SUMMARY
Poly(ADP-ribose) polymerase-1 (PARP-1) creates the posttranslational modification PAR from substrate NAD+ to regulate multiple cellular processes. DNA breaks sharply elevate PARP-1 catalytic activity to mount a cell survival repair response, whereas persistent PARP-1 hyperactivation during severe genotoxic stress is associated with cell death. The mechanism for tight control of the robust catalytic potential of PARP-1 remains unclear. By monitoring PARP-1 dynamics using hydrogen/deuterium exchange-mass spectrometry (HXMS), we unexpectedly find that a specific portion of the helical subdomain (HD) of the catalytic domain rapidly unfolds when PARP-1 encounters a DNA break. Together with biochemical and crystallographic analysis of HD deletion mutants, we show that the HD is an autoinhibitory domain that blocks productive NAD+ binding. Our molecular model explains how PARP-1 DNA damage detection leads to local unfolding of the HD that relieves autoinhibition, and has important implications for the design of PARP inhibitors.
INTRODUCTION
There is ample and growing evidence that the PARP family of proteins regulate a broad spectrum of biological pathways using the ADP-ribose posttranslational modification synthesized from NAD+ (Ryu et al., 2015; Vyas et al., 2013; Ame et al., 2004). PARP-1 is the founding PARP family member and has long been appreciated as a key player in the cellular response to genotoxic stress, where it links DNA damage detection to the acute production of poly(ADP-ribose) (PAR), thereby initiating the recruitment of a number of DNA repair factors to sites of damage (D’Amours et al., 1999). PARP-1 is involved in various DNA repair pathways including base excision repair (BER), homologous recombination (HR), non-homologous end joining (NHEJ), and nucleotide excision repair (NER) (De Vos et al., 2012; Robu et al., 2013). Although best known for facilitating DNA repair, PARP-1 activity has key roles in other cellular processes including gene expression, regulation of chromatin structure, and cell fate decisions (Gibson and Kraus, 2012).
A dramatic burst in PARP-1 mediated PAR production is a hallmark of the cellular response to DNA damage and contributes to the efficiency of repair and cell survival. However, elevated or persistent levels of DNA damage can stimulate PARP-1 PAR production to the point of exhausting the cellular energy pools of NAD+ and influencing cell death decisions (Fouquerel and Sobol, 2014). Recent studies indicate that PARP-1 hyperactivation can lead to a form of cellular necrosis, termed parthanatos, through a PAR-dependent inhibition of glycolysis (Andrabi et al., 2014; Fouquerel et al., 2014). The dramatic consequences of PARP-1 hyperactivation are also observed in human diseases like Cockayne Syndrome and Xeroderma Pigmentosum A, where sustained PARP-1 activation induces NAD+ depletion and leads to mitochondrial defects through inactivation of SIRT-1 (Fang et al., 2014; Scheibye-Knudsen et al., 2014). In contrast, PARP-1 activity is maintained at much lower levels during normal unstressed cellular conditions, but still at a level that allows PARP-1 to function in other important biological pathways. The range of PARP-1 activities required for biological function, along with the cell fate consequences associated with chronic hyperactivation, necessitate tight regulation of PARP-1 catalytic activity. However, many questions remain regarding how PARP-1 activity is regulated during both stressed and unstressed cellular conditions. The use of PARP inhibitors for cancer treatment underscores the need to understand PARP-1 activity and regulation at a molecular level.
PARP-1 has a modular six-domain architecture (Fig. 1A). N-terminal F1 and F2 zinc-finger domains (also known as Zn1 and Zn2) bind to a variety of DNA structures in a sequence-independent manner, including double-strand breaks (DSB), single-strand breaks (SSB), extensions, hairpins and cruciforms (D’Silva et al., 1999; Eustermann et al., 2011; Langelier et al., 2011a; Lonskaya et al., 2005; Pion et al., 2003). The F3 zinc-binding domain (also known as Zn3) has a unique structure and makes contributions to DNA binding and PARP-1 interdomain contacts that are critical for DNA damage-dependent catalytic activity (Langelier et al., 2012; Langelier et al., 2010; Langelier et al., 2008). An automodification domain (AD) contains a BRCA1 C-terminus (BRCT) fold and linker regions that contain major sites targeted for PARP-1 automodification (Chapman et al., 2013; Sharifi et al., 2013; Gagné et al., 2015). The WGR domain participates in DNA binding and forms interdomain contacts essential for DNA damage-dependent activation. The catalytic domain (CAT) contains two subdomains: a helical domain (HD) that is conserved in DNA damage-dependent PARPs 1, 2, and 3, and the ADP-ribosyltransferase (ART) domain that contains the active site and a fold that is conserved in all PARP family members (Hottiger et al., 2010).
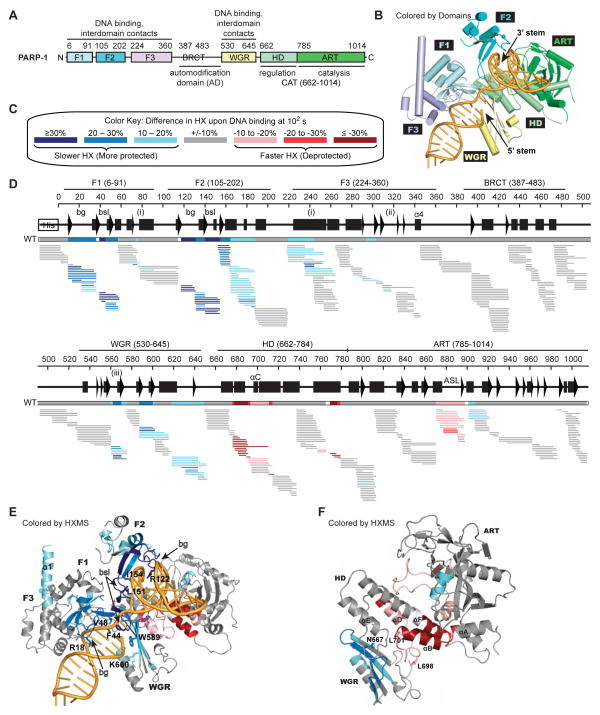
Figure 1. PARP-1 DNA damage detection leads to major decreases in HX in DNA-binding domains, and a dramatic increase in HX in the HD.
(A) PARP-1 domain architecture.
(B) Combined model of the crystal structure of the PARP-1 (F1, F3, WGR–CAT)/DNA complex, and the NMR structure of F1–F2 in complex with an SSB (Eustermann et al., 2015).
(C) Color key for the binning of HX differences. Percent difference is calculated by subtracting the percent deuteration of PARP-1 in complex with DNA from that of PARP-1 alone.
(D) Percent difference in HX is calculated for each peptide (represented by horizontal bars) at the 100 s time point and plotted using the color key in (C). The consensus behavior at each PARP-1 residue is displayed in the horizontal bar below the secondary structure annotation. White coloring indicates a gap in peptide coverage. Code for structural elements: bg, backbone grip; bsl, base stacking loop; (i), F1–F3 interface; (ii), F3–WGR–HD interface; (iii), WGR–HD interface. These peptides were identified in a single experiment, for which we have corresponding peptide data across all of the time points (see Fig. S3A). When available, we present the data for all measurable charge states of the 393 unique peptides.
