Abstract
Yolk provides an important source of nutrients during the early development of oviparous organisms. It is composed mainly of vitellogenin proteins packed into membrane-bound compartments called yolk platelets. Catabolism of yolk is initiated by acidification of the yolk platelet, leading to the activation of Cathepsin-like proteinases, but it is unknown how this process is triggered. Yolk catabolism initiates at cellularization in Drosophila melanogaster embryos. Using maternal shRNA technology we found that yolk catabolism depends on the Tor pathway and on the autophagy-initiating kinase Atg1. Whereas Atg1 was required for a burst of spatially regulated autophagy during late cellularization, autophagy was not required for initiating yolk catabolism. We propose that the conserved Tor metabolic sensing pathway regulates yolk catabolism, similar to Tor-dependent metabolic regulation on the lysosome.
KEY WORDS: Yolk, Tor, Drosophila
Summary: The conserved Tor and autophagy metabolic sensing pathways are required for a burst of spatially regulated yolk catabolism during Drosophila cellularization.
INTRODUCTION
Oviparous (non-placental) embryos are closed systems that are provisioned with nutrients during oogenesis. Storage molecules supply energy and biosynthetic precursors required for early embryogenesis. These include protein in yolk platelets, as well as glycogen and lipid droplets (Gutzeit et al., 1994; Koch and Spitzer, 1982). In insects, amphibians and many other species, yolk platelets are membrane-bound compartments that contain a dense aggregate of specific yolk proteins, primarily vitollegenins, which form in the oocyte by endocytosis of these proteins from maternal supplies (Koch and Spitzer, 1982).
Regulation of yolk catabolism is not well understood. A leading hypothesis is that yolk degradation is triggered by decreasing pH in yolk platelets (Fagotto, 1995). Supporting this, yolk platelets become acidic during early development in Xenopus laevis (Fagotto and Maxfield, 1994). In Ornithodoros moubata (tick) eggs, Cathepsin-like proteinases within yolk platelets are activated in an acid-dependent manner (Fagotto, 1990a,b). Similarly, in Drosophila melanogaster, activation of a Cathepsin B proteinase correlates with catabolism of yolk platelets during embryogenesis (Medina et al., 1988). Although these studies revealed that embryogenesis is subject to enzymatic regulation, upstream cell signaling factors that regulate yolk catabolism during embryogenesis have not yet been identified.
The Tor pathway is a potential candidate for regulating yolk catabolism during embryogenesis because it is known to maintain nutrient level homeostasis through regulation of metabolic pathways in many systems (Kim et al., 2013b). Tor is a serine-threonine kinase regulated by amino acid abundance, the AMP/ATP sensor AMPK, and other growth signaling pathways (Bolster et al., 2002; Garami et al., 2003; Hara et al., 1998). In turn, Tor controls a number of processes in order to adjust metabolic activity to match available nutrient supplies, including protein translation, glucose import and autophagy (Buller et al., 2008; Hara et al., 1998; Kanazawa et al., 2004). Under nutrient-rich conditions, Tor localizes to the outer surfaces of lysosomes (Dibble et al., 2012). This recruitment is initiated by amino acids, which recruit Tor to the lysosome through Rag GTPases and regulate its kinase activity (Sancak et al., 2008).
An important downstream target of Tor is Atg1, which Tor phosphorylates and inhibits under nutrient-rich conditions. Atg1, known as ULK1 in humans, is a serine-threonine kinase whose activation triggers formation of autophagosomes (Matsuura et al., 1997). Atg1/ULK1 is thought to initiate autophagosome formation through phosphorylation of a number of autophagy pathway proteins including the AMBRA1–PIK3C3 complex and membrane-recruiting protein Atg9 (Di Bartolomeo et al., 2010; Papinski et al., 2014). Additionally, Atg1/ULK1 negatively regulates Tor through phosphorylation, whereby mutual inhibition between Tor and Atg1 creates a negative feedback loop (Dunlop et al., 2011). Atg1/ULK1 has also been implicated in potentially non-autophagic roles. In Caenorhabditis elegans, mutation of the gene encoding the Atg1 homolog UNC-51 causes defects in axonal elongation and accumulation of membrane and vesicles (Ogura, 2006). Similarly, mutation of Atg1 in Drosophila causes defects in vesicular transport along neurons (Mochizuki et al., 2011).
Here, we report the requirement of Tor for activation of the Cathepsin-like proteinase that promotes yolk catabolism in D. melanogaster embryos. Additionally, we reveal that catabolism depends on Atg1, but is independent of autophagy. These findings shed light on how a conserved metabolic sensing pathway has been opted to regulate metabolite provision in early embryos, which are closed to nutrient import from the environment.
RESULTS
The Tor pathway regulates yolk catabolism in early Drosophila embryogenesis
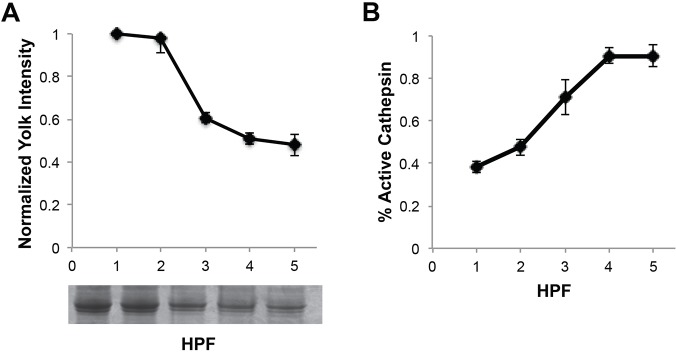
In Drosophila, three vitellogenins make up ∼20% of the total protein in the early embryo (Bownes and Hames, 1977; Warren and Mahowald, 1979). To measure vitellogenins, we subjected total embryonic extracts from ten embryos to SDS-PAGE and stained with Coomassie Blue. We found that vitellogenin levels, measured by densitometry of bands running at 45, 46, and 47 kDa, decreased starting at 2-3 h post fertilization (cellular blastoderm stage) (Fig. 1A). This finding is in agreement with previous reports of the timing of vitellogenin catabolism and activation of a yolk-bound Cathepsin B-like proteinase in Drosophila embryos (Bownes and Hames, 1977; Medina et al., 1988). Measurement of total vitellogenin in this manner is only semi-quantitative, because it is sensitive to loss of protein during sample preparation and variability in embryo size. To more quantitatively assess yolk catabolism we measured total Cathepsin B-like proteinase enzymatic activity with a standard fluorogenic peptide substrate. This activity was shown to coincide with yolk degradation in Drosophila (Medina et al., 1988). Cathepsins are activated at low pH, and have a variable optimum pH range for proteinase activity. We determined that Cathepsin B-like proteinase enzymatic activity in Drosophila embryonic extract exhibits maximal activation when pre-treated at pH 3.5, and has an optimal activity range of pH 4.5-5.5 (Fig. S1). These values are similar to those previously reported by Fagotto (1990b). A previous study by Medina et al. (1988) reported that 93% of Cathepsin B-like proteinase activity was in the insoluble yolk fraction during this developmental time window.
Fig. 1.
