT events.
Keywords: Clavicipitaceae, HGT, Loline, Ergot alkaloids, Phylogenetics, Phylome
Abstract
Clavicipitaceae is a fungal group that comprises species that closely interact with plants as pathogens, parasites or symbionts. A key factor in these interactions is the ability of these fungi to synthesize toxic alkaloid compounds that contribute to the protection of the plant host against herbivores. Some of these compounds such as ergot alkaloids are toxic to humans and have caused important epidemics throughout history. The gene clusters encoding the proteins responsible for the synthesis of ergot alkaloids and lolines in Clavicipitaceae have been elucidated. Notably, homologs to these gene clusters can be found in distantly related species such as Aspergillus fumigatus and Penicillium expansum, which diverged from Clavicipitaceae more than 400 million years ago. We here use a phylogenetic approach to analyze the evolution of these gene clusters. We found that the gene clusters conferring the ability to synthesize ergot alkaloids and loline emerged first in Eurotiomycetes and were then likely transferred horizontally to Clavicipitaceae. Horizontal gene transfer is known to play a role in shaping the distribution of secondary metabolism clusters across distantly related fungal species. We propose that HGT events have played an important role in the capability of Clavicipitaceae to produce two key secondary metabolites that have enhanced the ability of these species to protect their plant hosts, therefore favoring their interactions.
1. Introduction
Clavicipitaceae is a fungal clade within the Sordariomycetes. Species belonging to this clade are known to interact mainly with plants and insects. Interactions with plants range from parasitic or pathogenic, to symbiotic relationships (Schardl et al., 2013c). Their potential to interact with plants has been associated to the ability of these fungi to produce certain secondary metabolites, among which several types of alkaloids are considered of key importance. Indeed the production of alkaloids, which are toxic to mammals and insects, has been proposed to provide plant protection against herbivores (Schardl et al., 2004, Wäli et al., 2013). Alkaloids produced by Clavicipitaceae species include, among others, ergot alkaloids, lolines and indole-terpenes (Schardl et al., 2013a). Different species, and even different strains, of Clavicipitaceae are able to synthesize different combinations of these compounds though they rarely produce all of them (Schardl et al., 2013b).
Probably the best known alkaloid class produced by Clavicipitaceae are the ergot alkaloids. These compounds are toxic to humans and livestock and have been the cause of many epidemics during human history (Lee, 2009). However, the incidence of ergotism in humans is currently very low and usually related to overdose of drugs derived from ergot alkaloids (Strickland et al., 2011). Contrary to the situation in humans, livestock is still often exposed to toxic alkaloids produced by Clavicipitaceae. Indeed pasture grasses are often colonized by endophytic fungi that are able to synthesize ergot alkaloids. In the United States annual losses in cattle production due to ergot alkaloid intoxication are estimated to be of the order of 1 billion dollars (Strickland et al., 2011). In contrast, loline alkaloids have rarely been linked to intoxications in mammals. Instead, they are broad spectrum insecticides (Schardl et al., 2007).
The gene clusters coding for the enzymes for the synthesis of ergot alkaloids and loline were discovered in Claviceps purpurea (Haarmann et al., 2005, Tudzynski et al., 1999) and in Epichloë uncinata (Spiering et al., 2005), respectively. Subsequent sequencing of numerous additional Clavicipitaceae genomes has shown that there is a tight relationship between the presence of the cluster and the production of these metabolites (Schardl et al., 2013b). Cluster presence and absence is very variable across species, and even across different strains of the same species. For instance, of the two sequenced Epichloë festucae genomes, only one contains the loline cluster and is able to produce loline, while the other contains a cluster to synthesize indole-diterpenes (Schardl et al., 2013b). Differences in gene order and gene content can also be observed within a cluster. This high variability notwithstanding, there is generally a conserved core set of genes that are necessary to form the first stable compound of the pathway. This core set of genes is present in all species able to synthesize the compound. In addition to these core genes, there can be a variable number of accessory genes that participate in forming derivative compounds from the first stable compound. It is the variation in these accessory genes which provides the bulk of variability among the compounds synthesized by the clusters (Schardl et al., 2013b).
The synthesis of alkaloid compounds is not limited to Clavicipitaceae species. Ergot alkaloid production, for instance, has been detected in the distantly related Aspergillus fumigatus (Li and Unsöld, 2006, Panaccione and Coyle, 2005). A cluster containing 14 genes of which 8 were homologous to the genes found in the C. purpurea gene cluster was found in A. fumigatus (Coyle and Panaccione, 2005). As suggested by the differences in the specific gene content of the clusters, A. fumigatus and Clavicipitaceae species synthesize only a few common ergot alkaloid compounds, while most compounds are specific for each group of species. Despite this, it is thought that the two groups of species use orthologous genes to catalyze the first steps in the pathway and that differences affect later steps. More recently, an homologous loline cluster was found in the genome of Penicillium expansum (Ballester et al., 2015). Horizontal gene transfer (HGT) is known to play a role in the evolution of genetic clusters involved in the production of secondary metabolites (Wisecaver and Rokas, 2015). This process could also explain the appearance of similar gene clusters across these two distant groups of species. In order to assess whether HGT played a role in the evolutionary history of the gene clusters responsible to synthesize loline and ergot alkaloids, we performed a comprehensive phylogenetic analysis.
2. Materials and methods
2.1. Fungal genomes included
We downloaded 15 Clavicipitaceae genomes from NCBI (http://www.ncbi.nlm.nih.gov/). The latest version of their proteomes was downloaded from the University of Kentucky (http://www.endophyte.uky.edu/) (Schardl et al., 2013b). Two additional Clavicipitaceae were included from Uniprot. In addition 18 other fungal genomes were selected for comparative purposes. Among these genomes there were four additional Sordariomycetes, eight Eurotiomycetes, including P. expansum and A. fumigatus, two Leotiomycetes, two Dothideomycetes and one outgroup species. Details are found in Supplementary Table S1.