(E, F) Consensus HXMS data from (D) is mapped onto the structure of the DNA binding interfaces in (E), and the WGR and CAT domains alone in (F).
See also Figures S1–S2.
Four domains of PARP-1 are strictly required for DNA damage-dependent catalytic activity (F1, F3, WGR, CAT) (Altmeyer et al., 2009; Ikejima et al., 1990; Langelier et al., 2011a; Langelier et al., 2008; Tao et al., 2008). F2 is important for binding and activation by DNA single-strand breaks (SSB) (Ikejima et al., 1990) and for recruitment to sites of DNA damage (Ali et al., 2012). The crystal structure of the essential PARP-1 domains in complex with a DNA double-strand break (DSB), together with biochemical and biophysical analysis, have established that F1, F3, and WGR domains collectively bind to DNA damage and organize PARP-1 into a collapsed conformation that creates a network of essential interdomain contacts (Langelier and Pascal, 2013; Langelier et al., 2012). The PARP-1/DSB DNA crystal structure featured a distorted HD compared to the crystal structures of the PARP-1 CAT in the absence of DNA and regulatory domains. Specifically, two leucine residues (termed the “leucine switch”) were extracted from the HD hydrophobic core. Mutations designed to mimic these destabilizing changes in the HD hydrophobic core increased PARP-1 activity in the absence of DNA, indicating that alterations to the HD are required for PARP-1 activation. However, the mutations did not recapitulate the full activation of wild-type PARP-1 by DNA damage. Both activating mutations and PARP-1 interaction with DNA lead to a decrease in thermal stability, which suggests a change in PARP-1 dynamics associated with activation. However, the PARP-1/DSB DNA structure provides a static snapshot and there are no direct measurements of PARP-1 dynamics reported in the absence or presence of DNA. Importantly, it has remained unclear how the observed structural changes (i.e. “leucine switch”) or potential changes in protein dynamics might influence PAR synthesis activity in the ART, which exhibited no changes in conformation in the PARP-1/DSB DNA structure.
Here, we use hydrogen-deuterium exchange coupled with mass spectrometry (HXMS) to measure the dynamics of full-length PARP-1. Unexpectedly, we find that binding to damaged DNA coincides with >10,000-fold faster exchange behavior within specific portions of the HD, indicating that the impacted helices are conformationally destabilized as part of PARP-1 catalytic activation. We further show that the HD is inhibitory to PARP-1 catalytic activity, with the HD deletion acting like a constitutively hyperactive PARP. Furthermore, the autoinhibitory role of the HD is conserved in PARP-2 and PARP-3, the other DNA damage-dependent PARPs. Crystal structures of PARP-1 and PARP-2 CAT domains harboring HD deletions and bound to PARP inhibitors reveal how unfolding of a specific region of the HD allows NAD+ to bind in a productive conformation in the catalytic active site, and thereby lead to efficient poly(ADP-ribose) synthesis.
RESULTS
HXMS analysis of PARP-1 dynamics
We conducted HXMS analysis of full-length PARP-1 in the absence or presence of DNA damage using a dumbbell DNA ligand containing a single nucleotide gap as a model of SSB damage (see accompanying Eustermann et al., 2015 and Supplemental Experimental Procedures). Amide protons are protected from HX when secondary structure engages them in hydrogen bonds, for example within an α-helix or the interior of a β-sheet (Englander, 2006). Thus, HXMS can report on protein structure states and has proven useful to study protein dynamics (Englander, 2006), particularly when atomic-level coordinates are available for data interpretation (Bassett et al., 2012; DeNizio et al., 2014; Hoofnagle et al., 2001; Lee et al., 2004; Sekulic et al., 2010; Falk et al., 2015). PARP-1 binds to the DNA dumbbell as a monomer with F1 and F2 domains each engaging a different DNA end at the break (Eustermann et al., 2015), and with the F3, WGR, and CAT domains assembled as observed in the PARP-1/DSB crystal structure (Langelier et al., 2012) and consistent with NMR analysis of interdomain contacts on an SSB (Eustermann et al., 2015) (Fig. 1B). PARP-1 alone or PARP-1 plus DNA was incubated in D2O exchange buffer over a wide range of time points spanning 101 to 105 s. HXMS experiments were thus designed to measure the dynamics of PARP-1 in the steady state bound to DNA damage, or in the absence of DNA damage. Reaction time points were quenched, peptides were generated by proteolysis, and the extent of deuterium incorporation into each peptide was analyzed by MS (Fig. S1A). For each peptide, changes in HX in the presence of DNA damage were calculated by subtracting the percent deuteration of PARP-1 with DNA from that of PARP-1 alone. The changes in HX were then mapped onto the combined NMR/crystal structure model for PARP-1 bound to SSB DNA (Eustermann et al., 2015).
DNA binding was predicted to slow the rate of HX in peptides at or near the DNA binding interface, and indeed, peptides throughout the multi-domain DNA binding interface (F1, F2, F3, and WGR domains) were slower to exchange in the PARP-1/SSB DNA damage complex (Fig. 1C–E; Fig. S1B; Fig. S2A–C). The greatest protection from exchange (e.g. >30% less HX at 100 s) occurred in peptides in close proximity to DNA. With DNA damage present, these peptides took 100–1000 times longer to achieve the same level of deuterium incorporation observed in the absence of DNA damage. The greatest DNA-induced protection from HX is found in F1 and F2 (Fig. 1D–E; Fig. S2A–C), in regions including the base-stacking loop and the backbone grip, two structural features known to interact with DNA and to contribute to DNA binding affinity (Langelier et al., 2011a, 2012; Eustermann et al., 2011, Eustermann et al., 2015). In the WGR, the β-sheet region that contacts DNA near the 5′ end (including R587, W589, Y570 and K621; Langelier et al., 2012), and the region near residue K600, both showed protection from HX in the presence of DNA (Fig. 1D–E). HX protection was also observed at PARP-1 domain interfaces; for example, the contacts formed between F1 and F3 (Fig. 1D–E: α1 of F3, residues 78–91 of C-terminal helix of F1; also Fig. S2E–G), and an interface between the WGR (N567; peptide 562–569) and αE of the HD (Fig. 1D, 1F and Fig. S3A; peptide 740–750). The protection from HX at domain interfaces typically involved a small number of residues per peptide, in comparison to the broader protection observed at the protein-DNA interfaces; however, this observation is consistent with the extent of the domain contacts observed in the PARP-1/DSB crystal structure (Langelier et al., 2012). The mechanism of DNA SSB recognition and the process that drives the PARP-1 domain assembly that controls allosteric communication to the catalytic domain are reported in a separate NMR study by Neuhaus and colleagues (Eustermann et al., 2015).
A DNA-dependent increase in HX protection was observed at later time points for the AD of PARP-1 (Fig. S2H). The AD includes residues that are targeted for automodification (Chapman et al., 2013; Sharifi et al., 2013; Gagné et al., 2015); therefore we speculate that the HX protection originates from contacts formed with the CAT as the AD is positioned for modification (Fig. S2K). A CAT region proximal to the active site (Fig. 1D, 1F; Fig. S2K; Fig. S3A, residues 900–920) also shows a similar and corresponding increase in HX protection over the same time scale, consistent with our data interpretation and the compacted conformation of PARP-1 on DNA damage.