Yolk catabolism is correlated with Cathepsin B-like proteinase activation. (A) Vitellogenin levels of control shRNA embryos normalized to 1 HPF yolk intensity based on Coomassie Blue staining reveal that vitellogenin levels decrease from 2-3 h post fertilization. (B) The percentage of active Cathepsin B-like proteinase in control shRNA embryos increases over the first 5 h of development. Yolk concentration and Cathepsin B-like proteinase enzyme activity are negatively correlated (Pearson's correlation, r=−0.98, P=0.004). HPF, hours post fertilization. Data are represented as the s.d. of three biological replicates.
To measure the extent to which yolk catabolism has been activated, we measured Cathepsin B-like proteinase activity in embryonic lysate without acid pre-treatment at pH 3.5, and normalized it to activity after pre-activation. This procedure, which was devised by Fagotto in tick embryos, measures fractional activation and also corrects for variability in embryo size and lysate preparation (Fagotto, 1990b). The fraction of Cathepsin B-like proteinase that was activated was measured over the first 5 h of development. In control embryos, 50% of total Cathepsin B-like proteinase activity was already activated in 0-2.5 h embryos, whereas 100% was activated in 2.5-5 h embryos (Fig. 1B). The Cathepsin B-like proteinase activity negatively correlated with a decrease in total vitellogenin (r=−0.98, P=0.004; Fig. 1B). The fraction of Cathepsin B-like proteinase already activated at early stages is higher in Drosophila embryos (∼50%) than that reported in tick (∼2%) (Fagotto, 1990b). This difference might reflect more rapid development in Drosophila, where yolk is utilized within 24 h, as compared with tick eggs which consume yolk over a period of 15 days (Fagotto, 1990a).
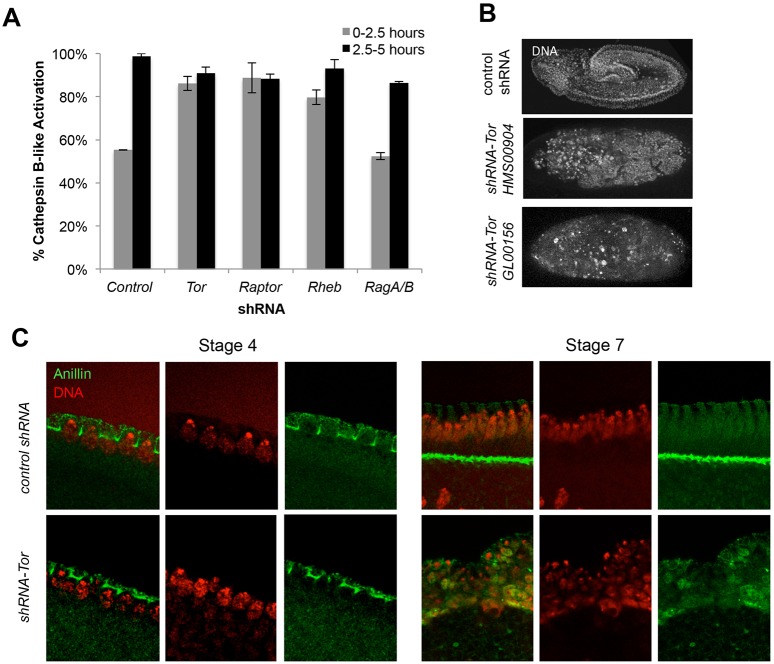
To determine whether Tor was involved in yolk catabolism, we generated Tor deficient embryos using the maternal-Gal4/UAS-shRNA system to generate females loaded with maternal short hairpin RNAs (shRNAs) targeting Tor (Ni et al., 2011; Sopko et al., 2014), referred to as shRNA-Tor embryos. Knockdown efficiency of all shRNA lines used in this paper were quantified by RT-PCR, as reported in Table S1. Note that Tor activity in the germ line is essential for growth and survival (LaFever et al., 2010; Sun et al., 2010). However, by using a maternal Gal4 driver that induces shRNA expression outside the germline stem cell compartment during stage 1 of oogenesis, we were able to bypass the early germline defects (Fig. S2D) (Yan et al., 2014). shRNA-Tor embryos were smaller than control shRNA embryos and showed significant DNA fragmentation post-cellularization, which to our knowledge has not previously been reported as a consequence of Tor mutation or chemical inhibition (Fig. 2B,C; Fig. S2A). Phospho (Thr398) S6k was undetectable in both shRNA-control embryos and shRNA-Tor embryos by immunoblotting, suggesting that the normal regulation of translation through S6k by Tor might not occur in embryos during this early period when they are maternally loaded with ribosomes (Fig. S2B). By introducing an EGFP-tagged Histone-2Av (His2Av-EGFP) into the shRNA-Tor line we were able to visualize the clumping and misorganization of DNA following syncytial divisions, which was not seen in shRNA-control embryos (Movies 1, 2). The same phenotype was observed using two shRNAs targeting different regions of the Tor transcript, reducing the possibility that the shRNA-Tor phenotype is a result of off-target effects (Fig. 2B). TUNEL staining of shRNA-Tor embryos was positive for nicked DNA after cellularization, suggesting that the fragmented DNA might be apoptosis-related (Fig. S3).
Fig. 2.
Tor is necessary for yolk catabolism. (A) The percentage of active Cathepsin B-like proteinase in shRNA-expressing embryos pre-cellularization (0-2.5 h) and post-cellularization (2.5-5 h). Data are represented as the s.d. of three biological replicates. (B) Phenotype of stage 7 shRNA-Tor embryos using two different hairpins, showing that shRNA-Tor embryos display significant DNA fragmentation post-cellularization. (C) Staining of stage 4 (left) and stage 7 (right) shRNA-control and shRNA-Tor embryos with the cellularization marker anillin, showing clumping of nuclei around the periphery of the embryos with no clear cellularization in shRNA-Tor embryos.
Most Cathepsin B-like proteinase activity was already activated in 0-2.5 h shRNA-Tor embryos and did not change significantly between 0-2.5 and 2.5-5 h (Fig. 2A, Fig. S2C). Thus, Tor is necessary for regulating yolk catabolism. Tor is part of a complex, Torc1, which includes Raptor, LST8 and Pras40 (Loewith et al., 2002). In shRNA-raptor embryos, we observed similar, high and unchanging, Cathepsin B-like proteinase activity to that of shRNA-Tor embryos (Fig. 2A). Torc1 activity also requires the GTPase, Rheb (Inoki et al., 2003; Tee et al., 2003). In Rheb-depleted embryos, similar to shRNA-Tor and shRNA-raptor embryos, Cathepsin B-like proteinase activity was prematurely elevated and does not significantly increase further post-cellularization (Fig. 2A). Depletion of the GTPase-activating protein RagA-B, required for recruitment of Tor to the lysosome, did not affect the kinetics of Cathepsin B-like proteinase activity (Fig. 2A). Knockdown of Rag A-B or its heterodimer partner RagC-D was shown to only decrease Tor activity by 50% in Drosophila S2 cells (Demetriades et al., 2014), and so it is possible that co-depletion of other Rags together would have a stronger effect.
