Abstract
Lamprey is one of only two living jawless vertebrates, a group that includes the first vertebrates. Comparisons between lamprey and jawed vertebrates have yielded important insights into the origin and evolution of vertebrate physiology, morphology and development. Despite its key phylogenetic position, studies of lamprey have been limited by their complex life history, which makes traditional genetic approaches impossible. The CRISPR/Cas9 system is a bacterial defense mechanism that was recently adapted to achieve high-efficiency targeted mutagenesis in eukaryotes. Here we report CRISPR/Cas9-mediated disruption of the genes Tyrosinase and FGF8/17/18 in the sea lamprey Petromyzon marinus, and detail optimized parameters for producing mutant F0 embryos. Using phenotype and genotype analyses, we show that CRISPR/Cas9 is highly effective in the sea lamprey, with a majority of injected embryos developing into complete or partial mutants. The ability to create large numbers of mutant embryos without inbred lines opens exciting new possibilities for studying development in lamprey and other non-traditional model organisms with life histories that prohibit the generation of mutant lines.
KEY WORDS: Lamprey, CRISPR, Vertebrate, Evolution
Summary: CRISPR/Cas9 technology is used to effectively disrupt the genes encoding tyrosinase and FGF8/17/18 in the sea lamprey Petromyzon marinus.
INTRODUCTION
The first vertebrates had simple cartilaginous head skeletons that lacked proper joints. Today, only two groups of jawless (agnathan) vertebrates survive: lampreys and hagfish. Comparisons between lamprey, hagfish and jawed vertebrates (gnathostomes) have been used for over a century to identify ancestral vertebrate features and deduce potential gnathostome novelties. However, due to the inaccessibility of hagfish, which live and spawn on the ocean floor (Ota et al., 2007), most studies of agnathan biology have focused on lampreys. Work on lamprey embryos, larvae and adults has provided important insights into the evolution of key vertebrate features including the adaptive immune system (Pancer et al., 2004), the endocrine system (Sower et al., 2009; Close et al., 2010), regenerative capacity (Smith et al., 2011), the head skeleton (McCauley and Bronner-Fraser, 2006) and large-scale gene duplications (Smith et al., 2013). There is also much interest in understanding lamprey biology with the goal of conserving native lamprey species and controlling invasive lampreys (reviewed by Sower, 2003).
Among lamprey species, the sea lamprey Petromyzon marinus has been used extensively to study the evolution of vertebrate development (Beamish, 1980; McCauley and Kuratani, 2008; Piavis, 1971). Several features make the sea lamprey particularly well-suited for such work. During their summer spawning season, large numbers of sexually mature adults can be trapped in shallow freshwater streams. Ripe, captured females can be held and manually stripped of tens of thousands of eggs over the course of several days (Nikitina et al., 2009). Once fertilized, lamprey embryos develop more slowly than most gnathostomes, allowing for fine-grained staging. Lamprey embryos are also similar in size to amphibian embryos, making them amenable to microsurgical grafting, vital dye labeling, and microinjection.
Lamprey embryos have been injected with both synthetic mRNA and DNA to achieve gain-of-function phenotypes and analyze cis-regulatory sequences (Parker et al., 2014; Sauka-Spengler et al., 2007). Translation-blocking morpholinated antisense oligonucleotides (morpholinos) have also been used to test the function of genes involved in early developmental process, such as neural crest specification (McCauley and Bronner-Fraser, 2006; Sauka-Spengler et al., 2007). However, morpholino-mediated gene knockdown is not a permanent perturbation in any organism, making analyses of later developmental functions difficult. This difficulty is pronounced in lampreys given their relatively long developmental time compared with other model vertebrates (∼4 days until gastrulation, ∼6 days until neural crest migration). Furthermore, the effectiveness of different morpholinos can vary dramatically in lamprey, with a large proportion producing no effect or early embryonic death (Lakiza et al., 2011; Nikitina et al., 2011; Sauka-Spengler et al., 2007) (our unpublished results).
The CRISPR/Cas system is a bacterial immune response mechanism that targets and degrades foreign DNA (Barrangou et al., 2007). Bacteria deploying CRISPR/Cas transcribe RNAs from genomic regions called clustered regularly interspaced short palindromic repeats (CRISPRs), some segments of which correspond to sequences present in pathogenic bacterial viruses. Upon infection, these small RNAs direct sequence-specific cleavage of the viral genome by the CRISPR-associated nuclease Cas9. The specificity of the CRISPR/Cas mechanism has made it a powerful tool for targeted mutagenesis of eukaryotic genomes (Mali et al., 2013). To do this, a synthetic RNA called a guide RNA (gRNA) is designed against a target sequence in the host genome immediately upstream of an endogenous protospacer-adjacent motif (PAM; 5′-NGG-3′). The gRNA is then introduced into the cell together with Cas9 mRNA or protein, allowing the Cas9 protein and gRNA to form a complex that finds and cuts the target sequence. Once cleaved by the gRNA-guided Cas9 enzyme, the DNA is repaired by endogenous repair mechanisms, leading to deletions and/or insertions via non-homologous end joining. CRISPR/Cas9 mutagenesis has been successfully applied to several traditional developmental model organisms, including Drosophila melanogaster (Bassett et al., 2013), Mus musculus (Wang et al., 2013), Danio rerio (Hwang et al., 2013), Xenopus tropicalis (Guo et al., 2014; Nakayama et al., 2013), nematodes (Lo et al., 2013), sea anemone (Ikmi et al., 2014) and seed plants (Belhaj et al., 2013). Although versatile, CRISPR/Cas9-mediated mutagenesis is not equally efficient in all organisms. Not surprisingly, model vertebrates such as zebrafish and Xenopus tropicalis seem to require different concentrations of mRNA and gRNA for efficient F0 gene knockdown (Jao et al., 2013; Guo et al., 2014).