2.2. Phylogenetic analysis of alkaloid gene clusters
The proteins encoded in the gene clusters that synthesize ergot alkaloids and loline were downloaded from UniProt. The list of genes can be found in Supplementary Table S2. For each gene in these two clusters, a blast search against a database including the complete UniProt (UniProt Consortium, 2015) database and the proteomes of 15 Clavicipitaceae species was performed. The sequences of the first 150 hits were downloaded. In the case of lpsA, lpsB and lpsC proteins with a sequence length over 5000 were discarded from the analysis. For each group of homologous sequences a maximum likelihood tree was reconstructed. This was done using the same pipeline described for phylome reconstruction in Huerta-Cepas et al. (2011). Briefly, the homologous sequences were aligned using three different alignment programs (MUSCLE v3.8 (Edgar, 2004), MAFFT v6.712b (Katoh et al., 2005) and Kalign (Lassmann and Sonnhammer, 2005)). Alignments were done in forward and reverse (Landan and Graur, 2007). The six resulting alignments were then used to create a consensus alignment with M-COFFEE (Wallace et al., 2006). The alignment was trimmed using trimAl v1.4. (Capella-Gutiérrez et al., 2009) (consistency-score cut-off 0.1667, gap-score cut-off 0.9). The resulting alignment was then used to reconstruct a maximum likelihood tree. First the evolutionary model best fitting the data was chosen by reconstructing neighbor joining trees as implemented in BiONJ (Gascuel, 1997) and assessing the likelihood using seven different models. The best model according to the AIC criterion (Akaike, 1973) was selected and used to reconstruct a maximum likelihood tree as implemented in PhyML v3.0 (Guindon et al., 2010). In all trees four rate categories were used and invariant positions were inferred from the data. Bootstrap supports were calculated for each tree. Trees were rooted preferentially at a species that did not belong to the Pezizomycotina and was far related to the event of interest. When that was not possible due to a lack of suitable homolog a Pezizomycotina species belonging preferentially to Dothideomycetes or Leotiomycetes was chosen, always selecting leaves as far related as possible to the seed sequence. Trees were then analyzed manually to assess the consistency between the topology and the known species topology. Trees can be found in Supplementary Figs. S1–S23.
2.3. Species tree reconstruction
In order to reconstruct the species tree we reconstructed a phylome so that we could obtain the genes that had one to one orthologs in all the species considered. For each gene encoded in the genome of E. festucae E2368 a homology search was performed against a database that contained 35 fungal species (see Supplementary Table S1). Results were filtered according to an e-value and an overlap threshold (e-value < 1e−05 and overlap > 0.5). A maximum of 150 homologous sequences was taken. Then, for each group of homologs a maximum likelihood tree was reconstructed using the same methodology detailed above. Data produced during phylome reconstruction was stored at phylomeDB (Huerta-Cepas et al., 2014) (http://phylomedb.org/) with phyID code 125. Trees were then scanned using ETE v2.2 (Huerta-Cepas et al., 2010) to search for trees that were single copies in the 35 species. 906 such trees were selected and their alignments, as reconstructed in the phylome, were concatenated into a single multiple sequence alignment that contained 657,273 amino acids. Then RAxML v8.0.3 (Stamatakis et al., 2005) was used to reconstruct the species tree. Rapid bootstrap, as implemented in RAxML was used to calculate branch support. In addition, the phylome was also used to infer a supertree using duptree (Wehe et al., 2008). This algorithm looks for the species tree that infers the least number of duplication events when reconciling it to the gene trees generated in the phylome. Both methods produced identical trees.
A larger species tree was reconstructed in order to more accurately calculate the number of gene loss events. 268 fully sequenced genomes with their proteome predictions were downloaded from NCBI. A super tree approach was used to obtain the most likely topology for the species tree. Firstly, random groups of 15 species were selected, a blast search was performed between the species and best bidirectional hits were selected. Genes that had one hit per species were chosen to build a concatenated gene tree. At most 100 genes were selected. The species tree was reconstructed using the same methodology explained above. This was repeated over 50,000 times obtaining a total of 52,527 trees. A super tree was reconstructed using duptree and including all the small species trees.
2.4. Topology comparison
For the genes predicted to have undergone a HGT an alternative constrained topology was reconstructed using RaxML v8.0.3 (Stamatakis et al., 2005). For each, tree two constrains were reconstructed, one where all Sordariomycetes were monophyletic and one where all Eurotiomycetes were monophyletic (see Supplementary Figs. S24–S38). Internal nodes were completely collapsed. RAxML was then used to reconstruct two alternative topologies using the constrains. The likelihoods per site for each tree were calculated using phyML allowing for branch and rate optimization but fixing the topologies. The evolutionary model used in each case was the same as the one selected during tree reconstruction. CONSEL (Shimodaira and Hasegawa, 2001) was then used in order to compare the original topology to each of the alternative topologies and to assess whether the alternative topologies, which did not support a HGT event, were supported by the data. In all cases except for dmaW the alternative topologies were significantly rejected (see Supplementary Table S3). In the case of dmaW, the topology where Sordariomycetes were monophyletic was significantly rejected but the one where Eurotiomycetes were monophyletic could not be rejected.
2.5. Independent gene loss events calculation
We counted the number of loss events of the loline and ergot alkaloid gene clusters needed to explain the distribution of the clusters across the phylogenetic tree. The loline cluster was found in P. expansum and in Clavicipitaceae. The common ancestor of these two groups of species was mapped on the species tree reconstructed above and the total number of losses in monophyletic groups was computed using ETE v2.2 (Huerta-Cepas et al., 2010). Losses after the acquisition of the gene cluster were not considered. For the ergot alkaloids gene cluster three groups were considered: the base of Eurotiomycetidae, the base of Clavicipitaceae and G. lozoyensis. The number of losses was calculated as explained above.