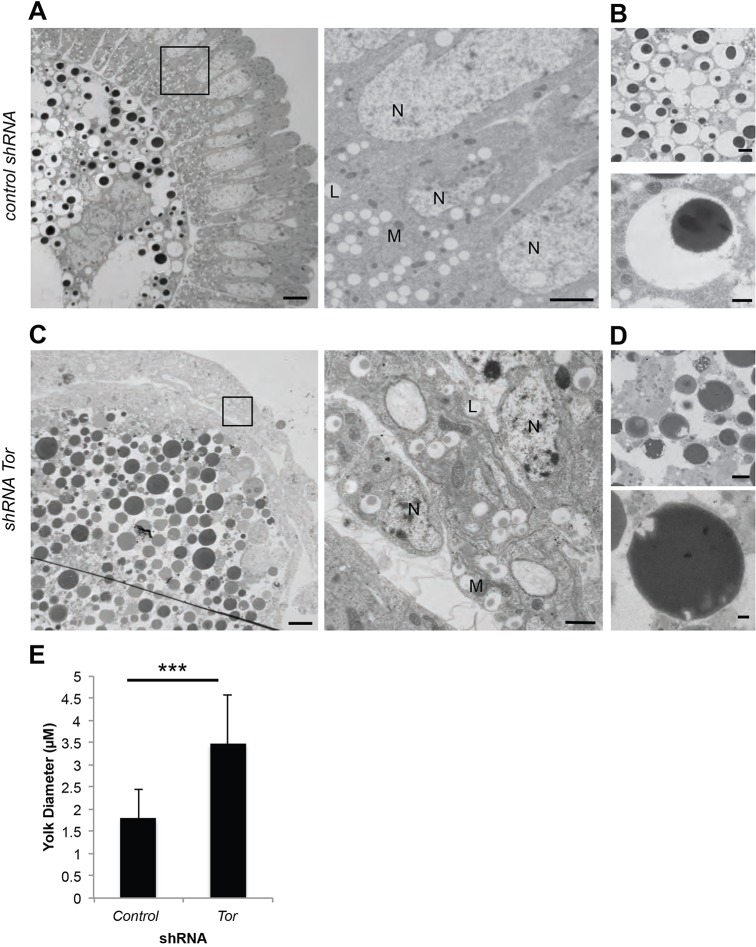
To determine whether the Tor-depleted embryos showed altered yolk platelet morphology, we examined them by thin section electron microscopy (EM). Prior to cellularization the shRNA-Tor embryos exhibited normal yolk morphology (Fig. S4A). However, during and after cellularization, shRNA-Tor embryos displayed abnormal yolk morphologies, specifically, decondensation of the vitellogenin mass as compared with control embryos, and appearance of electron-lucent areas within this mass (Fig. 3A-D). The diameter of the vitellogenin mass was also significantly larger than in control embryos (Fig. 3E). This morphology, combined with early activation of Cathepsin B-like proteinase, might indicate premature initiation of yolk catabolism.
Fig. 3.
Depletion of Tor leads to aberrant cellularization and abnormal yolk morphologies. (A-D) EM of shRNA-control (A) and shRNA-Tor (C) embryos post-cellularization, and yolk platelets in shRNA-control (B) and shRNA-Tor (D) embryos reveals abnormal yolk morphology in shRNA-Tor embryos compared with control embryos; specifically, decondensation of the vitellogenin mass and appearance of electron-lucent areas within this mass. (E) Diameter of yolk in control versus shRNA-Tor embryos (µm) also revealed a significant increase in the diameter of the vitellogenin mass in shRNA-Tor embryos. The longest diameter of the darkly stained EM portion of the yolk was measured as it represents the most energy-dense portion of the yolk platelets. Data are represented as the s.d. of the average of >100 yolk platelets counted from EM sections from three individual embryos; ***P<0.01. L, lipid droplet; M, mitochondria; N, nuclei. Scale bars: 10 µm in A (left), C (left); 2 µm in A (right), B (upper), D (upper); 1 µm in C (right); 500 nm in B (lower), D (lower).
EM also confirmed more general organizational defects seen at the light level. In control embryos, we observed normal membrane ingression around the nuclei after cellularization, whereas in shRNA-Tor embryos we observed clumping of nuclei around the periphery of the embryos with no clear cellularization (Fig. 2C). Additionally, swaths of multivesicular, endocytic-like compartments and potential autophagosomes were present throughout the disorganized cytoplasm (Fig. S4B). We conclude that Tor, the Torc1 complex member Raptor, and the lysosomal activating GTPase Rheb are required for normal yolk catabolism, and that Tor is also required for normal cytoplasmic morphology.
Atg1 promotes spatially regulated autophagy
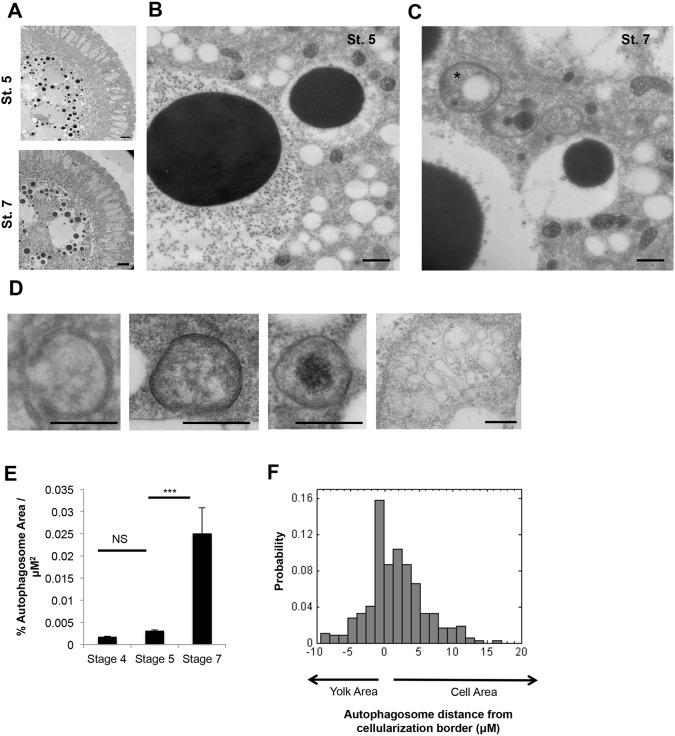
One important role of Tor is to regulate autophagy, which serves a major role in catabolizing macromolecules to provide nutrients to cells during starvation. Autophagy has been show to be involved in degradation of whole organelles including peroxisomes, mitochondria and lipid droplets (Hutchins et al., 1999; Kissova et al., 2004; Singh et al., 2009). We therefore suspected an involved role of autophagy in yolk catabolism. We started by characterizing autophagy during early development in shRNA-control embryos. In Drosophila, autophagy has been studied in specific tissues later in development, for instance in the developing fat body (Scott et al., 2004), but its activity during early developmental stages has not been characterized. By EM, we failed to observe autophagosomes during the syncytial divisions. Shortly after cellularization (by stage 7), abundant autophagosomes appeared, which were characterized by double bilayered compartments, 0.5-1 µm in diameter, often wrapped around mitochondria or lipid droplets (Fig. 4A-D). We carefully inspected micrographs for the presence of ribosomes on these membranes, to distinguish rough ER from autophagosome membranes. Multivesicular vacuoles that might represent late stage autolysosomes appeared at the same time (Fig. 4D). We also imaged the autophagosome marker, mCherry-Atg8a by immunofluorescence and observed formation of abundant puncta consistent with autophagosomes appearing shortly after the onset of cellularization (Fig. S5). Both EM and immunofluorescence revealed that autophagy is subject to tight spatial regulation in early Drosophila embryos. Autophagosomes formed primarily within a thin border, 5-7 µm wide, on both sides of the ingressing cellularization front, which was also observed with the mCherry-Atg8a reporter (Fig. 4F; Fig. S5B). Very few autophagosomes were observed in the central and apical regions of the blastomeres, or within the yolk mass.