With the aim of improving the tractability of the sea lamprey for developmental genetic studies, we tested the efficacy of the CRISPR/Cas9 system in sea lamprey embryos and larvae. As a proof-of-principle, we targeted two genes with easily scored knockdown phenotypes, but different levels of pleiotropy: Tyrosinase (Tyr) and FGF8/17/18. The enzyme tyrosinase is needed for melanin synthesis in all vertebrates. Successful mutagenesis of the lamprey Tyr gene should reduce pigmentation but have no other effects on development. By contrast, gene expression and pharmacological inhibition have shown that the FGF8/17/18 gene family is involved in several conserved developmental processes, including mesoderm specification, somitogenesis, CNS patterning, pharyngeal segmentation and skeletal differentiation (Abzhanov and Tabin, 2004; Jandzik et al., 2014; Mason et al., 2000; Reifers et al., 1998; Sun et al., 1999). Depending on the timing and penetrance of CRISPR/Cas9-mediated mutagenesis in lamprey embryos, we expected mutation of the single lamprey FGF8/17/18 homolog to produce a range of phenotypes mimicking the effects of pharmacological FGF signaling inhibition. We found that CRISPR/Cas9-mediated mutagenesis of both genes was highly effective, validating the method in this evolutionarily informative species.
RESULTS
Tyrosinase expression during sea lamprey development
Lamprey possesses neural crest-derived melanocytes that emerge from the dorsal neural tube around stage (st.) 25.5. The lamprey eye begins depositing melanin as the larva approaches st. 27. At st. 26.5 we observed Tyr mRNA expression in presumptive neural crest-derived melanocytes, and in the eye, presaging melanin deposition in the retina (Fig. 1A,B).
Fig. 1.
Tyr expression and disruption via CRISPR/Cas9. Lateral views with anterior to left in all panels. (A) A st. 26.5 sea lamprey larva showing Tyr expression in neural crest-derived melanocytes near the pharynx and retinal expression in the presumptive eye (arrow). (B) Lateral flank view of a st. 26.5 larva showing neural crest-derived melanocytes in the dorsal neural tube and on the dorsal surface of the yolk. (C,C′) Brightfield (C) and red channel fluorescence (C′) images showing the absence of pigment cells in an embryo that is positive for the LRD lineage tracer. (D-G) Example larvae of each mutant class at st. 30. Pharyngeal arches are labeled by number (1-9); e, eye; he, heart; mo, mouth; wt, wild type.
Optimization of Tyr mutagenesis
We synthesized two different gRNAs targeting an exon of lamprey Tyr homologous to exon 1 of human TYR. We then co-injected 500 pg Cas9 with 200 pg Tyr gRNA 1 and 200 pg Tyr gRNA 2, as described for Xenopus tropicalis (Guo et al., 2014). Using these quantities, 37% of larvae displayed an obvious reduction in pigmentation of 10% or more. We then increased the amount of Cas9 mRNA to 1 ng and the amount of gRNA to 400 pg Tyr gRNA 1 and 400 pg Tyr gRNA 2. While the occurrence of albino or nearly albino larvae was highest with this treatment (59%), we also observed a severe developmental delay or abnormal morphology in 24% of the larvae. We then injected individuals with the same amount of Cas9 mRNA (1 ng), but reduced the gRNA concentration to that of the initial trial (400 pg). This treatment produced a high frequency of albino/nearly albino (21%) and partially pigmented (44%) larvae, and reduced larval deformity to the level typical for healthy uninjected controls (∼5-8%) (Table 1; see Fig. 1D-G for scoring examples).
Table 1.
Mutagenesis efficiency (% affected) versus deformities (%) of Tyr gRNA and Cas9 mixtures
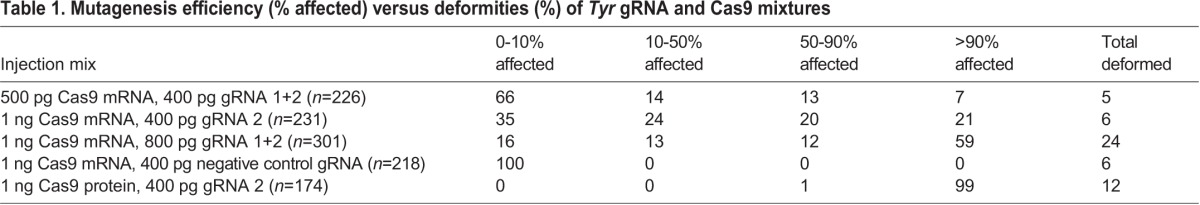
After determining the highest-tolerated RNA quantities for injection, we sought to test the relative effectiveness of the two Tyr gRNAs. We injected zygotes from a single fertilization event with 400 pg of each gRNA singly, along with 1 ng Cas9 mRNA to directly compare the efficiency of each gRNA (g1, n=58; g2, n=66). After scoring, a Wilcoxon rank-sum test was performed to test for differences in the ability of each of these gRNAs to produce a mutant phenotype; this resulted in a P-value of 0.7865, indicating that their activity is very similar. Prior to scoring, deformed embryos were removed from this experiment only.