HXMS detects local unfolding of the HD subdomain
The presence of DNA damage is allosterically signaled to the CAT through interactions between the DNA-binding regulatory domains (F1, F3, and WGR) and the HD subdomain of the CAT (Langelier et al., 2012). We thus analyzed the extent to which HD dynamics and structure are impacted when PARP-1 binds to a DNA break. In striking contrast to the slower HX observed in the PARP-1 DNA-binding domains, the HD showed a dramatic increase in HX in the presence of DNA (Fig. 1D, 1F). Indeed, the changes in HX incurred by the HD are among the largest of any region of PARP-1, with many amide hydrogen positions in helix αB and the C-terminal end of αF exchanged by the 10 s time point (Fig. S1B, magenta boxes; Fig. S3A) and essentially complete exchange by 100 s in peptides unequivocally mapped to these helices (Fig. 2A). HX at such early time points is not possible for amide hydrogens involved in the bonds that form a stable helix; thus, the HXMS data is more consistent with complete melting of these helical regions or with extremely rapid sampling of unfolded states. The rapid exchange of hydrogens starkly contrasts with the much slower exchange observed for the very same regions in the absence of DNA damage, where the same exchange required as much as 10,000 times longer to occur, consistent with these hydrogens contributing to stably folded helices (Fig. 2A–E; Fig. S1B; Fig. S3B–C). Other notable HX increases occurred in helices αC and αD of the HD (Fig. 1D; Fig. S3A). Collectively, HXMS analysis indicated that αB and the C-terminal end of αF of the HD populate an unfolded conformation, or rapidly sample an unfolded conformation, upon PARP-1 activation by DNA damage.
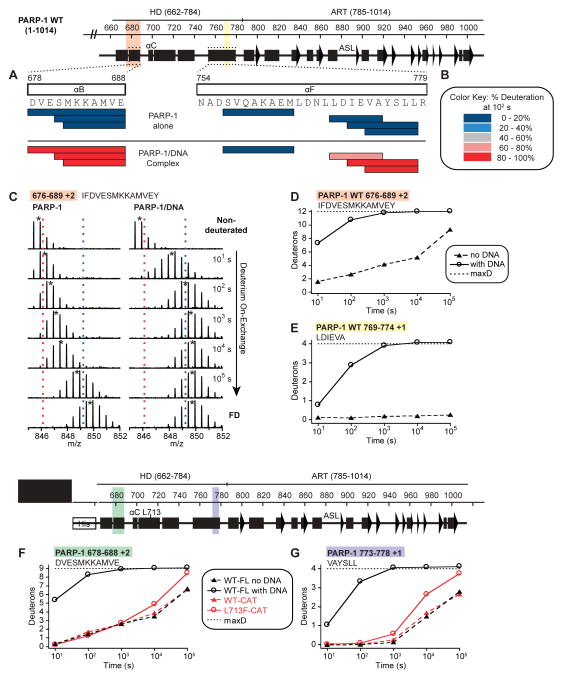
Figure 2. Regions of the HD αB and αF helices unfold when PARP-1 is bound to DNA damage.
(A) The percentage deuteration of peptides (represented by horizontal bars) in which all measureable residues unequivocally map within either αB or αF helices is plotted for PARP-1 alone and the PARP-1/DNA complex at the 100 s time point using the color key in panel (B).
(B) Color key for the binning of percentage deuteration.
(C) Raw MS data of a representative peptide from αB of PARP-1 HD. Centroid values are indicated with an asterisk. Red and blue dotted lines serve as guides for visualizing differences. FD represents the fully deuterated condition used for normalization due to back-exchange.
(D, E) HXMS of representative peptides from the αB in (D), and the C-terminal end of αF in (E) for PARP-1 alone and PARP-1 in complex with DNA. The calculated maximum number of exchangeable deuterons (maxD) is indicated.
(F, G) HXMS of representative peptides from the αB in (F), and the C-terminal end of αF in (G) for PARP-1 CAT WT and PARP-1 CAT L713F, drawn in red lines. The same peptides from FL PARP-1 WT are plotted for comparison (black lines).
See also Figures S3–S4.
To further investigate the relationship between HD unfolding and PARP-1 catalytic activity, we examined the HX behavior of a PARP-1 mutant with elevated DNA-independent activity. L713 is located in the HD hydrophobic core (Fig. S4B). Mutation L713F elevates PARP-1 activity in the absence of DNA damage (Langelier et al., 2012; Miranda et al., 1995). We could thus analyze potential changes in the dynamics of the CAT domain alone, without the regulatory domains of PARP-1. We observed an overall increase in the rate of HX throughout the HD of the L713F mutant compared with that of CAT WT (Fig. 2F–G; Fig. S4A–B). The earliest increases in HX are centered on the L713F mutation in helix αD (Fig. S4A). At longer exchange times (104 to 105 s), the increases in HX occur in the same helices that were observed to unfold in full-length PARP-1 in the presence of DNA damage (i.e. αB and C-terminus of αF). The catalytic activity of CAT L713F is approximately 20-fold higher than that of WT (Langelier et al., 2012), but not nearly the 1,000-fold stimulation of full-length PARP-1 observed in the presence of DNA damage (Langelier et al., 2010). Correspondingly, the HX behavior of the HD is at an intermediate state of conformational destabilization, indicating a relationship between the extent of HD unfolding and the level of PARP-1 activation.
HD unfolding requires interdomain communication
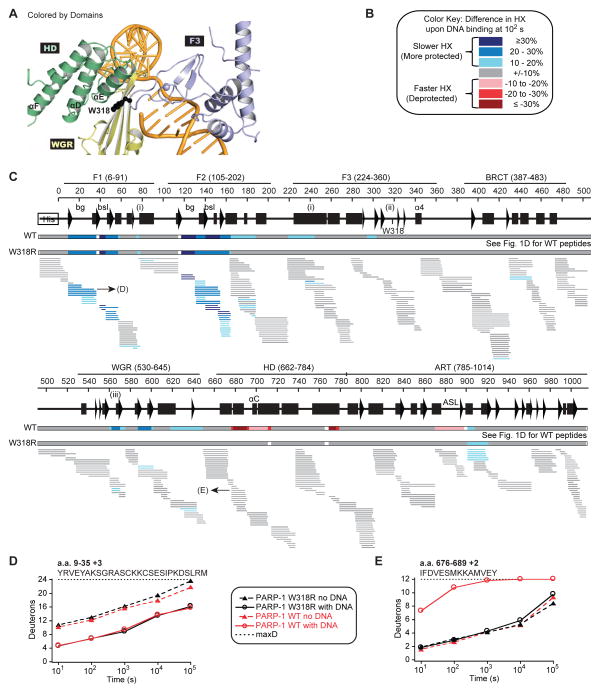
We predicted that disruptions to the allosteric communication between PARP-1 domains should maintain the HD in the folded and thus inactive state. The W318R mutant of PARP-1 binds to DNA damage with an affinity similar to that of PARP-1 WT, yet has no detectable DNA damage-dependent activity (Langelier et al., 2010; Steffen et al., 2014). W318 is positioned on an F3 loop that interacts with WGR residue K633 and HD residue R735 in αE (Fig. 3A). The W318R mutation disrupts a critical interface between F3, WGR, and HD domains and thus compromises allosteric communication (Langelier et al., 2012). Using HXMS, we measured the dynamics of PARP-1 W318R alone or in complex with DNA damage, and then analyzed the differences in HX for each peptide (Fig. 3C). Most strikingly, although the HX of WT and W318R are similar in the absence of DNA (Fig. S1B; Fig. S5A), the HD of W318R displays HX kinetics consistent with a folded domain, regardless of the presence or absence of DNA damage (Fig. 3C, 3E; Fig. S5A). The rapid HX observed with PARP-1 WT in αB, αC, αD, and αF is entirely absent in the W318R mutant, clearly indicating that there is no detectable DNA-induced HD conformational destabilization in the W318R mutant. In contrast, F1 and F2 of W318R show a level of protection similar to that observed for WT (Fig. 3C–D; Fig. S5B–E), confirming that W318R is engaged on the DNA break and consistent with the DNA binding capacity of this mutant (Langelier et al., 2010; Steffen et al., 2014). Interestingly, the HX protection observed at the F1–F3 interface for WT in the presence of DNA is also observed in the W318R mutant, signifying that this interface does form in the context of the mutant (Fig. 3C, residues 78–90 and 235–243). However, the overall extent of protection observed for F3 is reduced in W318R compared to WT, suggesting that F3 is less stably bound in the complex in the W318R mutant. Moreover, the HX protection observed in the WGR domain upon DNA binding is decreased in the W318R mutant, both in the WGR region that contacts DNA (residues 585–600) and the regions involved in interactions with F3 (residue K633) and the HD (residue 567) (Fig. 3C). These data indicate that despite the proper engagement of F1 and F2 on DNA, the W318R mutant perturbs the chain of PARP-1 allosteric communication, most prominently at the stage of positioning the WGR domain on DNA. Consequently, the appropriate WGR–HD interactions that lead to HD unfolding are disturbed, thus shutting down DNA damage-dependent PARP-1 activation.