Fig. 4.
Autophagy is initiated after cellularization and is spatially regulated. (A-C) EM images of shRNA-control embryos at stage 5 (pre-cellularization) and stage 7 (post-cellularization) (A), with magnification of stage 5 (B) and stage 7 (C) embryos, showing the appearance of autophagosomes after cellularization, and an autophagosome containing a lipid droplet (asterisk in C). (D) From left to right, examples of a phagopore, an autophagosome containing no organelles, an autophagosome containing mitochondria, and a multivesicular, endocytic-like structure. (E) Percentage of autophagosomes per µm2 in pre- and post-cellularization embryos, confirming that autophagosome appearance occurs post-cellularization. Autophagosomes were counted per stage in three biological replicates. Data are represented as the s.d. of three biological replicates; ***P<0.01. NS, not significant. (F) Cumulative distribution histogram of autophagosomes location within stage 7 embryos, showing that autophagosomes formed primarily on either side of the ingressing cellularization front. Autophagosomes were classified as located either in the yolk area (negative) or cell area (positive) based on their proximity to the cellularization border (F). Scale bars: 10 µm in A; 500 nm in B-D.
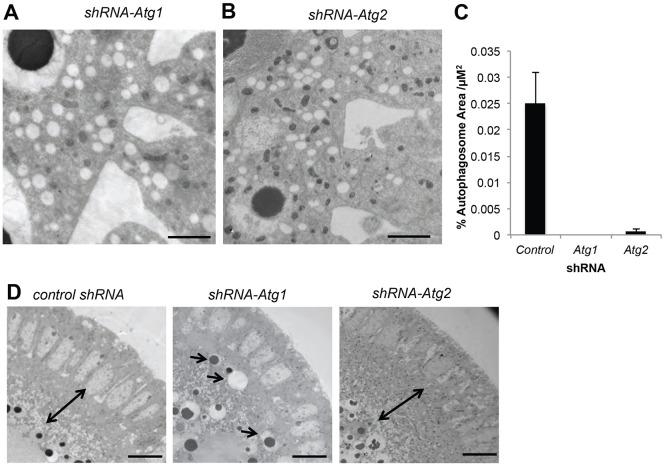
To test if the double bilayer-bound structures were indeed autophagosomes, and also evaluate the role of the autophagy pathway in their formation, we used EM to examine Atg1-deficient embryos generated by maternally expressing shRNA. We observed a drastic reduction in formation of autophagosomes at cellularization in these embryos (Fig. 5A,C). This was also observed using the UAS-mCherry-GFP-Atg8a reporter combined with an shRNA against Atg1 (Fig. S6). Additionally, when we knocked down a downstream component of the autophagy pathway, Atg2, autophagosomes at cellularization were also absent (Fig. 5B,C; Fig. S7). Interestingly, shRNA-Atg1 embryos also exhibited disorganization of organelles. In control embryos and shRNA-Atg2 embryos a layer of lipids and mitochondria forms between nuclei and yolk. In shRNA-Atg1 embryos this was not present (Fig. 5D). However, based on staining with the cellularization marker anillin, shRNA-Atg1 embryos appear to cellularize normally (Fig. S8). We conclude that autophagy is activated at the cellular blastoderm stage, that it triggers formation of abundant autophagosomes in a selective region spanning the cellularization front, and Atg1 also controls organelle patterning in an autophagy-independent manner.
Fig. 5.
Atg1 is necessary for autophagy after cellularization. (A-C) EM of stage 7 shRNA-Atg1 (A) and shRNA-Atg2 (B) embryos, and quantification of the percent autophagosomes per µm2 in shRNA-control versus shRNA-Atg1 and shRNA-Atg2 stage 7 embryos (C) reveals a drastic reduction in formation of autophagosomes at cellularization in shRNA-Atg1 and shRNA-Atg2 embryos. Data are represented as the s.d. of three biological replicates. (D) Stage 5 embryos in shRNA-control, shRNA-Atg1 and shRNA-Atg2 embryos show a layer of lipids and mitochondria forming between nuclei and yolk in control embryos and shRNA-Atg2 embryos (double-headed arrow) that is not present in shRNA-Atg1 embryos. Arrows indicate displaced organelles in shRNA-Atg1 embryos. Scale bars: 10 µm in D; 2 µm in A,B.
Atg1, but not autophagy, is required for normal yolk catabolism
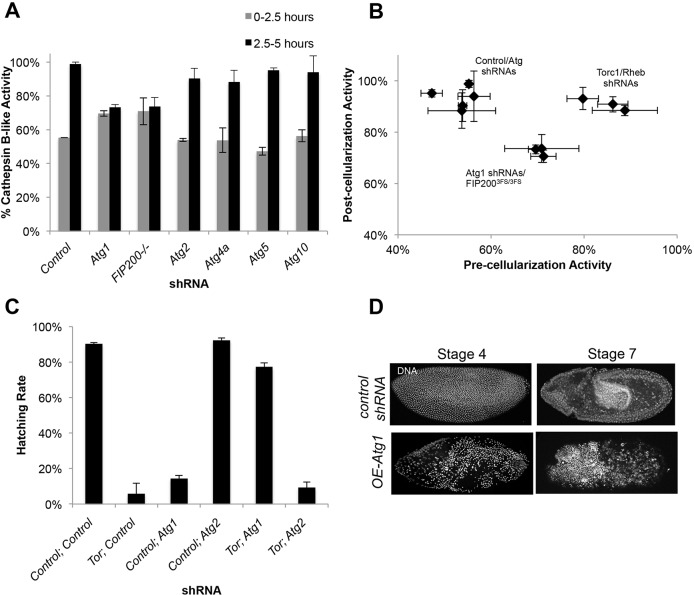
Given that both yolk catabolism and autophagy are initiated during the cellular blastoderm stage, and both are Tor-regulated, we next investigated whether autophagy is required for yolk catabolism. We measured Cathepsin B-like proteinase activity in embryos expressing shRNA targeting Atg1 and Atg2, which by EM were both deficient in autophagosomes (Fig. 5C). Surprisingly, Atg1, but not Atg2, was necessary for timely activation of Cathepsin B-like proteinase activity (Fig. 6A). Additional shRNAs against other autophagy proteins, Atg4a, Atg5 and Atg10, also had no effect on Cathepsin B-like proteinase enzyme activity compared with control embryos. We also tested Fip20035S/3F5 (Atg17) mutant embryos. Fip200 forms a complex with Atg1 and is required for autophagy (Hara et al., 2008). Fip200 homozygous mutants survive until the postnatal (P) 15 pupal stage, but die before eclosion (Kim et al., 2013a). Fip20035S/3F5 embryos showed similar Cathepsin B-like proteinase activity to shRNA-Atg1 embryos, demonstrating that the Atg1 complex, not just Atg1 kinase, is required for yolk catabolism. Interestingly, the Cathepsin B-like proteinase basal activity of shRNA-Atg1 and Fip20035S/3F5 embryos was higher than control shRNA embryos, but not as high as the basal activity of shRNA-Tor embryos. When both pre-and post-cellularization activity were plotted on a 2D graph, embryos deficient for Atg1 complex components (Atg1 and Fip200) show different activation levels compared with both the control embryos and Tor pathway knockdowns (Fig. 6B). Coomassie staining of shRNA-Atg1 compared with control showed no change in yolk vitellogenins between 0 and 2.5 hours post-fertilization (HPF) and 2.5-5 HPF, similar to shRNA-Tor embryos (Fig. S9). However, based on this staining we can only measure relative levels over time, we cannot determine whether total yolk levels differ. Based on these findings, we determined that the Atg1 complex is necessary for yolk catabolism, but autophagy is not required. Additionally, Atg1 knockdown affects yolk catabolism in a distinctly different manner from knockdown of components in the Tor pathway.