2.6. Gene order
For each of the 35 completely sequenced species used in the phylome, their genomes were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/). If the proteome was available, it was also downloaded. For species where the proteome was predicted, a blastP search was performed using the cluster genes as query and the position of the best hits was obtained from the data. In those cases where there was not a predicted proteome uploaded in NCBI, exonerate (Slater and Birney, 2005) was used to locate the proteins within the genome. Identity between proteins of the reference gene cluster and the homologous proteins was calculated using trimAl (Capella-Gutiérrez et al., 2009).
2.7. Divergence times calculation
r8s-PL (Sanderson, 2003) was used to calculate divergence times among the considered species. The species tree calculated from the phylome was calibrated using the node where Sordariomycetes and Leotiomycetes diverged. The calibration time was set at 306.20 MyA as reported in Timetree website (www.timetree.org) (Kumar and Hedges, 2011). The PL method, as implemented in r8s, was used to calculate the divergence times at all the branches in the species tree. Cross-validation was used to calculate the optimal smoothing parameter needed in r8s to calculate divergence times.
3. Results and discussion
3.1. Phylogenetic reconstruction of genes in the loline and ergot alkaloid gene clusters
We first reconstructed the phylogeny of each gene described as part of the gene clusters of loline and ergot alkaloids using a maximum likelihood approach. The clusters in the reference species were formed by 11 and 14 genes respectively (see Supplementary Table S2 for a complete list of genes), and were used as a seed in the process of phylogenetic reconstruction. First, homologs of proteins encoded by these genes were detected by similarity searches in UniProt and other protein databases (see Section 2). These homologs were aligned and a single phylogenetic tree was reconstructed for each individual seed gene (see Section 2). We then inspected the trees manually to search for signs of HGT events by comparing the gene trees to the currently accepted taxonomy. A topology test was run for each tree that presented indications of HGT in order to ensure that two alternative topologies where Sordariomycetes and Eurotiomycetes were monophyletic respectively, were not supported by the data (see Section 2 and Supplementary Table S3).
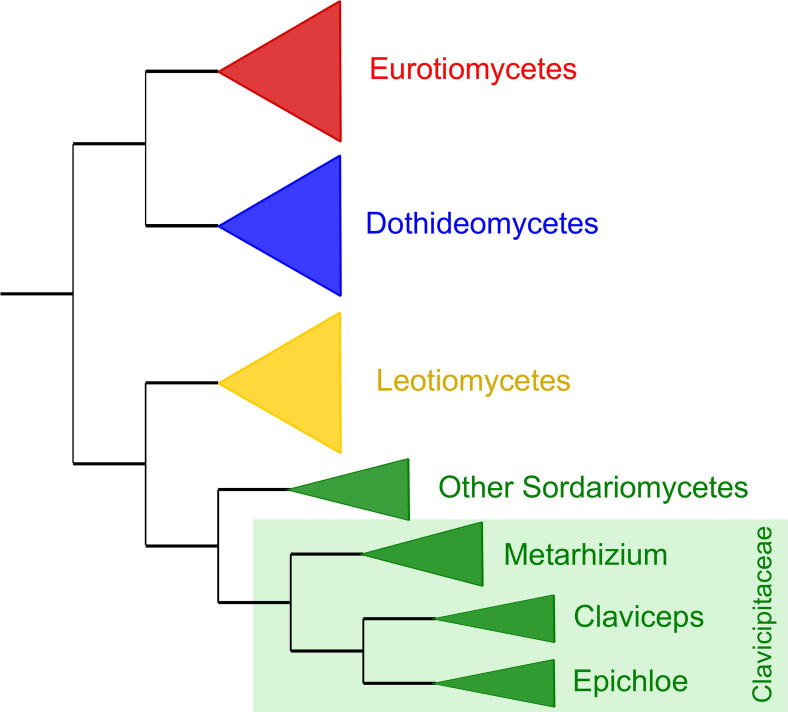
A schematic tree depicting the four main fungal clades in Pezizomycotina, namely Eurotiomycetes, Sordariomycetes, Leotiomycetes and Dothideomycetes, is shown in Fig. 1. All individual gene trees can be found in Supplementary Figs. S1–S23.
Fig. 1.
Schematic species tree depicting the relationships between the main groups considered.
3.2. A Loline alkaloid gene cluster found in Clavicipitaceae and in Eurotiomycetes
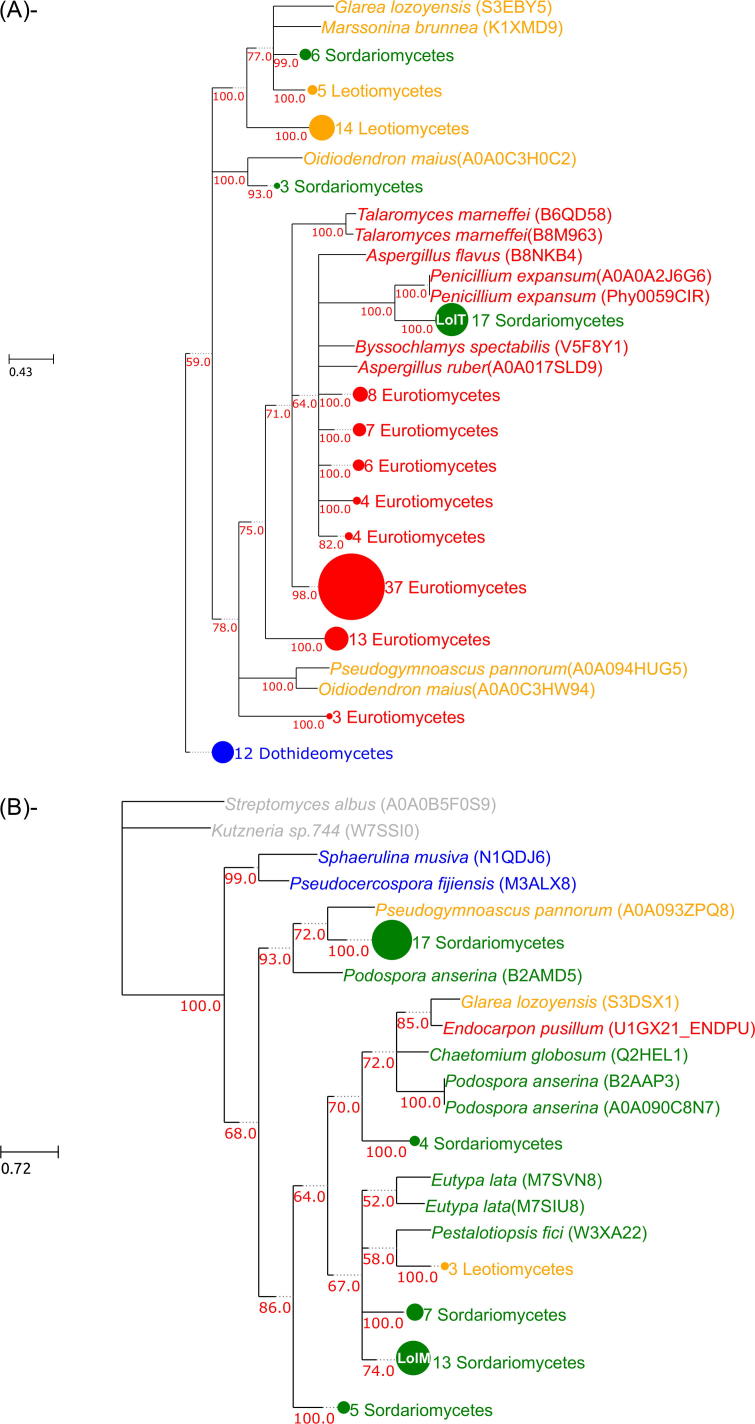