To verify disruption of the Tyr locus, two nearly albino individuals injected with Tyr gRNA 1 and two nearly albino embryos injected with Tyr gRNA 2 were pooled and genotyped. Of the seven unique alleles observed, one was wild type, five contained a deletion, and one contained both a deletion and an insertion (Fig. S2A). To look for a correlation between the prevalence of mutant alleles and the phenotype generated, we genotyped an individual positive for the lineage tracer LRD from each of the four mutant categories (Fig. 2). We found that the individual that appeared to have a wild-type level of pigmentation (but was LRD positive) displayed 0/14 mutated sequences, the individual showing ∼25% reduction in pigmentation had 5/13 mutant alleles (two of which were frameshifts), the individual with ∼75% reduction in pigmentation showed 9/10 mutant alleles (five of which were frameshifts), and the completely albino individual showed 13/13 mutant alleles (nine of which were frameshifts).
Fig. 2.
Genotyped Tyr loci of injected individuals. All sequences show the non-template strand in 5′ to 3′ orientation with respect to the Tyr gene. The target sequence and PAM are shown with orange and red highlighting, respectively, in the wild-type (wt) sequence. Deletions relative to wt are shown as red dashes, while polymorphisms and insertions are shown in red text (despite some inserted letters matching the wt sequence). All inferred insertions are stacked on the left side of the deletion region. (A) Sequencing of an injected, LRD-positive individual showing no reduction in pigmentation returned 14/14 wt sequences. (B) Sequencing of a Tyr mutant displaying ∼25% pigmentation reduction returned 5/13 mutated sequences, two of which were frameshifts. (C) Sequencing of a Tyr mutant displaying ∼75% pigmentation reduction returned 9/10 mutated sequences, five of which were frameshifts. (D) Sequencing of a completely albino individual returned 13/13 mutated sequences, ten of which were frameshifts.
Recently, Cas9 protein has become commercially available. To test whether an injection solution using Cas9 protein instead of mRNA is capable of generating a higher frequency of mutant lamprey, we injected 400 pg Tyr gRNA 2 in combination with 1 ng Cas9 protein. This resulted in an astounding 99% completely albino individuals (173/174), but a higher proportion (12%) of developmentally perturbed lamprey (21/174).
Mutagenesis of FGF8/17/18
As with Tyr, we found that injection of 1 ng Cas9 mRNA and 800 pg FGF8/17/18 gRNA caused high mortality. However, at lower RNA concentrations (500 pg Cas9 mRNA, 400 pg total gRNA), survival was similar to that of uninjected sibling controls, and surviving embryos displayed phenotypes consistent with FGF8/17/18 loss of function (Jandzik et al., 2014; Reifers et al., 1998; Crump et al., 2004). The earliest and most common mutant phenotype was a reduction in head size (Table 2, Fig. 3). This effect was observed at a high frequency in embryos injected with Cas9 mRNA and either FGF8/17/18 gRNA alone or in combination (FGF8/17/18 gRNA 1+gRNA 2). A Wilcoxon rank-sum test revealed that these gRNAs differ significantly in their ability to produce a mutant phenotype (g1, n=170; g2, n=224; P=0.0108). Mutant scores for each gRNA can be seen separately in Table 2.
Table 2.
Phenotypes (mutant class 1-5) of FGF8/17/18 gRNA + Cas9-injected larvae
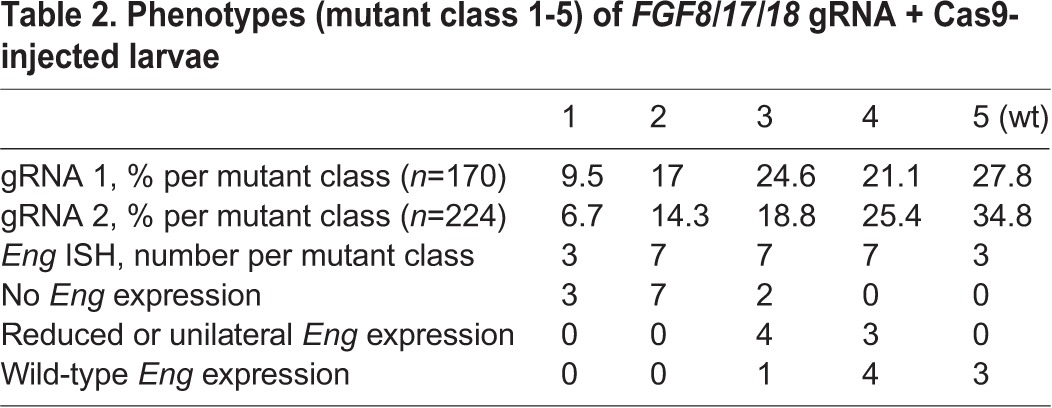
Fig. 3.
Mutant classes used for phenotypic analysis of FGF8/17/18 CRISPR/Cas9-mediated mutants. (A) Brightfield image of sibling lamprey embryos. These example embryos of classes 1-4 were injected with 500 pg Cas9 mRNA and 200 pg each of two FGF8/17/18 gRNAs. The embryo of class 5 is uninjected. (B) Red channel fluorescence showing LRD deposition in embryos. See Table 2 for information on the distribution of observed mutant classes in FGF8/17/18 experiments.