Fig. 6.
Atg1, but not autophagy, is required for yolk catabolism and is regulated in a Tor-dependent feedback loop. (A) Quantification of percentage of active Cathepsin B-like proteinase in embryos pre-cellularization (0-2.5 h) and post-cellularization (2.5-5 h) shows that Atg1, but not Atg2, is necessary for timely activation of Cathepsin B-like proteinase activity. (Note: Fip2003F5/3F5 are mutant embryos, not shRNA.) (B) A 2D plot of pre- versus post-Cathepsin B-like proteinase activation in shRNA-expressing embryos segregates into three different clusters, demonstrating that control embryos, embryos deficient for Atg1 complex components (Atg1 and Fip200), and Tor pathway knockdown embryos all show different activation levels. (C) Hatching rates of double shRNA knockdowns show that depletion of Atg1 rescued the hatch rate defects of shRNA-Tor embryos, but the same rescue was not demonstrated with expression of shRNA-Atg2. Data are represented as the s.d. of either biological triplicates (Cathepsin B-like proteinase activity) or biological duplicates (hatching rates). (D) Overexpression of Atg1 reveals a similar phenotype to control shRNA embryos.
Atg1 knockdown rescues shRNA-Tor-expressing embryos
Atg1 and Tor have previously been shown to mutually inhibit each other (Dunlop et al., 2011), so we hypothesized that knocking down Tor leads to over-activation of Atg1, and vice versa. To test this, we constructed a line simultaneously targeting Atg1 and Tor (shRNA-Tor; shRNA-Atg1). Strikingly, depletion of Atg1 rescued both the morphology and hatch rate defects of shRNA-Tor embryos (Fig. 6C; Fig. S10A). The shRNA-Atg1; shRNA-Tor double knockdown embryos were compared with single shRNA-Tor and shRNA-Atg1 knockdown combined with control shRNAs to control for any effects of combining multiple shRNAs (Table S1). Additionally, when we overexpressed Atg1 we observed a similar phenotype to shRNA-Tor embryos, including positive TUNEL staining (Fig. 6D; Fig. S10B). To determine whether the rescue was a result of a decrease in autophagy or an alternate activity of Atg1, we looked at the effect of knocking down Atg2. Expression of shRNA-Atg2 failed to rescue shRNA-Tor (Fig. 6C). Altogether, the double shRNA knockdowns demonstrates that Tor and Atg1 interact in a manner separate from autophagy, and that Atg1 has a separate function other than autophagy, which could be regulation of Tor.
DISCUSSION
Our study revealed new aspects of Tor/Atg1 biology, as well as providing progress on how yolk catabolism is regulated. The dramatic Tor knockdown phenotype, with profound disorganization of the embryo, and its nearly complete rescue by Atg1 knockdown, are novel results from our study. Several controls, including measurement of mRNA levels, use of multiple shRNAs, and failure to rescue Tor depletion by Atg2 knockdown, attest to the specificity of these effects. These findings complement and extend previous studies on Tor and Atg1 function in later Drosophila development that relied on somatic mutations, where perdurance of maternally loaded protein complicates analysis (Lee et al., 2007; Scott et al., 2004). Previous studies examining rescue of Tor mutants by knocking down or mutating Atg1 reported mixed results: Scott et al. (2004) observed that a zygotic Tor−/−; Atg1−/− mutant was less viable than the zygotic Tor−/− mutant. However, Lee et al. (2007) found that homozygous Tor−/− larvae, which usually die at second/early third instar larval stage, developed through mid-late third instar larval stage in a heterozygous Atg1 mutant background. Given that Atg1 and Tor are in a negative feedback loop, one possibility is that simultaneously decreasing the activity of Tor and Atg1 prevents over-activation of either kinase. The remaining amount of Tor and Atg1 protein still present might then be able to elicit a normal phenotype. Both knockdown of Tor and overexpression of Atg1 resulted in positive TUNEL staining, suggesting apoptotic-related cell death (Figs S3, S10). Previous work overexpressing Atg1 in the wing imaginal disks and fat body also reported cell death and positive TUNEL staining (Scott et al., 2007). Interestingly, the work by Scott et al. (2007) was able to reduce Atg1-induced cell death through inhibition of autophagy. Our study found that shRNA-Tor embryos can be rescued through inhibition of Atg1, but we were not able to rescue shRNA-Tor embryos by inhibiting another downstream autophagy gene, Atg2, suggesting that Atg1 and Tor might have additional functions in the early embryo (Fig. 6). Particularly, Atg1 has multiple roles that include initiating autophagy and promoting yolk catabolism. Both turn on dramatically at the end of cellularization, but depend on different downstream components as autophagy, but not yolk catabolism, are blocked by knockdown of Atg2.
An interesting finding of our work is the timing and spatial regulation of autophagy during cellularization. In exploring the role of Atg1 in promoting autophagy in the early embryo, we documented formation of abundant autophagosomes spanning the basal region of blastomeres at the end of cellularization, coinciding with the initiation of zygotic transcription. In mouse embryos, autophagosomes also form around the period of zygotic transcription, after the first cleavage division (Tsukamoto et al., 2008). Given this timing, autophagosomes might play a role in degradation of maternal proteins during the mid-blastula transition, and/or in providing nutrients to the developing embryo by catabolism of cytosol, organelles, lipid droplets and glycogen granules. Consistent with a metabolic role, we frequently observed mitochondria and lipid droplets inside autophagosomes by EM (Fig. 4C,D). Autophagosomes were only formed within a narrow 6 µm-wide zone spanning the basal region of blastomeres (Fig. 4F). The significance of the high degree of spatial regulation is unclear. It might serve to specifically degrade molecules that would otherwise impede gastrulation, or to protect apically localized factors such as mRNAs. More detailed analysis of maternal proteins could potentially address these questions. We additionally observe the misorganization of organelles in shRNA-Atg1 embryos that is not seen in shRNA-control or other autophagy-deficient embryos. Previous studies have shown that Atg1 phosphorylates a myosin light chain kinase, which could potentially affect organelle distribution through disrupting actin-associated myosin II (Sopko et al., 2014; Tang et al., 2011).