The Loline gene cluster is formed by, at most, 11 genes, of which 8 are core genes (lolA, lolC, lolD, lolE, lolF, lolO, lolT and lolU) and 3 are accessory genes (lolP, lolM and lolN). Our phylogenetic analyses showed that all core genes, with the exception of lolA and lolU, grouped with homologs of the Eurotiomycetes species P. expansum. In addition, the genes lolC, lolD, lolO and lolT were grouped within a larger group of Eurotiomycetes species (see Supplementary Figs. S2, S3, S8 and S10). As an illustrative example we discuss the lolT tree, which is depicted schematically in Fig. 2A. As shown in the image, 17 sequences representing homologs of lolT in Clavicipitaceae species group closely to P. expansum, and this group is embedded within a much larger group of Eurotiomycetes species among which there are representatives of most of the currently sequenced taxonomic groups within this class. LolE and lolF (see Supplementary Figs. S4 and S5) did not show such close relation to Eurotiomycetes beyond their grouping with P. expansum. In all cases two alternative topologies where Sordariomycetes and Eurotiomycetes were monophyletic were discarded (see Supplementary Table S3). Bootstrap analysis showed that the grouping between Clavicipitaceae and P. expansum was highly supported (100%) in all cases. Of the two remaining early-pathway genes, lolA presents a puzzling tree topology (see Supplementary Fig. S1) where the Clavicipitaceae lolA genes appear as sister group to a large group of Sordariomycetes, Leotiomycetes, Dothideomycetes and Eurotiomycetes homologs. Such pattern could be the result of an ancient duplication that occurred before the four main clades separated and where two copies of the gene have only been conserved in some Clavicipitaceae. It is also possible that long branch attraction (LBA) has distorted the topology. A topology test showed that a tree where the Clavicipitaceae grouped with the other Sordariomycetes was not discarded by the data. This, coupled with a relatively low bootstrap support (72%) points to the possibility of LBA. LolU, on the other hand, is grouped with other Sordariomycetes, in a tree that is mostly congruent with the species tree (see Supplementary Figs. S11). Like lolU, the three trees for the accessory genes appear to be more congruent to the species tree, see Fig. 2B for an example.
Fig. 2.
Example trees for the genes in the loline cluster. A. lolT tree presents a topology where Clavicipitaceae are grouped with Eurotiomycetes. B. lolM tree which presents a topology congruent with the species tree. For both trees circles represent collapsed nodes of the tree and are sized according to the number of leaves that diverged from the node. Colors belong to the different taxonomic groups: green – Sordariomycetes, yellow – Leotiomycetes, red – Eurotiomycetes, blue – Dothideomycetes and gray – Prokaryotes. Bootstrap supports lower than 75 are indicated on the tree. Nodes with a support below 33 are collapsed. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
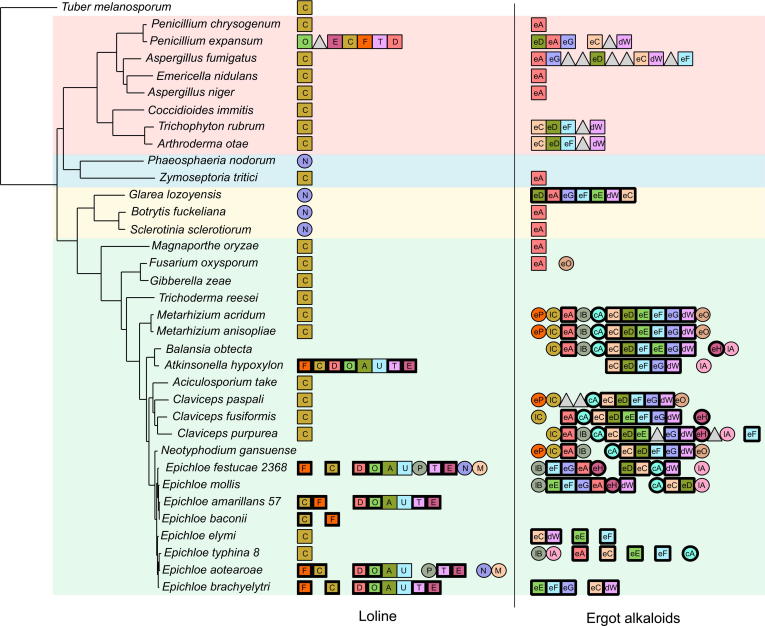
In addition to the information provided by phylogenetic trees of individual genes, we also considered the conservation of the gene order within the cluster in a group of 35 completely sequenced Pezizomycotina species. This set of species includes representatives of the four most sequenced groups of Pezizomycotina species: Eurotiomycetes, Dothideomycetes, Leotiomycetes, and Sordariomycetes. In order to provide an evolutionary context, we reconstructed the evolutionary relationships of these 35 species based on a gene concatenation approach that included 906 single-copy, widespread genes. The inferred species tree (Figs. 3 and S39) was fairly congruent with previously published trees (Schardl et al., 2013b).