Figure 3. Disruption of PARP-1 interdomain communication prevents HD unfolding.
(A) Location of residue W318 (shown as sticks) at the interface of the F3, WGR, and HD domains.
(B) and (C) Same as Fig. 1C and 1D for PARP-1 W318R. When available, we present the data for all measurable charge states of the 391 unique peptides. A horizontal bar representing PARP-1 WT data from Fig. 1D is shown for comparison.
(D, E) HXMS of specific W318R peptides for the F1 domain in (D), and αB in (E), drawn in black lines. The same peptides from PARP-1 WT are plotted for comparison (red lines). The calculated maximum number of exchangeable deuterons (maxD) is indicated.
See also Figure S5.
Notably, ART region 900–920 showed an increase in protection from HX in the presence of DNA in both PARP-1 WT and the W318R mutant (Fig. 3C). Similarly, the increase in HX protection observed for PARP-1 WT with DNA in the AD region at later time points is preserved in the W318R mutant (Fig. S2H–I). These observations are consistent with the W318R mutant maintaining the same global positioning of domains as seen with PARP-1 WT. As mentioned previously, we interpret these complementary changes in HX to reflect the AD being positioned next to the ART for modification when PARP-1 binds to DNA (Fig. S2K). Consistently, the activating L713F mutation of the CAT domain alone did not show increased ART protection at 900–920 relative to CAT WT (Fig. S4A–B), since the regulatory domains and AD are excluded from this analysis. Thus, PARP-1 W318R is capable of assembling onto DNA similarly to WT; however, the major deficiency is a breakdown in the interdomain communication that can no longer lead to HD unfolding and catalytic activation.
The HD is an autoinhibitory domain
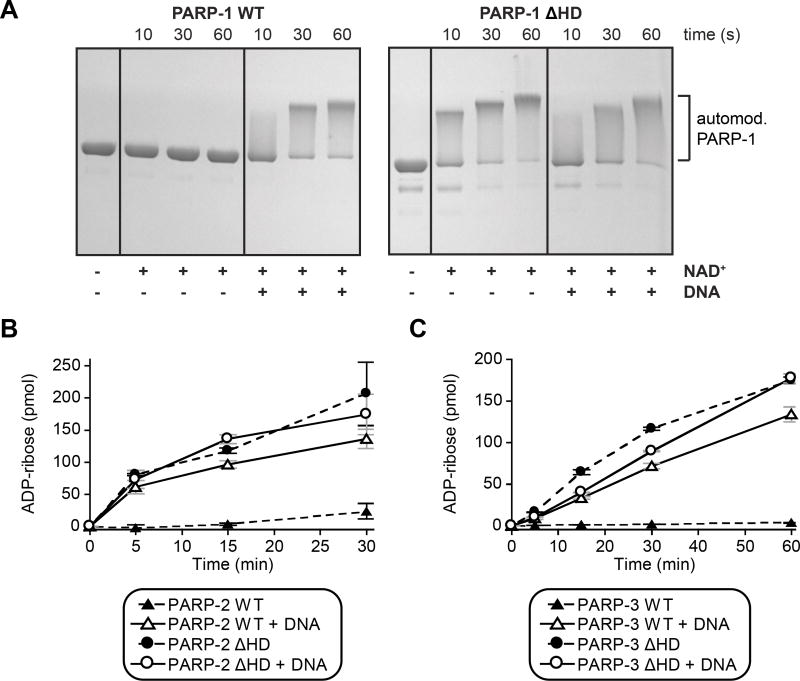
The local HD unfolding suggested that certain regions of the HD are not required for PARP-1 catalytic activity, at least not in the folded conformation observed in available crystal structures. However, there was a possibility that the altered HD conformation could still play an active role in promoting PARP-1 activity. We deleted the HD of PARP-1 (PARP-1 ΔHD) to clarify its role. PARP-1 ΔHD completely removes helices αD, αE, and αF, while still allowing αA to contribute to the fold of the ART, and allowing αB and αC to provide connector residues to the ART. PARP-1 ΔHD activity was measured in an SDS-PAGE assay that monitors PARP-1 automodification in the presence of NAD+ as a decrease in electrophoretic migration. Remarkably, the HD deletion fully recapitulates the effect of DNA binding since the activity of PARP-1 ΔHD in the absence of DNA is very similar to the activity of PARP-1 WT in the presence of DNA (Fig. 4A). Moreover, addition of DNA to PARP-1 ΔHD does not lead to a further increase in catalytic activity, indicating that HD deletion completely bypasses the DNA binding requirement. Notably, deletion of the HD in full-length PARP-1 presented major production issues, which we now interpret as a toxic effect of the overactive ΔHD protein in E. coli, most likely caused by a depletion of NAD+. We overcame this limitation by preparing PARP-1 ΔHD using sortase-mediated joining of two PARP-1 fragments (see Supplemental Experimental Procedures).
Figure 4. The HD is an autoinhibitory domain of DNA damage-dependent PARPs.
(A) Catalytic activity of PARP-1 FL WT and PARP-1 ΔHD (1 μM) in the absence or presence of DNA (1 μM) measured using the SDS-PAGE automodification assay.
(B, C) Catalytic activity of PARP-2 and PARP-3 WT and ΔHD mutants (PARP-2: 20 nM, DNA: 20 nM; PARP-3: 60 nM, DNA: 480 nM) measured using the colorimetric assay. Experiments were performed in triplicate and the averages and standard deviations are shown.
See also Figure S6.