A major finding of our work concerns regulation of yolk catabolism by the Tor pathway and Atg1. First a caveat; for molecular analysis we used activity levels of Cathepsin-B like proteinase enzyme activity as a surrogate for measuring catabolism itself. In repeated experiments we found direct measurement of vitellogenin was inaccurate, mainly because it was hard to normalize, and thus did not take into account variation in embryo size. For Cathepsin B-like proteinase activity we normalized measured activity to maximal activity following an acid treatment. This approach was developed by Fagotto (1990a,b), and we found it gave consistent measurements. This is the first molecular clue as to how yolk catabolism is triggered, and also an important new function for Tor and Atg1. Studies from a variety of organisms suggest that yolk platelets already include Cathepsin-like proteinases that are capable of catabolizing vitellogenins, but these are stored as pro-enzymes, and must be activated by acidification of the yolk platelet from a resting pH of ∼5.5 to ∼4.5 to trigger catabolism in oocytes (Fagotto and Maxfield, 1994; Medina et al., 1988). This process is controlled throughout development such that each lineage in the embryo can trigger yolk catabolism at the appropriate time, and thus maintain the appropriate amount of nutrients prior to hatching and feeding (Jorgensen et al., 2009). How the embryo regulates the timing of yolk catabolism had not been elucidated, and it has been unclear if degradation is triggered as a developmental event, or in response to nutrient demand in different lineages. Given that the Tor pathway measures metabolism and regulates catabolism in response in other systems, we speculate that Tor inactivation triggers yolk catabolism in response to nutrient demand, though we cannot rule out developmental regulation of the Tor-Atg1 circuit. Atg1 integrates input from the energy sensor AMPK to activate downstream autophagy (Kim et al., 2011). Given that autophagy and yolk catabolism occur at similar times in the Drosophila embryo, Atg1 might potentially act as a hub to coordinate autophagy and yolk catabolism simultaneously. Previous work has also demonstrated that Atg1 can inhibit activity of the MAP kinase ERK in neurons, and that this activity is required for protein composition at the synapses (Wairkar et al., 2009). Therefore, Atg1 could also affect yolk catabolism indirectly through additional downstream signaling pathways.
Our finding of an essential role of Tor in promoting yolk catabolism opens the door to molecular analysis. Given that Rheb is also required for yolk catabolism, a lack of amino acids might activate Tor on the surface of yolk platelets, as it does on the surface of lysosomes in somatic cells. Proteomics of yolk platelets from Xenopus laevis identified Rheb on the yolk membrane, supporting the hypothesis that Tor and Rheb might be localized to the yolk platelet membrane similar to their localization to the lysosome membrane (Jorgensen et al., 2009). Based on Fagotto's ‘sleepy lysosome’ hypothesis, we propose that the Atg1-Tor signaling axis detects amino acid demand (and/or is subject to developmental regulation) and then acts to decrease the pH of yolk platelets, which in turn activates a Cathepsin proteinase (Fig. 7) (Fagotto, 1995). An important link connecting Atg1-Tor signaling and regulation of lysosomal pH might be the regulation of H+-ATPase on the lysosome membrane. It has been demonstrated that H+-ATPase interacts with both the Ragulator and the RAGs, and that the interactions are strengthened by amino acid starvation and weakened by amino acid stimulation (Bar-Peled et al., 2012; Zoncu et al., 2011). There is also evidence for Tor affecting lysosome pH, namely that suppression of Tor activity through starvation or Tor-specific catalytic inhibitors leads to a drop in pH, and consequently lysosome activation (Zhou et al., 2013). However, regulation of H+-ATPases is generally a poorly understood phenomena. In yeast, the vacuolar H+-ATPase can reversibly disassemble and reassemble the V0 and V1 complex in order to regulate activity. For example, intact, active V-ATPase complexes are disassembled into free V1 and V0 complexes in response to glucose deprivation, then can reassemble upon re-addition of glucose (Kane, 1995). However, a clear mechanism linking H+-ATPase regulation and metabolic signaling has not been identified in multicellular organisms. Yolk might provide a particularly good system for understanding the link between the Tor pathway and pH regulation of organelles. Compared with lysosomes, where the pH is thought to be more tightly regulated, yolk platelets have a much wider pH range (pH 4.5-4.8 versus pH 4.5-6.5, respectively) and are much larger (0.1-1 μm versus 1.5-3 μm, respectively) (Fagotto and Maxfield, 1994; Ohkuma and Poole, 1978). Developing an in vitro system where one could chemically perturb Tor or Atg1 activity and then quantitatively measure the pH of yolk platelets could be one way to answer this question. Exactly how Tor and Atg1 turn on yolk acidification is an important future question, which might have implications for control of lysosomal activity more generally.
Fig. 7.

Model of yolk catabolism. Atg1 is necessary for both autophagy, via downstream Atg proteins, and yolk catabolism. Its role in yolk catabolism could be via direct regulation of Tor. Tor, its Torc1 complex member Raptor, and its activating GTPase Rheb are also required for yolk catabolism, suggesting that they might be localized on the yolk platelet membrane similar to how they localize to the lysosome membrane. An important missing link might be regulation of the H+-ATPase by the Tor-Atg1 circuit.
MATERIALS AND METHODS
Fly stocks
The UAS-shRNA and Gal4 fly stocks used in this study were obtained from the TRiP and Bloomington's Drosophila Stock Center (BDSC). For maternal UAS-shRNA expression, we used either the maternal triple-driver MTD-Gal4 (BDSC 31777), a dual MAT-Gal4 (constructed from BDSC 7062 and BDSC 7063), and a single MAT-Gal4 (BDSC 7063). mRNA knockdown efficiencies and primers are listed in Table S1. For a control shRNA we used a hairpin against white mRNA (HMS00017), except for with the double knockdowns, in which we used hairpins against EGFP because they were available on both chromosomes (BDSC 41550, BDSC 41551). To visualize histones we inserted P{w[+mC]=His2Av-EGFP.C}2/SM6a into shRNA-Tor and control shRNA-expressing lines (BDSC 24163). The FIP2003F5/3F5 line was graciously provided by Hong-Wen Tang (Tang et al., 2011). The UAS-HA-Atg1 overexpression construct was assembled by amplifying Atg1 from BDSC 51654 and inserting it into pVALIUM20 with an HA sequence on the N-terminus (TRiP, Harvard Medical School). Transgenic lines were generated by Genetic Services (Cambridge, MA, USA).
Embryo collections and hatching rates
Eggs were collected from shRNA/Gal4 females crossed to shRNA males. Flies were fed molasses and yeast paste. All embryos were collected at 27°C and dechorionated by soaking in 50% bleach for 3-5 min followed with extensive washing in distilled water. Embryos were washed in PBS with 0.1% Triton X-100 and staged on a dissecting microscope. Embryos were snap-frozen in liquid nitrogen and stored at −80°C until use. For hatching rates, 20 females and ten males were placed in vials at 27°C for 18 h. Embryos were collected from the vials and plated on molasses agar plates. The plates were incubated at 27°C for 24 h and hatched embryos were counted. All hatching rates were measured in biological duplicates.