Fig. 3.
Left panel shows a Maximum likelihood phylogeny of 34 species based on the concatenation of 167 genes. Leaves are colored according to the main taxonomic groups within Pezizomycotina: Leotiomycetes (yellow), Sordariomycetes (green), Dothideomycetes (blue) and Eurotiomycetes (red). The two panels on the right detail the composition of the two gene clusters discussed in each species. Each gene in each cluster is represented by a square, a circle or a triangle. Squares represent core genes, circles represent accessory genes, whereas triangles represent genes that are not known to be part of the original gene cluster. Spaces between genes indicate that the two genes have more than three non related genes between them or that they belong to different contigs. Homologous genes have the same color. Genes with an identity below 50% to the original gene from the cluster have been excluded from the image. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As seen in Fig. 3, only P. expansum and some of the Clavicipitaceae species conserved the loline cluster. In all the other species no homologs with more than 50% identity to the proteins encoded in the reference cluster could be found, either forming a cluster or dispersed in the genome. If we consider proteins with a lower identity (see Fig. S39) those proteins never are found in a cluster and are likely paralogous genes. The gene cluster found in P. expansum contained six of the eleven genes that form the Clavicipitaceae loline cluster (lolC, lolD, lolE, lolF, lolO and lolT). The fact that two core genes (lolA and lolU) are missing from the P. expansum gene cluster and that an additional Non Ribosomal Peptide Synthesis protein is found instead suggests that P. expansum likely lacks the ability to produce loline, but rather the cluster present in this species is most probably involved in the production of a different kind of alkaloid. This is congruent with the fact that the production of loline was not reported for any of the 58 species of Penicillium tested by Frisvad and collaborators, including P. expansum (Frisvad et al., 2004).
The cluster responsible for the synthesis of loline in Clavicipitaceae species has an homologous cluster in P. expansum. Gene order between the two gene clusters is not conserved but their phylogenetic trees suggest a close relationship between the origin of both clusters.
3.3. Ergot alkaloid gene clusters
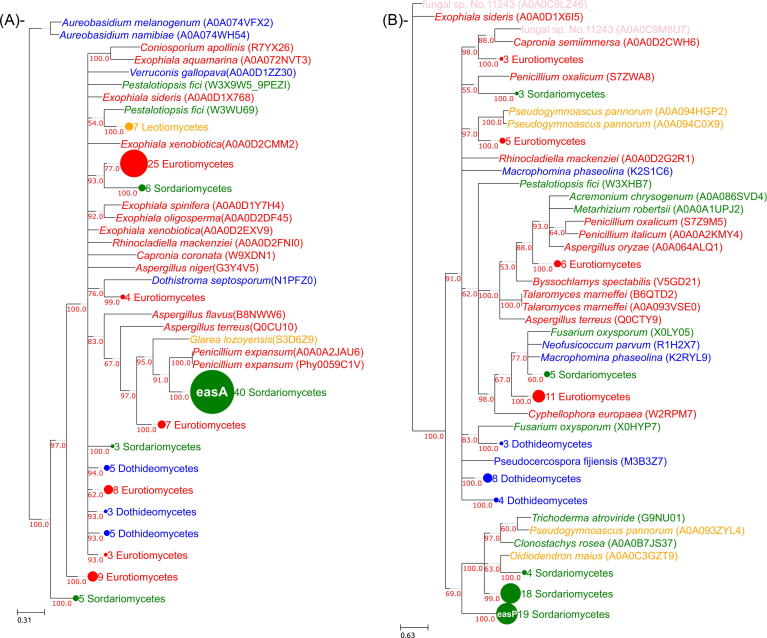
Up to 14 genes can be involved in the synthesis of ergot alkaloids and derivatives in Clavicipitaceae. As in the case of the loline biosynthetic pathway, genes in the ergot alkaloid synthesis pathway are divided into core and accessory genes. The core genes encode enzymes needed to synthesize the ergoline ring system, the skeleton of ergot alkaloids, and include dmaW, easA, easC, easD, easE, easF and easG. Accessory genes code for enzymes that introduce subsequent chemical modifications: cloA, easH, easO, easP, lpsA, lpsB and lpsC. Of these genes, it is unknown whether easO and easP participate in the synthesis of ergot alkaloids, yet they are part of the cluster. Our phylogenetic analyses showed that, as in the case of the loline cluster, there are two distinct topologies associated to the two different groups. The core genes (see Supplementary Figs. S13–S19) display a unique tree topology where Clavicipitaceae group with Glarea lozoyensis (Leotiomycetes) and different Eurotiomycetes species. Fig. 4A shows a scheme of the easA tree where the Clavicipitaceae species group with P. expansum and G. lozoyensis followed by a larger group of Eurotiomycetes. The grouping with P. expansum is not consistent among all trees, and different species groups can be found as the closest Eurotiomycetes relatives. Among them are the genes that belong to the ergot alkaloids cluster described in A. fumigatus (Robinson and Panaccione, 2012). Like in the case of the loline cluster, a topologies more consistent with the species tree were discarded in all cases (see Supplementary Table S3). Bootstrap values in this case are a bit more diverse, ranging from 100% in easC to 72% in easD. The accessory genes, on the other hand, show a diversity of gene tree topologies. easO and easP (see Fig. 4B and Supplementary Figs. S21 and S22) present a topology congruent with the species tree, lpsA, lpsB and lpsC (see Supplementary Fig. S23) are paralogs and are associated with a few Aspergillus sequences. Finally, easH and cloA (see Supplementary Figs. S12 and S20) seem to follow the same phylogenetic pattern described for the core genes and are found branching out from within a group of Eurotiomycetes sequences.