We also deleted the HD in the isolated CAT domain in order to perform a quantitative catalytic analysis of the HD deletion using a colorimetric assay. The colorimetric assay requires the presence of a His-tag, which is removed during the sortase reaction and thus absent in full-length PARP-1 ΔHD. Initial rates of reactions were compared for CAT ΔHD and full-length PARP-1 WT in the presence of DNA (Fig. S6A–B). Our results clearly indicate that deleting the HD of PARP-1 increases the reaction rate to the same level reached when PARP-1 binds to DNA damage. Thus, the HD acts as an autoinhibitory domain that greatly subdues ART catalytic activity. Taken with the HXMS analysis, the local unfolding of the HD effectively “deletes” the structure and relieves the autoinhibitory effect.
PARP-2 and PARP-3 share with PARP-1 a common allosteric mechanism of activation in response to DNA damage that involves WGR–HD contacts (Langelier et al., 2014). We tested if the HD of PARP-2 and PARP-3 also has an inhibitory effect on catalytic activity, similar to that observed for PARP-1. HD deletions of PARP-2 and PARP-3 were tested in a colorimetric assay that measures the incorporation of ADP-ribose into PAR using biotinylated NAD+ (Langelier et al., 2011b). Deletion of the HD of PARP-2 and PARP-3 led to a significant increase in DNA-independent activity (Fig. 4B, 4C), thus indicating that the HD has an autoinhibitory effect on each of the DNA damage-dependent PARPs.
Disruption of HD–ART contacts leads to PARP-1 activation
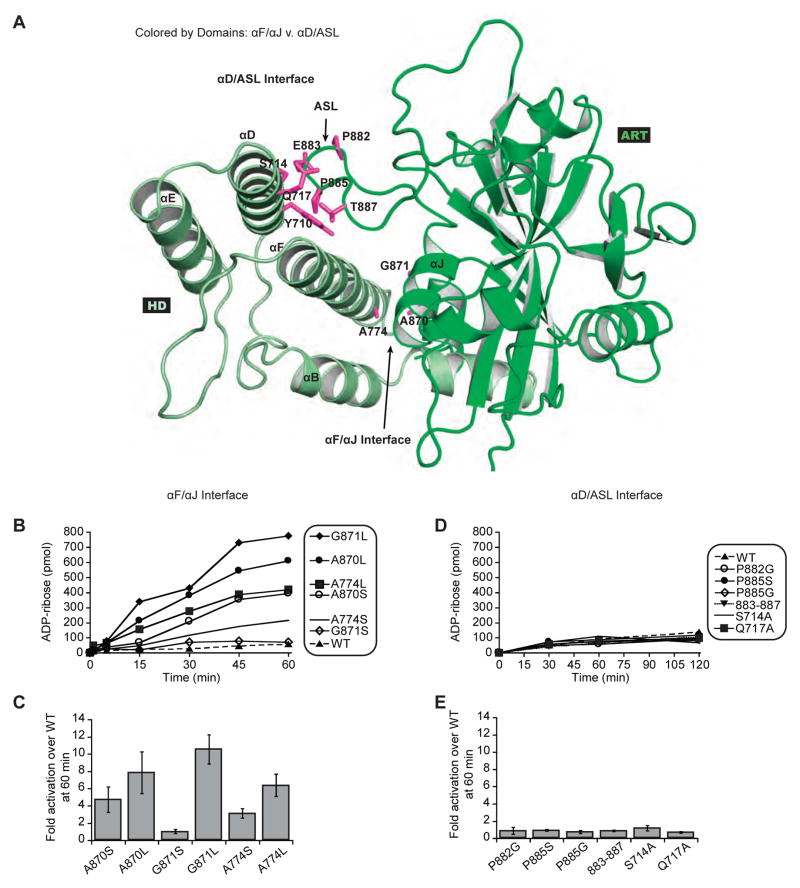
We investigated the mechanism of HD inhibition by exploring the location of HD–ART contacts that might be responsible for holding PARP-1 at a low level of activity in the absence of DNA damage. The HD has two major interaction points with the ART: αD of HD contacts the ART active site loop (ASL; also known as D-loop), and αF of HD contacts αJ of the ART (Fig. 5A). These two ART regions exhibited HX increases in the presence of DNA, albeit subtle compared to the massive changes observed for HD (Fig. 1D, 1F; Fig. S3A). The HD–ART interface between αF and αJ is structurally well conserved between the three DNA-dependent PARPs. In contrast, the HD–ART interface between αD and the ASL is not structurally conserved between PARPs 1, 2, and 3. Further, the most significant increases in HX were located in HD helices αB and αF, in a region adjacent to helix αJ of the ART (Fig. 1F). We thus hypothesized that contacts between αJ and αF are disrupted in order to relieve HD inhibition of PARP-1. Mutations were made at the αF–αJ interface with the expectation that disruptive changes would relieve HD inhibition, leading to mutants with an increase in DNA-independent activity. Mutations were designed to replace small non-polar amino acid sidechains (Ala, Gly) with polar or bulky sidechains (Ser, Leu), thereby disrupting the local network of contacts. As hypothesized, mutations at the point of contact between αF and αJ led to an increase in DNA-independent activity, 3- to 10-fold greater than that of PARP-1 WT (Fig. 5A–C). This level of activation represents approximately 10% of PARP-1 WT activity in the presence of DNA, indicating that point mutants alone cannot fully recapitulate the major structural changes expected based on HXMS analysis.
Figure 5. Disruption of HD–ART contacts mediated by αF/αJ increases PARP-1 DNA-independent activity.
(A) Location of mutations at the αF/αJ interface versus the ASL/αD interface.
(B) and (D) DNA-independent catalytic activity of PARP-1 FL WT and mutants (60 nM) using the colorimetric assay.
(C) and (E) Fold activation over PARP-1 FL WT at 60 minutes presented as an average of three independent experiments with the associated standard deviation.
See also Figure S6.
As a control set of mutations, we also made amino acid substitutions at the second HD–ART interface mediated by αD and the ASL. These contacts are primarily made through main chain interactions; thus, we created the P882G, P885G, and P885S mutants to disrupt the structure and flexibility of the ASL. We observed no effect on DNA-independent activity for these individual mutants (Fig. 5D–E). We further created a quadruple substitution mutant of the ASL (E883A/A884S/P885S/V887A), as well as mutations of αD at the contact site with ASL (S714A and Q717A). None of these mutations at the αD–ASL contact point led to an increase in PARP-1 DNA-independent activity, but rather maintained WT levels of activity in the absence of DNA (Fig. 5D–E). We did observe a reduction in DNA-dependent activity for some of the more severe mutants at the αD–ASL interface, despite similar DNA binding affinities (Fig. S6C–D). The observed deficiencies could reflect that αD–ASL contacts serve as an anchor point to counterbalance the major unfolding transition occurring in the other half of the HD in the presence of DNA.