Immunostaining and western blots
For immunostaining, collected embryos were fixed in a 1:1 dilution of methanol and heptane. Embryos were shaken vigorously to remove the perivitelline layer and then transferred to 100% methanol. Methanol was exchanged three times, followed by PBST (PBS+0.1% Triton X-100). Hoechst (1 mM) was added and then the embryos were gently rocked in the dark for 10 min. Embryos were washed with TBST then, transferred to a slide and mounted with ProLong Gold Mounting Reagent (Invitrogen). Embryos were imaged using a Nikon A1R point scanning confocal. For western blotting, embryos were placed directly in NuPage SDS loading buffer with 50 mM DTT and lysed using a plastic pestle. Phospho (Thr398) p70 s6K antibody (Cell Signaling, #9209) was used at 1:500.
TUNEL staining
TUNEL staining was carried out using TMR red labeling In Situ Cell Death Detection Kit (Roche). Embryos were fixed in a 1:1 mixture of 4% formaldehyde and heptane, washed twice with heptane, then transferred to a 1:1 mixture of methanol:heptane to remove the perivetelline membrane. Embryos were transferred to 100% methanol, then to 100% ethanol, then rehydrated in 3:1, 2:1, 1:1, 1:2 mixtures of ethanol:PBS, then finally transferred to PBS with 0.1% Triton X-100. Staining was carried out according the manufacturer's protocol (Roche).
Thin section electron microscopy
Embryos were dechorinated for 3 min in 50% bleach, fixed with gluteraldehyde, thin sectioned (∼60-70 nm sections) and stained with lead citrate using a previously described method (Rickoll, 1976). Sections were imaged on a Tecnai G2 Spirit BioTWIN. Autophagosomes were quantified by dividing the total sum of the area of double membrane structures that were fully enclosed per total area (∼25,000 µm2) of the section. Sections from three embryos were counted for each shRNA line and developmental time point. For the shRNA-control stage 7 embryos, ∼50-75 autophagosomes were measured for each replicate embryo.
Quantitative real-time PCR (qPCR)
Total RNA was isolated from ∼400 embryos collected from 0-4 h eggs from Mat-Gal4/shRNA females using Ambion Purelink RNA Mini Kit following manufacturer's protocol. cDNA was generated from 500 ng of purified RNA with the iScript cDNA Synthesis Kit (Bio-Rad). Samples were measured in triplicate using iQ SYBR Green Supermix (Bio-Rad) on a Bio-Rad CFX96 Real Time PCR machine. Transcripts were normalized using ribosomal protein L32, alpha-tubulin and Gapdh1 as reference genes, then compared with shRNA-white control embryos (Table S1). Primers were designed using FlyPrimerBank (Hu et al., 2013) (Table S2).
Yolk measurements
For Coomassie Blue staining, embryos were collected in 1-h increments from 0-5 h. Embryos were staged using a dissecting microscope and ten embryos were frozen in triplicate for each line and time point. Embryos were flash-frozen and stored at −80°C until use. Samples were lysed in NuPage LDS loading buffer with 50 mM DTT and a plastic pestle. Samples were loaded onto Bis-Tris gels (Bio-Rad) and stained with Coomassie Blue. The Cathepsin B-like proteinase assay was followed as described by Fagotto (1990a,b). Briefly, samples (50-150 embryos) were lysed in 100 µl 10 mM sodium acetate, pH 5.5 and split into two tubes. The non-acid treated sample was diluted 1:10 in 400 mM sodium acetate, pH 5.5, 4 mM EDTA and 4 mM DTT. Samples were incubated for 1 min at 30°C. Acid-treated samples were diluted 1:10 in 100 mM sodium formate, pH 3.5, 2 mM EDTA and 1 mM DTT, then incubated for 5 min at 30°C. A 20-50 µl aliquot of the samples was then added to the substrate assay buffer [100 mM sodium acetate buffer, pH 5.5, 0.05% Brij, 2 mM DTT, 1 mM EDTA, 5 μM Z-Phe-Arg-AMC (Bachem)] and incubated for 5 min at 30°C. The reaction was stopped with 100 mM monocholoracetate and 100 mM sodium acetate, pH 4.5. The samples were measured on a Perkin-Elmer Plate Reader using 360 nm excitation and 460 nm emission filters.
Acknowledgements
We are grateful to the many people who provided strains and antibodies for use in the this study, particularly Christine Field (Systems Biology Department, Harvard Medical School), Marianna Foos (Genetics Department, Harvard Medical School), the Harvard Transgenic RNAi Project, and the Bloomington Stock Center. Thanks to Jonathan Zirin (Genetics Department, Harvard Medical School) for thoughtful advice throughout the study. Thank you to the Nikon Imaging Center at Harvard Medical School for access to equipment and advice on microscopy experiments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
H.K. and T.M. designed the experiments. H.K. performed immunofluorescence and Cathepsin experiments. N.P. and R.S. provided genetic tools and feedback. R.S. measured and analyzed shRNA efficiencies. M.C. performed electron microscopy. H.K. wrote the manuscript.
Funding
This work was supported by the National Institutes of Health [GM39565 to T.M.]; H.K. was supported by a National Defense Science and Engineering Graduate (NDSEG) Fellowship. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.125419/-/DC1
References
- Bar-Peled L., Schweitzer L. D., Zoncu R. and Sabatini D. M. (2012). Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196-1208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolster D. R., Crozier S. J., Kimball S. R. and Jefferson L. S. (2002). AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J. Biol. Chem. 277, 23977-23980. 10.1074/jbc.C200171200 [DOI] [PubMed] [Google Scholar]
- Bownes M. and Hames B. D. (1977). Accumulation and degradation of three major yolk proteins in Drosophila melanogaster. J. Exp. Zool. 200, 149-156. 10.1002/jez.1402000118 [DOI] [PubMed] [Google Scholar]
- Buller C. L., Loberg R. D., Fan M.-H., Zhu Q., Park J. L., Vesely E., Inoki K., Guan K.-L. and Brosius F. C. III (2008). A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am. J. Physiol. Cell Physiol. 295, C836-C843. 10.1152/ajpcell.00554.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C., Doumpas N. and Teleman A. A. (2014). Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786-799. 10.1016/j.cell.2014.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo S., Corazzari M., Nazio F., Oliverio S., Lisi G., Antonioli M., Pagliarini V., Matteoni S., Fuoco C., Giunta L. et al. (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191, 155-168. 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C. C., Elis W., Menon S., Qin W., Klekota J., Asara J. M., Finan P. M., Kwiatkowski D. J., Murphy L. O. and Manning B. D. (2012). TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 47, 535-546. 10.1016/j.molcel.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop E. A., Hunt D. K., Acosta-Jaquez H. A., Fingar D. C. and Tee A. R. (2011). ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 7, 737-747. 10.4161/auto.7.7.15491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F. (1990a). Yolk degradation in tick eggs: I. Occurrence of a cathepsin L-like acid proteinase in yolk spheres. Arch. Insect Biochem. Physiol. 14, 217-235. 10.1002/arch.940140403 [DOI] [PubMed] [Google Scholar]
- Fagotto F. (1990b). Yolk degradation in tick eggs: II. Evidence that cathepsin L-like proteinase is stored as a latent, acid-activable proenzyme. Arch. Insect Biochem. Physiol. 14, 237-252. 10.1002/arch.940140404 [DOI] [PubMed] [Google Scholar]
- Fagotto F. (1995). Regulation of yolk degradation, or how to make sleepy lysosomes. J. Cell Sci. 108, 3645-3647. [DOI] [PubMed] [Google Scholar]
- Fagotto F. and Maxfield F. R. (1994). Changes in yolk platelet pH during Xenopus laevis development correlate with yolk utilization. A quantitative confocal microscopy study. J. Cell Sci. 107, 3325-3337. [DOI] [PubMed] [Google Scholar]
- Garami A., Zwartkruis F. J. T., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L. and Thomas G. (2003). Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11, 1457-1466. 10.1016/S1097-2765(03)00220-X [DOI] [PubMed] [Google Scholar]