Fig. 4.
Example trees for the genes in the ergot alkaloid cluster. A. easA tree where the Clavicipitaceae species group with Eurotiomycetes and one Leotiomycetes. B. Tree in which easP clusters within a group of other Sordariomycetes and Leotiomycetes. Tree image follows the same format as Fig. 2.
The gene order for ergot alkaloid synthesis cluster is fairly conserved within Clavicipitaceae (Fig. 3). There are also some signs of clustering in different Eurotiomycetes species, the largest cluster being the one found in A. fumigatus. Arthroderma otae and Trichophyton rubrum have both a small cluster of five genes which are homologous to genes in the Clavicipitaceae gene cluster. Curiously the gene order for these four genes is conserved in Clavicipitaceae. The production of ergot alkaloids has been confirmed in several Eurotiomycetes species though the final compounds differ depending on the species which is in accordance to the differences within the gene cluster.
The last species predicted to have the ergot alkaloid gene cluster is the Leotiomycetes G. lozoyensis. It is unknown if this species is able to produce ergot alkaloids. The gene order is most similar to the one found in P. expansum. This is congruent with the fact that the two species tend to appear close in the trees.
3.4. Horizontal gene transfer versus vertical descent
The existence of clusters of genes present in a small number of distantly related species can be explained either through horizontal gene transfer, by vertical inheritance of an ancestral cluster followed by losses of the cluster or by the recurrent formation of the cluster through reordering of ancestral genes. We tried to asses which of the three evolutionary scenarios was more likely to have resulted in the discussed presence/absence patterns of the ergot alkaloids and loline gene clusters. The third method, formation of the cluster from recurrent genes, is the less likely since it would imply that the genes still had orthologs in the other species but were not found close in the genome. The phylogenetic trees show that most Pezizomycotina species do not have orthologs of the cluster genes in their genomes. The variability in the gene order found in the conserved clusters could be a point in favor for this hypothesis, but gene order is known to change in secondary metabolite clusters and therefore a lack of conservation should not discard the notion of a common origin of the clusters.
It is difficult to distinguish between horizontal gene transfer and vertical descent followed by gene loss. Not much is known about how HGT events occur in fungi, yet more and more examples of clear cases of HGT events involving fungi have been brought to light (Coelho et al., 2013, Marcet-Houben and Gabaldón, 2010, Moran and Jarvik, 2010). This points to the fact that fungi are able to incorporate foreign DNA into their genomes even if the exact mechanisms have not been elucidated. One method used to distinguish between HGT and vertical descent is to calculate the number of independent losses that should have occurred to explain the observed patterns of gene presence and absence. The higher the number of loss events, the more likely the HGT event is thought to be. We inferred the number of times our two gene clusters were lost using a species tree that included 268 fully sequenced fungal genomes (see Section 2). We estimate that, assuming only vertical descent, there have been at least 23 independent loss events of the loline cluster and 16 of the ergot alkaloids gene cluster. This number of losses is much higher than the values that have been previously considered as thresholds for the acceptance of HGT (Marcet-Houben and Gabaldón, 2010, Snel et al., 2002). The fact that the gene clusters responsible for the synthesis of secondary metabolites are not essential to the fungi could explain the sparse distribution of the presence/absence of the gene clusters. Yet, it would also mean that the gene clusters needed to be conserved in all the common ancestors leading to the lineages that contain the gene clusters.
As discussed above, we have two secondary metabolism gene clusters that are present only in distant lineages. The inferred number of gene losses is far above the normally considered acceptable thresholds for the acceptance of an HGT event. This leads to the possibility that HGT was involved in the evolution of these gene clusters. The phylogenetic trees derived for the genes in the loline gene cluster show that it is likely that the cluster was transferred from Eurotiomycetes species to Clavicipitaceae. According to the trees, the putative donor species would be P. expansum.
The phylogenetic trees for the genes involved in the synthesis of ergot alkaloids show the presence of at least two HGT events. There are three groups of distantly related species involved: the Eurotiomycetes, the Clavicipitaceae and finally the Leotiomycetes G. lozoyensis. As shown in the trees, the Eurotiomycetes were likely the original donors, as they tend to occupy more basal positions in the trees. We can then hypothesize different scenarios. On one hand there could have been two HGT events from Eurotiomycetes to the two other groups. On the other one there could have been one single transfer from Eurotiomycetes to one of the other two groups and then a transference between the Leotiomycetes and Clavicipitaceae species. At the moment we are unable to assess which of the two scenarios is more likely. It is possible that a larger taxon sampling will be able to elucidate this matter.