The HD blocks productive NAD+ binding
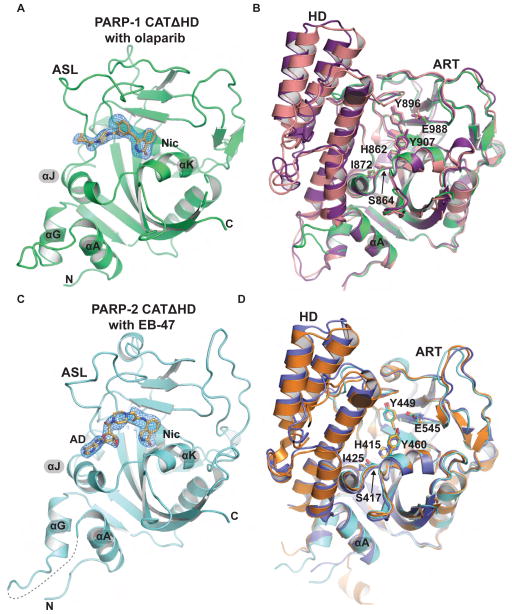
Our biochemical data indicated that the HD–ART interface mediated by αF and αJ is disrupted during PARP-1 activation. αJ contributes to the base of the NAD+ binding site, thus we anticipated that the disruption of αF–αJ contacts could release potential conformational restraints placed on αJ, and thereby allow a different and more active conformation of αJ. Alternatively, the folded conformation of the HD could position αF such that it interferes with ART activity without the requirement for a change in αJ position. To distinguish between these possibilities, we crystallized the CAT domains of PARP-1 and PARP-2, each bearing HD deletions (CATΔHD). As expected, both PARP-1 and PARP-2 CATΔHD exhibited a substantial increase in catalytic activity compared to their respective WT versions (Fig. S6E–F). PARP-1 CATΔHD crystals were grown in the presence of the inhibitor olaparib, and PARP-2 CATΔHD crystals were grown in the presence of the inhibitor EB-47. The crystals of PARP-1 CATΔHD and PARP-2 CATΔHD diffracted to 2.6 Å and 2.7 Å, respectively, and the structures were determined by molecular replacement (Fig. 6A, 6C; Table 1).
Figure 6. Crystal structure analysis of constitutively hyperactive PARP-1 and PARP-2.
(A) Crystal structure of PARP-1 CATΔHD (green) bound to olaparib (PDB code 5ds3). The weighted FO-FC difference electron density prior to modeling olaparib is shown, with the final olaparib coordinates overlayed. The locations of the nicotinamide binding pocket (Nic) and the active site loop (ASL) are labeled.
(B) Structural alignment of a PARP-1 catalytic domain crystal structure (purple, 1a26), the catalytic domain extracted from the PARP-1/DNA crystal structure (pink, 4dqy), and the PARP-1 CATΔHD/olaparib structure (green). Key NAD+ binding site residues are drawn as sticks and labeled.
(C) Crystal structure of PARP-2 CATΔHD (cyan) bound to EB-47 (PDB code 5dsy). The weighted FO-FC difference electron density prior to modeling EB-47 is shown, with the final EB-47 coordinates overlayed. The locations of the nicotinamide binding pocket (Nic) and the adenosine binding site (AD) are labeled.
(D) PARP-2 catalytic domain crystal structures (orange, 1gs0; blue, 3kjd) aligned to the PARP-2 CATΔHD/EB-47 structure (cyan).
Table 1.
Crystallographic statistics
| Data Collectiona | ||
|---|---|---|
| PARP-1 ΔHD with olaparib | PARP-2 ΔHD with EB-47 | |
| Space Group | P6122 | P212121 |
| Unit Cell Dimensions |
a=b=94.0 Å, c=135.2 Å α=β=90°, γ=120° |
a=92.2 Å, b=119.9 Å, c=120.7 Å α=β=γ=90° |
| 1 molecule / asymmetric unit | 4 molecules / asymmetric unit | |
| Wavelength (Å) | 1.12 | 1.12 |
| Resolution range (Å) | 20–2.6 (2.7–2.6) | 50–2.7 (2.8–2.7) |
| Completeness (%) | 99.7 (99.4) | 98.4 (87.2) |
| Average Redundancy | 13.8 (13.9) | 12.0 (11.5) |
| Mean (I/σI)b | 11.8 (1.4) | 11.0 (1.3) |
| Rmerge (%)b | 20.4 (198.0) | 25.8 (199.5) |
| Rpim (%)b | 5.6 (54.2) | 7.7(60.6) |
| Mean I CC(1/2)b | 0.997 (0.605) | 0.994 (0.492) |
| Model Refinementa | ||
| Resolution Range (Å) | 20–2.6 (2.66–2.60) | 50–2.7 (2.73–2.7) |
| Number of reflections | 10,627 (535) | 37,280 (3,524) |
| Rcrystc | 0.198 (0.392) | 0.193 (0.333) |
| Rfreec | 0.256 (0.422) | 0.247 (0.383) |
| Number of atoms / Average B-factor (Å2) | 2,017 / 28.5 | 8,090 / 55.1 |
| protein | 1,928 / 27.8 | 7,882 / 55.4 |
| inhibitor | 32 / 22.1 | 156 / 42.6 |
| solvent | 57 / 55.5 | 52 / 47.6 |
| Phi/Psi, most favored (%) | 93.3 | 97.6 |
| R.m.s.d. bond angles (°) | 1.625 | 1.331 |
| R.m.s.d. bond lengths (Å) | 0.013 | 0.010 |
Values in parentheses refer to data in the highest resolution shell.
As calculated in AIMLESS (Winn et al., 2011): Rmerge = ΣhklΣj|Ij − 〈I〉| / ΣhklΣj Ij. 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents; Rpim takes into account measurement redundancy when calculating Rmerge; Mean I CC(1/2) is the correlation between mean intensities calculated for two randomly chosen half-sets of the data.
Rcryst = Σhkl|Fobs − kFcalc|/ Σhkl|Fobs|. Rfree = Rcryst for 5% of reflections excluded from crystallographic refinement.
The PARP-1 CATΔHD structure revealed that the ART does not show significant structural changes in the absence of the inhibitory HD (Fig. 6B). Importantly, the position of αJ is unchanged when ΔHD and WT structures are compared (Fig. 6B). Olaparib is well ordered in the catalytic active site of PARP-1 (Fig. 6A) with the pyrimidine ring positioned within the nicotinamide binding pocket in a manner similar to that observed in a recent crystal structure of TNK2 (PARP-5b) in complex with olaparib (Narwal et al., 2012). The remainder of olaparib binds to PARP-1 in a different conformation than that seen in the TNK2/olaparib structure, and the difference in binding mode likely reflects the major differences between the ASL of TNK2 and PARP-1. Indeed, olaparib binding to TNK2 induces a conformational change in the ASL, where the ASL forms a large part of the interaction surface. In contrast, olaparib makes few contacts with the ASL of PARP-1.
The PARP-2 CATΔHD structure also indicates that HD deletion does not induce conformational changes in the ART that can explain the increase in catalytic activity (Fig. 6D). αJ of PARP-2 CATΔHD maintains a position identical to that seen in PARP-2 WT. There is a subtle change in the ASL conformation when PARP-2 CATΔHD is compared to WT, but the ASL makes a crystal contact that likely dictates the observed conformation. Further, alignment of several PARP-1/PARP-2 CAT WT structures indicates that the ASL is inherently mobile and adopts variable positions.