- Gutzeit H. O., Zissler D., Grau V., Liphardt M. and Heinrich U. R. (1994). Glycogen stores in mature ovarian follicles and young embryos of Drosophila: ultrastructural changes and some biochemical correlates. Eur. J. Cell Biol. 63, 52-60. [PubMed] [Google Scholar]
- Hara K., Yonezawa K., Weng Q.-P., Kozlowski M. T., Belham C. and Avruch J. (1998). Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484-14494. 10.1074/jbc.273.23.14484 [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S.-I., Natsume T., Guan J.-L. and Mizushima N. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497-510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Sopko R., Foos M., Kelley C., Flockhart I., Ammeux N., Wang X., Perkins L., Perrimon N. and Mohr S. E. (2013). FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 3, 1607-1616. 10.1534/g3.113.007021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins M. U., Veenhuis M. and Klionsky D. J. (1999). Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079-4087. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T. and Guan K.-L. (2003). Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829-1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Steen J. A. J., Steen H. and Kirschner M. W. (2009). The mechanism and pattern of yolk consumption provide insight into embryonic nutrition in Xenopus. Development 136, 1539-1548. 10.1242/dev.032425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T., Taneike I., Akaishi R., Yoshizawa F., Furuya N., Fujimura S. and Kadowaki M. (2004). Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J. Biol. Chem. 279, 8452-8459. 10.1074/jbc.M306337200 [DOI] [PubMed] [Google Scholar]
- Kane P. M. (1995). Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270, 17025-17032. [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B. and Guan K.-L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132-141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Park H. L., Park H.-W., Ro S.-H., Nam S. G., Reed J. M., Guan J.-L. and Lee J. H. (2013a). Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy 9, 1201-1213. 10.4161/auto.24811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. G., Buel G. R. and Blenis J. (2013b). Nutrient regulation of the mTOR Complex 1 signaling pathway. Mol. Cells 35, 463-473. 10.1007/s10059-013-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I., Deffieu M., Manon S. and Camougrand N. (2004). Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 279, 39068-39074. 10.1074/jbc.M406960200 [DOI] [PubMed] [Google Scholar]
- Koch E. A. and Spitzer R. H. (1982). Autoradiographic studies of protein and polysaccharide synthesis during vitellogenesis in Drosophila. Cell Tissue Res. 224, 315-333. 10.1007/BF00216876 [DOI] [PubMed] [Google Scholar]
- LaFever L., Feoktistov A., Hsu H.-J. and Drummond-Barbosa D. (2010). Specific roles of Target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development 137, 2117-2126. 10.1242/dev.050351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Kim S., Lee J., Park J., Lee G., Kim Y., Kim J.-M. and Chung J. (2007). ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 8, 360-365. 10.1038/sj.embor.7400917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P. and Hall M. N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457-468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y. and Ohsumi Y. (1997). Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245-250. 10.1016/S0378-1119(97)00084-X [DOI] [PubMed] [Google Scholar]
- Medina M., Leon P. and Vallejo C. G. (1988). Drosophila cathepsin B-like proteinase: a suggested role in yolk degradation. Arch. Biochem. Biophys. 263, 355-363. 10.1016/0003-9861(88)90646-7 [DOI] [PubMed] [Google Scholar]
- Mochizuki H., Toda H., Ando M., Kurusu M., Tomoda T. and Furukubo-Tokunaga K. (2011). Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS ONE 6, e19632 10.1371/journal.pone.0019632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Tao R., Handler D., Karpowicz P. et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405-407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K.-I. (2006). The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development 133, 3441-3450. 10.1242/dev.02503 [DOI] [PubMed] [Google Scholar]
- Ohkuma S. and Poole B. (1978). Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. USA 75, 3327-3331. 10.1073/pnas.75.7.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papinski D., Schuschnig M., Reiter W., Wilhelm L., Barnes C. A., Maiolica A., Hansmann I., Pfaffenwimmer T., Kijanska M., Stoffel I. et al. (2014). Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell 53, 471-483. 10.1016/j.molcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickoll W. L. (1976). Cytoplasmic continuity between embryonic cells and the primitive yolk sac during early gastrulation in Drosophila melanogaster. Dev. Biol. 49, 304-310. 10.1016/0012-1606(76)90278-5 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L. and Sabatini D. M. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496-1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O. and Neufeld T. P. (2004). Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167-178. 10.1016/j.devcel.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Scott R. C., Juhász G. and Neufeld T. P. (2007). Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1-11. 10.1016/j.cub.2006.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M. and Czaja M. J. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131-1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Foos M., Vinayagam A., Zhai B., Binari R., Hu Y., Randklev S., Perkins L. A., Gygi S. P. and Perrimon N. (2014). Combining genetic perturbations and proteomics to examine kinase-phosphatase networks in Drosophila embryos. Dev. Cell 31, 114-127. 10.1016/j.devcel.2014.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Quan Z., Zhang B., Wu T. and Xi R. (2010). TSC1/2 tumour suppressor complex maintains Drosophila germline stem cells by preventing differentiation. Development 137, 2461-2469. 10.1242/dev.051466 [DOI] [PubMed] [Google Scholar]
- Tang H.-W., Wang Y.-B., Wang S.-L., Wu M.-H., Lin S.-Y. and Chen G.-C. (2011). Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 30, 636-651. 10.1038/emboj.2010.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A. R., Manning B. D., Roux P. P., Cantley L. C. and Blenis J. (2003). Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259-1268. 10.1016/S0960-9822(03)00506-2 [DOI] [PubMed] [Google Scholar]
- Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A. and Mizushima N. (2008). Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117-120. 10.1126/science.1154822 [DOI] [PubMed] [Google Scholar]
- Wairkar Y. P., Toda H., Mochizuki H., Furukubo-Tokunaga K., Tomoda T. and Diantonio A. (2009). Unc-51 controls active zone density and protein composition by downregulating ERK signaling. J. Neurosci. 29, 517-528. 10.1523/JNEUROSCI.3848-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T. G. and Mahowald A. P. (1979). Isolation and partial chemical characterization of the three major yolk polypeptides from Drosophila melanogaster. Dev. Biol. 68, 130-139. 10.1016/0012-1606(79)90248-3 [DOI] [PubMed] [Google Scholar]
- Yan D., Neumuller R. A., Buckner M., Ayers K., Li H., Hu Y., Yang-Zhou D., Pan L., Wang X., Kelley C. et al. (2014). A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell 28, 459-473. 10.1016/j.devcel.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Tan S. H., Nicolas V., Bauvy C., Yang N. D., Zhang J., Xue Y. and Codogno P. (2013). Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 23, 508-523. 10.1038/cr.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y. and Sabatini D. M. (2011). mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678-683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]