To verify that animals displaying these phenotypes were alive and injected rather than exhibiting autofluorescence due to cell death, we documented the relative brightness on both GFP and LRD absorption channels. Dead embryos fluoresced more strongly on the GFP channel than injected embryos, whereas they fluoresced less strongly on the LRD channel (Fig. S3). This supports the idea that these strong, consistent phenotypes that were observed are due to actual genetic perturbations rather than a termination of development.
We further analyzed FGF8/17/18 gRNA+Cas9 mRNA-injected embryos and larvae for changes in gene expression by in situ hybridization (Figs 4 and 5). We examined three genes known to be dependent on FGF signaling in lamprey and gnathostomes: Engrailed (Eng), which is expressed in the midbrain-hindbrain boundary (Reifers et al., 1998); Ednra, an Endothelin receptor expressed in post-migratory neural crest cells (Jandzik et al., 2014); and SoxE1, a sox9 ortholog that marks prechondrocytes (Jandzik et al., 2014; McCauley and Bronner-Fraser, 2006). We also looked at the expression of Mef2, a marker of formed somites and pharyngeal mesoderm, to monitor somitogenesis and pharyngeal segmentation (Jandzik et al., 2014). Consistent with previous reports, expression of Eng (Fig. 4), Ednra (not shown) and SoxE1 (Fig. 5A-D) was lost or reduced in presumptive FGF8/17/18 mutants, while Mef2 expression revealed a loss of somites and disruptions in somite patterning, as seen in the zebrafish fgf8 mutant acerebellar (Reifers et al., 1998) (Fig. 5E-K). Mef2, Ednra and SoxE1 expression in the pharyngeal arches also revealed a loss of pharyngeal segmentation, a consequence of FGF8/17/18 loss of function in both gnathostomes and lamprey (Jandzik et al., 2014; Reifers et al., 1998; Crump et al., 2004). Occasionally, these disruptions were unilateral, presumably resulting from localization of the injection solution to one of two blastomeres when injections were performed at mid-cleavage (e.g. Fig. 4E). We also assayed HhA expression, a lamprey ortholog of sonic hedgehog, which is a marker of the zona limitans intrathalamica within the forebrain. As expected, HhA expression in this signaling center was unaffected in FGF8/17/18 mutant larvae, indicating that loss of Eng expression reflects a specific loss of FGF8/17/18 in the midbrain-hindbrain boundary, rather than a non-specific disruption of neural patterning (Fig. 5L,M).
Fig. 4.
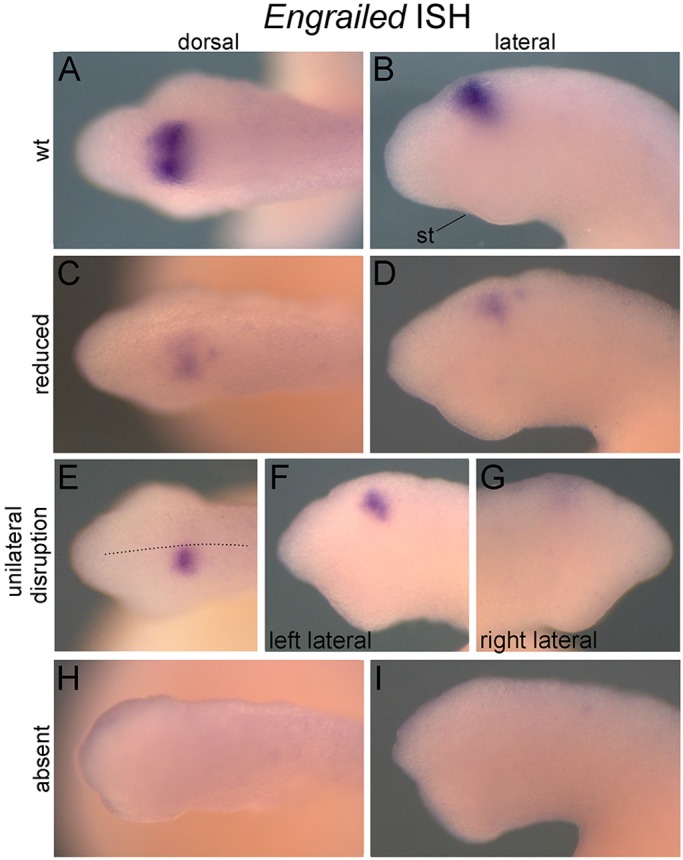
Eng ISH on FGF8/17/18 CRISPR/Cas9-mediated mutants at st. 23. Anterior is to the left in all panels except G, where it is to the right. (A,B) Wild-type Eng expression in the midbrain-hindbrain boundary of a control embryo. (C-I) Eng expression in FGF8/17/18 CRISPR/Cas9-mediated mutants. Reduction (C,D), unilateral disruptions (E-G), and total absence (H,I) of Eng expression were all observed (see Table 2 for counts). The dotted line (E) indicates the dorsal midline. st, stomodeum.
Fig. 5.