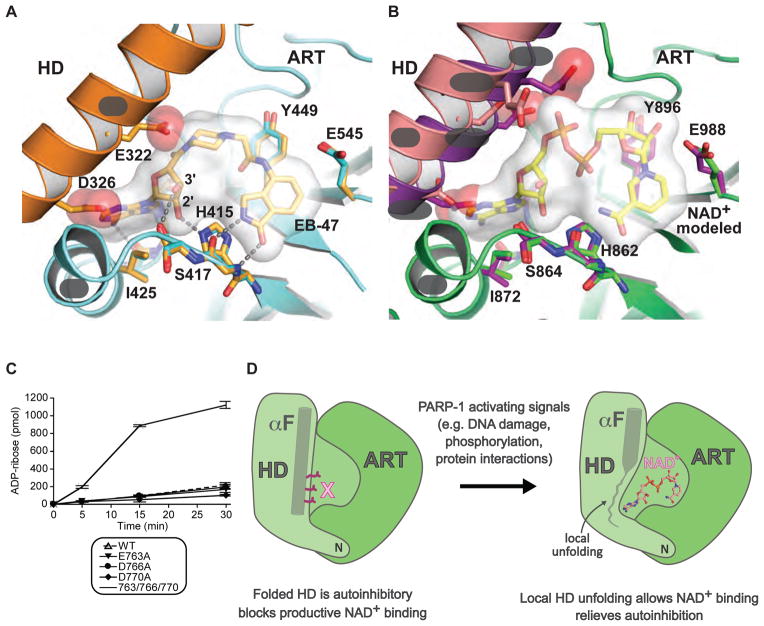
EB-47 is well ordered in the PARP-2 CATΔHD structure, with the isoindolinone group at one end occupying the nicotinamide binding site (Nic; Fig. 6C), and the adenosine group at the opposite end binding adjacent to αJ (Fig. 6C). EB-47 was designed to mimic the adenosine group of NAD+ (Jagtap et al., 2004). The PARP-2 CATΔHD complex with EB-47 displays a network of contacts with the adenosine group, most notably an interaction between the ribose 2′ hydroxyl and H415 (Fig. 7A). H415 forms part of the conserved PARP family triad H-Y-E (Gibson and Kraus, 2012), which collectively contribute to the PARP NAD+ binding site based on the related diphtheria toxin structure determined in complex with substrate NAD+ (Bell and Eisenberg, 1996); there are no structures available for PARP family members bound to NAD+. A recent crystal structure of TNK2 in complex with EB-47 provided a similar model for adenosine group interaction with a PARP catalytic domain (Haikarainen et al., 2013). The TNK2/EB-47 and PARP-2 CATΔHD/EB-47 structures both indicate that the adenine base is oriented in the anti conformation, similar to the orientation observed in an iota-toxin complex (Tsuge et al., 2008), and in contrast to the syn conformation observed in the Diphtheria toxin complex with NAD+, commonly used to model NAD+ interaction with PARPs.
Figure 7. The HD is an autoinhibitory domain that blocks productive NAD+ binding.
(A) The catalytic active site of PARP-2 CATΔHD/EB-47 structure. A complete HD-ART catalytic domain structure for PARP-2 (1gs0, in orange) was aligned to PARP-2 CATΔHD (5dsy, in cyan), highlighting the conserved position of key ART domain residues, and steric clashes introduced by the positioning of residues on αF.
(B) NAD+ was modeled in the active site of PARP-1 based on adenosine contacts observed in the PARP-2/EB-47 structure and nicotinamide contacts observed in toxin structures (e.g. Diptheria toxin). Key NAD+ binding residues are shown for PARP-1 CATΔHD (green, 5ds3), chicken PARP-1 (purple, 1a26), and the PARP-1/DNA complex (pink, 4dqy). αF residues that project toward the NAD+ binding site are drawn as sticks (D770, D766, E763/Q763; chicken PARP-1 has a Gln residue at position 763).
(C) DNA-independent catalytic activity of three single mutants and a combined triple mutant located on αF compared to WT PARP-1. Experiments were performed in triplicate and the averages and standard deviations are shown.
(D) Model for PARP-1 catalytic domain changes associated with activating signals.
We next analyzed whether the HD might interfere with ART function, and thereby lead to an inhibitory effect. Comparison of an HD-containing catalytic domain structure to the PARP-2 CATΔHD structure indicated that αF in the folded HD conformation sterically interferes with the adenosine group of EB-47 (Fig. 7A). Specifically, D326 clashes with the adenine base and E322 clashes with the piperazine/carbonyl portion of EB-47, which is expected to occupy the same space as the phosphate groups of a bound NAD+. E322 positioned near the phosphate groups could introduce an unfavorable interaction between negatively charged groups. The PARP-2 CATΔHD/EB-47 complex allowed us to model PARP-1 interaction with NAD+ based on the observed PARP-2 interactions with the adenosine group, and based on the positioning of the EB-47 isoindolinone group in the nicotinamide binding site. The model of PARP-1 bound to NAD+ indicates that the folded conformation of the HD positions αF to interfere with NAD+ binding (Fig. 7B). Most notably, three residues from the same face of αF project toward the NAD+ binding site – D770, D766, and E763 (Fig. 7B). D770 is positioned to interfere with binding of the adenine base, and the charged side chains of D766 and E763 are positioned to interfere with the phosphate groups of NAD+. Importantly, these residues are located on αF near the interface with αJ, and in a region that demonstrated a drastic increase in HX in the presence of DNA, even at the 10 s time point. Our model suggested that no single residue could account for the autoinhibitory role of the HD, but rather that the positioning of several residues on αF mediates autoinhibition, consistent with the required unfolding of the helix to relieve autoinhibition. We predicted that a combination of mutations that collectively relieves steric clashes could potentially overcome autoinhibition of the HD. We performed site-directed mutagenesis of PARP-1 residues predicted to interfere with NAD+ binding (D770, D766, E763), and assessed their activities in the absence of DNA. Alanine substitutions at each of these positions individually did not increase PARP-1 DNA-independent activity; however, the combination of the three mutations yielded a substantial increase in DNA-independent PARP-1 activity (Fig. 7C). The triple mutant did not reach the activity level obtained with an HD deletion (Fig. S6E, approximately 15% of the activity of PARP-1 CAT ΔHD), thus indicating that the truncation of these amino acid side chains does not achieve the same result as conformational destabilization of αF. Collectively, our results signify that PARP interaction with DNA damage induces HD unfolding, thereby locally disrupting αF structure and its autoinhibitory interactions near αJ, resulting in a fully competent binding site for NAD+ utilization in the poly(ADP-ribosyl)ation reaction (Fig. 7D).
DISCUSSION
PARP-1 catalytic activity is rapidly and acutely elevated upon engaging DNA damage during the cellular response to genotoxic stress. We have performed HXMS to evaluate the differences in the structure and dynamics of PARP-1 in the absence and presence of DNA damage. Strikingly, we observed local unfolding of the HD when PARP-1 is activated by DNA damage. The magnitude of the structural changes observed in solution was remarkable, with the HX behavior of HD helices indicating a stable fold in the absence of DNA, but an unfolded (or very rapidly unfolding) state when PARP-1 engages DNA damage. Rapid HX is not consistent with these helices changing position in space to a more ‘solvent accessible’ location. While HX results have been interpreted as related to surface exposure, the amide protons within a folded α-helix must await helical unfolding (or wait for it to rapidly sample unfolded states many times) in order to exchange with protons (or deuterons) in solvent (Englander, 2006). It is notable that for the N-terminal portion of helix αF, which is ‘solvent exposed’ before and after PARP-1 binding to DNA damage, there is essentially no rapid HX (Fig. 2A), consistent with our interpretation that this part of the helix remains stably folded while the C-terminal portion of helix αF unfolds when PARP-1 is bound to DNA damage.