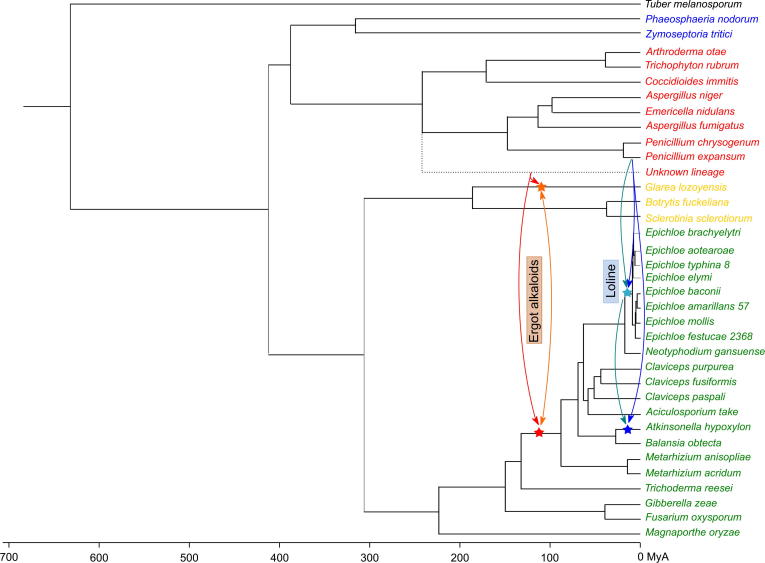
3.5. Timing of putative HGT events
Eurotiomycetes and Sordariomycetes diverged roughly 400 MyA (Kumar and Hedges, 2011). Despite this long period of time the two secondary metabolite clusters are at least partially conserved across species from within these two groups, whereas they are completely absent in sequenced species of other clades. We have shown that this uneven distribution could be the result of HGT. In order to exchange genetic material, two species need to have co-existed in the same place and time period. In order to assess the timing of the proposed HGT events, we inferred divergence times from molecular data (see Section 2 and Fig. 5). Our estimates place Clavicipitaceae as a relatively young group that diverged from other Sordariomycetes roughly 184 MyA, an estimate which is fairly congruent with the 173 MyA predicted by Shung and collaborators (Sung et al., 2008). The emergence of the ergot alkaloid cluster in Clavicipitaceae must have occurred between this divergence point and the split between Metarhizium and other Clavicipitaceae species, about 137 MyA. As stated above, the genes that have likely been transferred from Eurotiomycetes to Clavicipitaceae tend to group with different Eurotiomycetes in their phylogenies instead of a particular Eurotiomycetes clade. As some of these species diverged long before the Clavicipitaceae diverged the donor was likely an Eurotiomycetes species of a different lineage that has not been sequenced yet.
Fig. 5.
Chronogram showing the divergence times between the different species and inference of the timing of the HGT events discussed. The red star indicates where the ergot alkaloid gene cluster emerged in Clavicipitaceae, the red arrow represents the possible HGT direction. The blue stars and arrows show the two alternative hypothesis considered for the emergence of the loline gene cluster. The light colored arrow shows an initial transference from P. expansum to the base of Epichloe followed by a second transference from Epichloe to A. hypoxylon. The dark colored arrow shows the alternative hypothesis where first P. expansum transferred the loline cluster to A. hypoxylon and from this species it was transferred to the base of Epichloe.
The emergence of loline, in contrast, appears to be more recent, as it does not appear in Metarhizium species. This implies an HGT event that took place between 137 and 83 MyA. As shown in the trees, the loline genes that have been transferred consistently group with P. expansum. This time frame falls after the divergence between Penicillium and Aspergillus but not before the divergence between P. expansum and P. chrysogenum. This discrepancy in the data leads to the need to accept that this is a vertical event with the subsequent 23 loss events, consider the possibility that the donor was a more ancestral Penicillium species that has not been sequenced yet and that the loline cluster was present at the base of Penicillium and then lost or the need to invoke a second HGT. If we keep with the assumption that 23 loss events are less parsimonious than two HGT events then we can discard the first hypothesis. The second hypothesis would imply one single HGT event coupled with, at least, 7 loss events in Clavicipitaceae species and 6 loss events in Penicillium species. While not the most parsimonious explanation, that rate of loss events in secondary metabolism gene clusters makes it a reasonable explanation. The third alternative is based on the fact that most of the Clavicipitaceae species that have the loline gene cluster are from the genus Epichloe, with only one Clavicipitaceae species outside this genus having the cluster: Atkinsonella hypoxylon. The gene clusters between Epichloe and A. hypoxylon share the same gene order and their proteins share a higher percentage of identity than the average for the genomes (average of 90% identity between the proteins in the cluster versus an average of 82% between the complete proteomes). This could indicate that a second HGT event occurred between these two groups after an initial transference from P. expansum. It is unclear though to which of the two groups the original transfer was done. Species from the genus Epichloe diverged roughly 11.5 MyA, which is after the divergence between P. expansum and P. chrysogenum, therefore the HGT event could have been to both, the base of Epichloe or to A. hypoxylon specifically. We need more data to be able to distinguish between the two hypothesis or a deeper understanding on the likelihood that a secondary metabolism gene cluster is lost specifically in a large number of species.
4. Conclusions
Ergot alkaloids and loline are two of the key compounds synthesized by Clavicipitaceae species. They play an important role in the defense of their plant hosts by protecting them from herbivores, including mammals and insects. The origin of the genes that encode the proteins needed to synthesize these two compounds is diverse. We hypothesize that in both cases horizontal gene transfer events were involved in the acquisition of most of the key enzymes needed to synthesize the first stable intermediate from existing clusters in Eurotiomycetes species. A vertical inheritance of those clusters would imply a large number of independent gene losses, leading to a less parsimonious scenario. The core genes needed to synthesize ergot alkaloids were transferred at least two times with the main donor likely being an unknown lineage in Eurotiomycetes. The core genes transferred to Clavicipitaceae and the Leotiomycetes G. lozoyensis either in parallel or in two sequential events. The core genes for the loline cluster were also transferred twice, once to A. hypoxylon and another time to the base of Epichloe. The order of these transferences is still unclear. After the transference of the core set of genes, metabolic gene clusters were modified by the addition of genes from other sources. It might be argued that the original selective advantage of the transferred cluster was based on a distinct, but related, function as compared to their current role.
Acknowledgments
TG group research is funded in part by a grant from the Spanish Ministry of Economy and Competitiveness grants, ‘Centro de Excelencia Severo Ochoa 2013–2017’ SEV-2012-0208, and BIO2012-37161 cofounded by European Regional Development Fund (ERDF), a Grant from the Qatar National Research Fund grant (NPRP 5-298-3-086), and a grant from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC (Grant Agreement no. ERC-2012-StG-310325).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fgb.2015.12.006.
A. Supplementary data
References
- Akaike, H., 1973. Information theory and extension of the maximum likelihood principle. In: Proc. 2nd Int. Symp. Inf. Theory, pp. 267–281.