SoxE1, Mef2 and HhA ISH on FGF8/17/18 CRISPR/Cas9-mediated mutants. Anterior is to the left in all panels except H, where it is to the right. (A-D) SoxE1 ISH on wild-type (A,C) and FGF8/17/18 CRISPR/Cas9-mediated mutant (B,D) larvae. (B) A unilateral disruption of FGF8/17/18 resulted in the loss of SoxE1 expression in the posteriormost six pharyngeal arches (asterisks). (E-K) Mef2 ISH on wild-type (E) and FGF8/17/18 CRISPR/Cas9-mediated mutant (F-K) larvae. (F) Some highly affected mutant larvae showed a complete loss of pharyngeal arch SoxE1 expression (asterisks). (G,H) Some moderately affected mutant larvae showed a unilateral disruption of Mef2 expression in pharyngeal pouch mesoderm (asterisks in H). (I-K) Mef2 also revealed somite shape and spacing, which was sometimes slightly disrupted (J) or highly disrupted (K) compared with wild type (I). (L,M) HhA ISH on a wild-type larva (L) and a presumed FGF8/17/18 CRISPR-mediated mutant larva (M), both of which show proper expression of HhA in the zona limitans intrathalamica (arrows), indicating that this organization center has, as expected, remained unperturbed.
To verify disruption of the FGF8/17/18 locus in putative mutants, two embryos with reduced heads (mutant class 3) that were injected with Cas9 mRNA+FGF8/17/18 gRNA 1 alone and two embryos with reduced heads (mutant class 3) that were injected with Cas9 mRNA+FGF8/17/18 gRNA 2 alone were pooled and genotyped. Of the 12 alleles observed, six were wild type and six contained a deletion (Fig. S2B,C).
Negative controls
To ensure that our pigment and FGF-related phenotypes were specific to these perturbations rather than being a result of non-specific Cas9 or gRNA activity, a negative control gRNA was synthesized against Xenopus laevis tyrosinase (see Table S1 for sequence) and injected at 400 pg per embryo with 1 ng Cas9 mRNA (n=231). We found that 5.6% (13/231) of these negative control lamprey showed non-specific, inconsistent developmental defects (such as a severe developmental delay, or bending along the anteroposterior axis), supporting the specificity of the head-reduction phenotype of FGF8/17/18 mutants. Similarly, no control-injected embryo showed a reduction in pigmentation, ruling out the possibility that these levels of Cas9 mRNA or gRNA inherently disrupt melanin production.
DISCUSSION
Sea lampreys take 7-15 years to mature, can grow to almost 1 m in length, and require large, live prey as adults. As a result, the establishment of mutant or transgenic lines of this species is a practical impossibility. Here we present a proof-of-principle application of CRISPR/Cas9-mediated mutagenesis in the sea lamprey. Using our optimized parameters, we found that a majority of injected ‘F0’ embryos develop into complete or partial mutants with reproducible phenotypes and minimal confounding side effects. Remarkably, this efficiency approached 100% when using purified Cas9 protein in the injection mixture. The ability to generate large numbers of genotype-verifiable mutant F0 larvae from the embryos of wild-caught adults opens exciting new possibilities for studying the genetic bases of development in this phylogenetically important species.
Our study suggests that the sea lamprey has a combination of features that make it well-suited to CRISPR/Cas9-based genetic manipulation. Like amphibian embryos, sea lamprey embryos are easy to inject and can tolerate large quantities of synthetic RNA and protein. Having more Cas9 protein and gRNA per cell not only increases mutagenesis efficiency, but opens up the possibility of mutagenizing multiple genes simultaneously. We also found that the nucleotide composition of the lamprey genome makes it relatively easy to design multiple, high-quality gRNAs for a given target exon. The most effective CRISPR gRNAs have a GC content of over 50% (Doench et al., 2014; Gagnon et al., 2014), and all target sites must incorporate a 3′ PAM signal (5′-NGG-3′). Because of this, genes that are very AT rich can lack appropriate target sites near PAM motifs. Sea lamprey exons have an average GC content of 61% (Smith et al., 2013). As a result, a typical lamprey exon will have several potential CRISPR/Cas9 target sites near PAM signals. This permits multiple gRNAs to be designed against a single target exon and co-injected, mitigating potential variation in the effectiveness of individual gRNAs. Having multiple gRNAs per gene also provides opportunities to control for off-target phenotypes. For example, if two or more different gRNAs (or gRNA cocktails) targeting the same gene have the same effect on development, it can be safely assumed that the phenotype is specific to the gene, even before genotyping.
Although the lamprey genome lends itself to CRISPR/Cas9-mediated manipulations, we did encounter some challenges with the current publicly available assemblies. Owing to large sequence gaps, many sea lamprey genes identified as cDNAs, or through transcriptome sequencing, are not present in the current version of the genome. Because the exon structure is unknown, CRISPR target sites for these genes run the risk of spanning exons and thus not being contiguously represented in the genome (which is required for recognition by the Cas9/gRNA complex). Fortunately, exon structure tends to be strongly conserved across animals (Rogozin et al., 2003) and thus the risk of spanning exons can be reduced by focusing on evolutionarily conserved exons. The incomplete genome also makes it impossible to scan for all potential off-targets, as is becoming standard practice for other model organisms. These issues should be resolved with future versions of the lamprey genome assembly.