- Ballester A.-R., Marcet-Houben M., Levin E., Sela N., Selma-Lázaro C., Carmona L., Wisniewski M., Droby S., González-Candelas L., Gabaldón T. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant-Microbe Interact. MPMI. 2015;28:232–248. doi: 10.1094/MPMI-09-14-0261-FI. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinform. Oxf. Engl. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M.A., Gonçalves C., Sampaio J.P., Gonçalves P. Extensive intra-kingdom horizontal gene transfer converging on a fungal fructose transporter gene. PLoS Genet. 2013;9:e1003587. doi: 10.1371/journal.pgen.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle C.M., Panaccione D.G. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol. 2005;71:3112–3118. doi: 10.1128/AEM.71.6.3112-3118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Smedsgaard J., Larsen T.O., Samson R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 2004;49:201–241. [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haarmann T., Machado C., Lübbe Y., Correia T., Schardl C.L., Panaccione D.G., Tudzynski P. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry. 2005;66:1312–1320. doi: 10.1016/j.phytochem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J., Capella-Gutierrez S., Pryszcz L.P., Denisov I., Kormes D., Marcet-Houben M., Gabaldón T. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 2011;39:D556–D560. doi: 10.1093/nar/gkq1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Capella-Gutiérrez S., Pryszcz L.P., Marcet-Houben M., Gabaldón T. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Dopazo J., Gabaldón T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Hedges S.B. TimeTree2: species divergence times on the iPhone. Bioinform. Oxf. Engl. 2011;27:2023–2024. doi: 10.1093/bioinformatics/btr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G., Graur D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol. Biol. Evol. 2007;24:1380–1383. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E.L.L. Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.R. The history of ergot of rye (Claviceps purpurea) I: from antiquity to 1900. J. R. Coll. Physicians Edinb. 2009;39:179–184. [PubMed] [Google Scholar]
- Li S.-M., Unsöld I.A. Post-genome research on the biosynthesis of ergot alkaloids. Planta Med. 2006;72:1117–1120. doi: 10.1055/s-2006-947195. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M., Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. TIG. 2010;26:5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Moran N.A., Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328:624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- Panaccione D.G., Coyle C.M. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 2005;71:3106–3111. doi: 10.1128/AEM.71.6.3106-3111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.L., Panaccione D.G. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia. 2012;104:804–812. doi: 10.3852/11-310. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinform. Oxf. Engl. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Schardl C.L., Florea S., Pan J., Nagabhyru P., Bec S., Calie P.J. The epichloae: alkaloid diversity and roles in symbiosis with grasses. Curr. Opin. Plant Biol. 2013;16:480–488. doi: 10.1016/j.pbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C.L., Grossman R.B., Nagabhyru P., Faulkner J.R., Mallik U.P. Loline alkaloids: currencies of mutualism. Phytochemistry. 2007;68:980–996. doi: 10.1016/j.phytochem.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Schardl C.L., Leuchtmann A., Spiering M.J. Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 2004;55:315–340. doi: 10.1146/annurev.arplant.55.031903.141735. [DOI] [PubMed] [Google Scholar]
- Schardl C.L., Young C.A., Hesse U., Amyotte S.G., Andreeva K., Calie P.J., Fleetwood D.J., Haws D.C., Moore N., Oeser B., Panaccione D.G., Schweri K.K., Voisey C.R., Farman M.L., Jaromczyk J.W., Roe B.A., O’Sullivan D.M., Scott B., Tudzynski P., An Z., Arnaoudova E.G., Bullock C.T., Charlton N.D., Chen L., Cox M., Dinkins R.D., Florea S., Glenn A.E., Gordon A., Güldener U., Harris D.R., Hollin W., Jaromczyk J., Johnson R.D., Khan A.K., Leistner E., Leuchtmann A., Li C., Liu J., Liu J., Liu M., Mace W., Machado C., Nagabhyru P., Pan J., Schmid J., Sugawara K., Steiner U., Takach J.E., Tanaka E., Webb J.S., Wilson E.V., Wiseman J.L., Yoshida R., Zeng Z. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013;9:e1003323. doi: 10.1371/journal.pgen.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C.L., Young C.A., Pan J., Florea S., Takach J.E., Panaccione D.G., Farman M.L., Webb J.S., Jaromczyk J., Charlton N.D., Nagabhyru P., Chen L., Shi C., Leuchtmann A. Currencies of mutualisms: sources of alkaloid genes in vertically transmitted epichloae. Toxins. 2013;5:1064–1088. doi: 10.3390/toxins5061064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinform. Oxf. Engl. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Slater G.S.C., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B., Bork P., Huynen M.A. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Spiering M.J., Moon C.D., Wilkinson H.H., Schardl C.L. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics. 2005;169:1403–1414. doi: 10.1534/genetics.104.035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinform. Oxf. Engl. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Strickland J.R., Looper M.L., Matthews J.C., Rosenkrans C.F., Flythe M.D., Brown K.R. Board-invited review: St. Anthony’s Fire in livestock: causes, mechanisms, and potential solutions. J. Anim. Sci. 2011;89:1603–1626. doi: 10.2527/jas.2010-3478. [DOI] [PubMed] [Google Scholar]
- Sung G.-H., Poinar G.O., Spatafora J.W. The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol. Phylogenet. Evol. 2008;49:495–502. doi: 10.1016/j.ympev.2008.08.028. [DOI] [PubMed] [Google Scholar]
- Tudzynski P., Hölter K., Correia T., Arntz C., Grammel N., Keller U. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. MGG. 1999;261:133–141. doi: 10.1007/s004380050950. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäli P.P., Wäli P.R., Saikkonen K., Tuomi J. Is the pathogenic ergot fungus a conditional defensive mutualist for its host grass? PLoS One. 2013;8:e69249. doi: 10.1371/journal.pone.0069249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace I.M., O’Sullivan O., Higgins D.G., Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehe A., Bansal M.S., Burleigh J.G., Eulenstein O. DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinform. Oxf. Engl. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- Wisecaver J.H., Rokas A. Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front. Microbiol. 2015;6:161. doi: 10.3389/fmicb.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.