A perennial challenge for understanding the function of developmental regulatory genes is that many are highly pleiotropic. In genetic model systems, strategies for temporally or spatially restricting expression of Cas9 enzyme or gRNA have been proposed as a way to circumvent this problem. We targeted the highly pleiotropic gene FGF8/17/18 to get a sense for how this issue might affect the creation and scoring of sea lamprey mutants. We found that although a large percentage of CRISPR/Cas9-mediated FGF8/17/18 mutants appeared to be near-complete nulls, many others had milder phenotypes consistent with mosaics or hypomorphs, frequently displaying unilateral disruptions in FGF-dependent processes. This difference in penetrance corresponded well to the intensity of lineage tracer fluorescence in the mutant embryos (Fig. 3), as reported in axolotl (Flowers et al., 2014). Going forward, sorting mutants according to lineage tracer intensity and distribution should allow researchers to separate and score complete versus partial mutant embryos. In addition, it should be possible to intentionally restrict CRISPR/Cas9 activity to particular embryonic domains by injecting single blastomeres at later cleavage stages (i.e. 8- or 16-cell stage). One strategy for this would be to inject Cas9 mRNA or protein into zygotes, followed by gRNA injection at later cleavage stages. Although there is no modern developmental fate map for lamprey, lamprey embryos are easy to inject, and thousands can be injected by a single worker each hour. Co-injections of gRNA with a lineage tracer should allow selection of embryos and larvae with the gRNA restricted to the desired tissue, facilitating the study of highly pleiotropic developmental regulators.
In this study, a pair of gRNAs designed against FGF8/17/18 showed different levels of activity, whereas the pair designed against Tyr did not. Variation in gRNA efficiency is well-documented (Hsu et al., 2013) and is likely to be due to the specific binding properties of each RNA. Individual genotyping of Tyr mutants revealed that there is likely to be a correlation between the number of mutated Tyr sequences and the level of albinism observed in a given individual (Fig. 2), although a true test of this would require much more genotyping of many more individuals. Furthermore, Tyr disruptions using gRNA 1 yielded up to 99% albino individuals when used in combination with the protein (see Table 1), suggesting that the targeted region of tyrosinase is catalytically important, such that a frameshift is not required to hinder melanin synthesis. If a frameshift were required, we would at best still expect ∼1/3 of all pigment cells in a given batch of larvae to retain proper melanin deposition.
MATERIALS AND METHODS
Lamprey spawning and husbandry
Adult spawning phase P. marinus were supplied by Hammond Bay Biological Station (Millersburg, MI, USA) in spring and kept in holding tanks until fully ripened. Eggs and sperm were stripped manually from ripe adults (Nikitina et al., 2009). For Tyr and FGF8/17/18 gRNA injections, six and seven different females, respectively, were used during eight and ten different fertilization events, respectively. Sperm from two to four males was typically combined per fertilization event, although some fertilization events had a single sire. Embryos and larvae were kept at 17-18°C either in a closed, recirculating system, or in single 0.5-1 liter dishes containing deionized water supplemented with 400-600 ppm artificial sea salt. Water was changed as necessary, and dead embryos were removed from each dish every day to prevent contamination. Staging was performed according to Tahara (1988). Injected embryos (including negative controls) typically exhibited a slight developmental delay; to account for this delay, all staging was performed based on morphological landmarks of unaffected embryos rather than actual age (usually resulting in a 0.5-1 day delay in fixation compared with wild-type siblings).
Identification and cloning of P. marinus Tyr
Using gnathostome tyrosinase protein sequences as a query, we searched the March 2007 P. marinus genome assembly for a Tyr homolog (Smith et al., 2013). We identified four putative Tyr exons on contig 46507, amplified one of these by PCR, and ligated it into the pJet1.2 vector (Thermo Fisher Scientific). Orthology of the protein encoded by this exon to gnathostome tyrosinase was confirmed by alignment and phylogenetic analysis using the maximum likelihood method in the software package MEGA6 (Tamura et al., 2013) (Fig. S1). The corresponding sequence was deposited in GenBank (accession number KR150760).
RNA design and synthesis
Human codon-optimized Streptococcus pyogenes Cas9 mRNA (Cong et al., 2013) was transcribed as previously described (Guo et al., 2014). After digestion with DNase, unincorporated nucleotides were removed using a Sephadex G-50 column (GE Healthcare). The mRNA was further purified by phenol/chloroform extraction and precipitation as per standard procedures. Precipitated mRNA was resuspended in RNase-free water for injection.
CRISPR/Cas9 target sites with the sequence 5′-GG(18N)NGG-3′ were chosen based on three criteria: (1) 50-80% GC content; (2) maximal proximity to the presumptive start codon; and (3) no off-target matches to the known P. marinus genome or EST databases showing more than 80% similarity by BLAST, or with fewer than three mismatches in the ten bases proximal to the PAM sequence. The sequences of all target sites are listed in Table S1. Complementary oligonucleotides (Life Technologies) corresponding to the selected genomic target sequences and containing 4 nt overhangs were annealed, phosphorylated in vitro, then ligated into the BsaI-digested DR274 plasmid (Hwang et al., 2013). gRNAs were produced as previously described (Blitz et al., 2013) using full or half-size reactions of the T7 High Yield Kit (New England Biolabs). gRNAs were purified for injection as described above for Cas9 mRNA. Two target sites within the same exon were chosen per targeted gene.
Preparation of protein
Cas9 protein was purchased from PNA Bio. Cas9 protein was resuspended as per the manufacturer's instructions, aliquoted, and stored at −70°C. 1 µg Cas9 protein was incubated with 400 ng gRNA (a 2.5:1 ratio by mass) for 10 min on ice, then brought up to 5 μl with LRD and nuclease-free water for injection (see below).
Microinjection
Microinjections were performed in embryo system water in a room kept at 19-20°C. Embryos were injected at the 1- to 2-cell stage with ∼5 nl injection solution containing 500 or 1000 pg Cas9 mRNA, 400 or 800 pg gRNA, and 10% lysinated Rhodamine-dextran (LRD). For injections with Cas9 protein we used 400 pg Tyr gRNA 2 plus 1000 pg Cas9 protein. For some injections, 125 pg GFP mRNA was included to test for degradation of injected RNAs. Embryos were sorted by fluorescence at st. 21-26, and LRD-negative embryos were discarded.
Phenotypic analyses of mutant embryos and larvae
To assess the efficiency of Tyr mutagenesis, we fixed Tyr gRNA+Cas9 mRNA/Cas9 protein-injected lamprey larvae at st. 30 (∼30 days post-fertilization). Larvae were compared with wild-type siblings and scored as having: (1) wild-type pigmentation; (2) <50% pigment reduction; (3) 50-90% pigment reduction; or (4) 90-100% pigment reduction. FGF8/17/18 gRNA+Cas9 mRNA-injected larvae were fixed when their injected, unaffected siblings reached st. 23.5 or st. 26.5 and scored for gross morphology. Phenotypic scoring for head size (by total estimated volume) was split into five mutant classes: (1) no or almost no head, where only a small bump is visible; (2) head is highly reduced with no discernible morphological landmarks; (3) head is 50-75% of its normal volume, with visible (but reduced) first stream neural crest mesenchyme; (4) head is slightly reduced in volume, with most head characters discernible, although the upper lip is usually truncated; (5) grossly wild type in appearance. Changes in the expression of Eng, Ednra, SoxE1 and Mef2 were visualized by in situ hybridization.
It should be noted that there was some variability in the severity and frequency of non-specific defects between injection batches due to differences in how eggs from different females tolerated injection. Females with particularly robust eggs showed survival and frequencies of developmental delay and deformity identical to uninjected wild-type embryos. By contrast, females with sensitive eggs showed a higher percentage of death and non-specific defects upon injection compared with their wild-type siblings. Such variation is not unexpected, given the genetic and environmental variation inherent in wild populations, and is effectively mitigated by injecting each gRNA into two or more females per season.
Genotyping
To confirm mutagenesis, we isolated genomic DNA from gRNA+Cas9 mRNA-injected embryos and larvae as per standard methods. The target loci were amplified by PCR and ligated into the pJet1.2 vector. Genotyping primer sequences are listed in Table S1. After transformation and plating as per standard methods, single clones were amplified, purified and sequenced.
In situ hybridization (ISH)
Riboprobe templates were amplified by PCR using primers incorporating an SP6 RNA polymerase site. Riboprobes were then synthesized with SP6 RNA polymerase, and ISH carried out as previously described (Cerny et al., 2010). ISH was performed in parallel on wild-type embryos and larvae to normalize signal development times. St. 25+ larvae probed for Tyr expression were bleached in 1×SSC/5% formamide/1% H2O2 to allow for visualization of signal in pigmented cells.
Acknowledgements
We thank Haley Stein and Cole Steinmetz for assistance with P. marinus husbandry; Andrew Hansen and Kathryn Hutton for assistance with genotyping; and the anonymous reviewers whose suggestions significantly improved the work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.M.M. and M.K. conceived the project. D.M.M., T.S., M.R. and D.J. designed experiments. T.S., M.R., D.J. and M.V.C. performed experiments. D.M.M., T.S. and M.R. wrote the manuscript.
Funding
T.S., M.R., D.J., M.V.C. and D.M.M. were supported by a National Science Foundation grant [IOS1257040] to D.M.M. M.K. was supported by National Institutes of Health grant [3R01GM074746]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.125609/-/DC1
References
- Abzhanov A. and Tabin C. J. (2004). Shh and Fgf8 act synergistically to drive cartilage outgrowth during cranial development. Dev. Biol. 273, 134-148. 10.1016/j.ydbio.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A. and Horvath P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709-1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P. and Liu J.-L. (2013). Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220-228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish F. W. H. (1980). Biology of the North American Anadromous Sea Lamprey, Petromyzon Marinus. Can. J. Fish. Aquat. Sci. 37, 1924-1943. 10.1139/f80-233 [DOI] [Google Scholar]
- Belhaj K., Chaparro-Garcia A., Kamoun S. and Nekrasov V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39 10.1186/1746-4811-9-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz I. L., Biesinger J., Xie X. and Cho K. W. Y. (2013). Biallelic genome modification in F 0 Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51, 827-834. 10.1002/dvg.22719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny R., Cattell M., Sauka-Spengler T., Bronner-Fraser M., Yu F. and Medeiros D. M. (2010). Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc. Natl. Acad. Sci. USA 107, 17262-17267. 10.1073/pnas.1009304107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close D. A., Yun S.-S., McCormick S. D., Wildbill A. J. and Li W. (2010). 11-Deoxycortisol is a corticosteroid hormone in the lamprey. Proc. Natl. Acad. Sci. USA 107, 13942-13947. 10.1073/pnas.0914026107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. G., Maves L., Lawson N. D., Weinstein B. M. and Kimmel C. B. (2004). An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development 131, 5703-5716. 10.1242/dev.01444 [DOI] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B. L., Xavier R. J. and Root D. E. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262-1267. 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers G. P., Timberlake A. T., Mclean K. C., Monaghan J. R. and Crews C. M. (2014). Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development 141, 2165-2171. 10.1242/dev.105072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., Pauli A., Montague T. G., Zimmerman S., Richter C. and Schier A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhang T., Hu Z., Zhang Y., Shi Z., Wang Q., Cui Y., Wang F., Zhao H. and Chen Y. (2014). Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development 141, 707-714. 10.1242/dev.099853 [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O. et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827-832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J.-R. J. and Joung J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikmi A., McKinney S. A., Delventhal K. M. and Gibson M. C. (2014). TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 5, 5486 10.1038/ncomms6486 [DOI] [PubMed] [Google Scholar]
- Jandzik D., Hawkins M. B., Cattell M. V., Cerny R., Square T. A. and Medeiros D. M. (2014). Roles for FGF in lamprey pharyngeal pouch formation and skeletogenesis highlight ancestral functions in the vertebrate head. Development 141, 629-638. 10.1242/dev.097261 [DOI] [PubMed] [Google Scholar]
- Jao L.-E., Wente S. R. and Chen W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904-13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakiza O., Miller S., Bunce A., Lee E. M.-J. and McCauley D. W. (2011). SoxE gene duplication and development of the lamprey branchial skeleton: insights into development and evolution of the neural crest. Dev. Biol. 359, 149-161. 10.1016/j.ydbio.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Lo T.-W., Pickle C. S., Lin S., Ralston E. J., Gurling M., Schartner C. M., Bian Q., Doudna J. A. and Meyer B. J. (2013). Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195, 331 10.1534/genetics.113.155382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M. and Church G. M. (2013). Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957-963. 10.1038/nmeth.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason I., Chambers D., Shamim H., Walshe J. and Irving C. (2000). Regulation and function of FGF8 in patterning of midbrain and anterior hindbrain. Biochem. Cell Biol. 78, 577-584. 10.1139/o00-073 [DOI] [PubMed] [Google Scholar]
- McCauley D. W. and Bronner-Fraser M. (2006). Importance of SoxE in neural crest development and the evolution of the pharynx. Nature 441, 750-752. 10.1038/nature04691 [DOI] [PubMed] [Google Scholar]
- McCauley D. W. and Kuratani S. (2008). Cyclostome studies in the context of vertebrate evolution. Zool. Sci. 25, 953-954. 10.2108/zsj.25.953 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Fish M. B., Fisher M., Oomen-Hajagos J., Thomsen G. H. and Grainger R. M. (2013). Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis 51, 835-843. 10.1002/dvg.22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina N., Bronner-Fraser M. and Sauka-Spengler T. (2009). Culturing lamprey embryos. Cold Spring Harb. Protoc. 2009, pdb.prot5122 10.1101/pdb.prot5122 [DOI] [PubMed] [Google Scholar]
- Nikitina N., Tong L. and Bronner M. E. (2011). Ancestral network module regulating prdm1 expression in the lamprey neural plate border. Dev. Dyn. 240, 2265-2271. 10.1002/dvdy.22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K. G., Kuraku S. and Kuratani S. (2007). Hagfish embryology with reference to the evolution of the neural crest. Nature 446, 672-675. 10.1038/nature05633 [DOI] [PubMed] [Google Scholar]
- Pancer Z., Amemiya C. T., Ehrhardt G. R. A., Ceitlin J., Gartland G. L. and Cooper M. D. (2004). Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174-180. 10.1038/nature02740 [DOI] [PubMed] [Google Scholar]
- Parker H. J., Bronner M. E. and Krumlauf R. (2014). A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 514, 490 10.1038/nature13723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piavis G. W. (1971). The Biology of Lampreys. London: Academic Press. [Google Scholar]
- Reifers F., Bohli H., Walsh E. C., Crossley P. H., Stainier D. Y. R. and Brand M. (1998). Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381-2395. [DOI] [PubMed] [Google Scholar]
- Rogozin I. B., Wolf Y. I., Sorokin A. V., Mirkin B. G. and Koonin E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512-1517. 10.1016/S0960-9822(03)00558-X [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Meulemans D., Jones M. and Bronner-Fraser M. (2007). Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell 13, 405-420. 10.1016/j.devcel.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Smith J., Morgan J. R., Zottoli S. J., Smith P. J., Buxbaum J. D. and Bloom O. E. (2011). Regeneration in the Era of functional genomics and gene network analysis. Biol. Bull. 221, 18-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Kuraku S., Holt C., Sauka-Spengler T., Jiang N., Campbell M. S., Yandell M. D., Manousaki T., Meyer A., Bloom O. E. et al. (2013). Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 45, 415-421. 10.1038/ng.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sower S. A. (2003). The endocrinology of reproduction in lampreys and applications for male lamprey sterilization. J. Great Lakes Res. 29, 50-65. 10.1016/S0380-1330(03)70477-8 [DOI] [Google Scholar]
- Sower S. A., Freamat M. and Kavanaugh S. I. (2009). The origins of the vertebrate hypothalamic-pituitary-gonadal (HPG) and hypothalamic-pituitary-thyroid (HPT) endocrine systems: new insights from lampreys. Gen. Comp. Endocrinol. 161, 20-29. 10.1016/j.ygcen.2008.11.023 [DOI] [PubMed] [Google Scholar]
- Sun X., Meyers E. N., Lewandoski M. and Martin G. R. (1999). Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 13, 1834-1846. 10.1101/gad.13.14.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y. (1988). Normal stages of development in the lamprey, Lampetra-Reissneri (Dybowski). Zool. Sci. 5, 109-118. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. and Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725-2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F. and Jaenisch R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910-918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]