HD unfolding was not anticipated since the helical character of the HD is consistently maintained in available crystal structures. The PARP-1/DNA crystal structure provided evidence of HD distortion (the “leucine switch”); however, αF and αB retained their helical folded conformation (Langelier et al., 2012). αF is moved slightly away from αJ in the structure, but in this position αF nevertheless still sterically blocks the adenosine group of NAD+ (Fig. 7B). The PARP-1/DNA crystal structure likely represents a snapshot of the HD unfolding transition and reports on an intermediate that was stable enough to crystallize. The fully activated, unfolded conformation of the HD has a low probability of being captured in a crystal lattice; thus highlighting the appropriateness of HXMS analysis. We have made several mutations to the HD that should clearly distort its conformation, but none of these have led to an increase in activity that equals that reached when PARP-1 interacts with DNA, except for the full deletion of the HD. The necessity to remove the contacting elements in total through engineered deletions is key to our understanding that a distorted but stable HD conformation cannot explain how relief from autoinhibition is achieved in PARP-1. Rather, the unfolding (or rapid sampling of unfolded states) of helices within the HD is required to relieve autoinhibition.
The constitutive DNA-independent activity of HD deletion mutants of PARPs 1, 2, and 3 indicate a conserved autoinhibitory role for the HD (Fig. 4), and x-ray structure analysis of PARP-1 and PARP-2 HD deletions indicated that the folded conformation of the HD blocks productive binding of NAD+ (Fig. 7). Binding to DNA damage organizes PARP-1 domains such that a series of interdomain contacts are formed with the HD, primarily through the WGR domain but also with contributions from F3 (e.g. N567 and W318 interacting with the αE helix of the HD). We envision that the native, folded HD conformation is distorted in order to properly form these contacts, and these structural distortions include the repositioning of residues that contribute to the folded core of the HD, most notably the “leucine switch” and other residues that normally stack against the interior face of the αF helix. These HD structural distortions, and potentially other distortions not captured in the crystal structure snapshot, lead to the unfolding we observe in αB, αD, and the C-terminal portion of αF. Our experiments collectively show in detail that the local unfolding allows NAD+ binding, culminating in the massive increase in catalytic activity of PARP-1 when it is bound to damaged DNA (Fig. 7D).
There is an interesting parallel between the DNA damage-dependent PARPs and other enzymes that robustly consume NAD+ such that inhibition is required to ensure cellular viability. For example, the pathogenic bacterium Streptococcus pyogenes produces both an NAD+ glycohydrolase toxin (SPN) with an ART-fold that can deplete cellular NAD+ and an immunity factor (IFS) that binds to SPN to block NAD+ access to the active site (Kimoto et al., 2006; Meehl et al., 2005; Michos et al., 2006; Smith et al., 2011). The toxicity of SPN in human cells is due to NAD+ and ATP depletion (Michos et al., 2006; Smith et al., 2011). Similarly, SPN is lethal to S. pyogenes in the absence of IFS, and recombinant SPN produced in the absence of IFS is toxic to E. coli. Expression of the PARP HD deletions required the presence of a PARP inhibitor, without which the proteins were not expressed, or in some cases caused the E. coli cultures to stop growing and/or lyse. These observations illustrate the need for an HD autoinhibitory domain to repress ART catalytic activity and preserve cell viability under normal conditions. Under conditions of genotoxic stress, HD inhibition is relieved in a controlled and temporary way to allow the cellular response to DNA damage to proceed.
The HD inhibits ART catalytic activity by blocking productive NAD+ binding, and the autoinhibited state leaves the DNA damage-dependent PARPs at a very low basal level of activity. All biological pathways that require elevated PARP-1 catalytic activity will therefore need to overcome this mechanism of autoinhibition, and there are examples of PARP-1 activating signals that stimulate more modest levels of activity than that observed with DNA damage. It is thus interesting to highlight that the extent of HD conformational destabilization correlates with the extent of catalytic activation (Fig. 2; Fig. 3). PARP-1 CAT L713F has partial activity relative to PARP-1 activated by DNA damage, and correspondingly the HD exhibited increased exchange, but not to the level of complete unfolding (Fig. 2F–G). Moreover, the W318R mutation completely shuts down DNA-dependent PARP-1 activity and also fully prevented HD unfolding (Fig. 3C, 3E). HD unfolding is thus critical for PARP-1 activation, and the extent of unfolding can act as a regulator of NAD+ access and therefore PARP-1 catalytic output. We expect that other PARP-1 activating signals (e.g. phosphorylation, interaction with other proteins) will act to either directly perturb HD structure, or perhaps alter HD structure by engaging the WGR domain and promoting its interaction with the HD (Fig. 7D).
HD unfolding changes the local environment of the NAD+ binding site, and thus the binding location of many of the clinically used PARP inhibitors, which all compete with NAD+ (Steffen et al., 2013). The PARP-1 and PARP-2 CAT HD structures, which mimic HD unfolding, will thus be useful for analysis of inhibitor interactions with the activated conformation of the DNA damage-dependent PARPs. One objective of designing PARP selective inhibitors has been to target the adenosine binding pocket and the structural variations observed among PARP family members, including the HD (Steffen et al., 2013). It will be important to evaluate whether these inhibitors engage dynamic features of the HD, such as αF, and how this will impact inhibitor efficacy.
EXPERIMENTAL PROCEDURES
Gene cloning and mutagenesis
The PARP-1 CATΔHD construct used for crystallization was created by replacing PARP-1 residues 678–787 with an 8-residue linker (GSGSGSGG) in a pET28 construct coding for PARP-1 residues 661–1011. The PARP-2 CATΔHD construct used for crystallization was created by deleting residues 241–334 from a pET28 construct coding for PARP-2 isoform 2 residues 216–570.
Protein Purification
PARP-1, PARP-2, and PARP-3 WT and mutants were expressed and purified as described previously using three chromatography steps (Ni2+ affinity, heparin-sepharose, and gel filtration) (Langelier et al., 2011b; Langelier et al., 2008; Langelier et al., 2014).
HX reactions
Deuterium on-exchange was performed at room temperature (RT) by adding 5 μL of each sample (~14 μg of CAT truncation constructs, ~5 μg of full-length PARP-1 constructs) to 15 μL of deuterium on-exchange buffer (10 mM HEPES, pD 7.0, 150 mM NaCl) for a final D2O content of 75%. A 20 μL aliquot was removed at each time point, and the reaction was quenched with 30 μL ice-cold quench buffer (1.66 M guanidine hydrochloride, 10% glycerol, and 0.8% formic acid, for a final pH of 2.4–2.5) and rapidly frozen in liquid nitrogen.
Protein fragmentation and MS
Each HX sample (50 μL) was thawed on ice and injected into an on-line temperature-controlled system, as previously described (DeNizio et al., 2014).
SDS-PAGE assay
The SDS-PAGE assay was performed as described previously (Langelier et al., 2008) using 1 μM protein, 1 μM DNA and 0.5 mM NAD+ where indicated.
Colorimetric Assay
The colorimetic assays were performed essentially as described for PARP-1, PARP-2, and PARP-3 (Langelier et al., 2014; Langelier et al., 2008; Langelier et al., 2011b).
Crystallization and Structure Determination
PARP-1 CATΔHD and PARP-2 CATΔHD were crystallized by hanging drop vapor diffusion. Diffraction data was collected at the SIBYLS beamline (ALS 12.3.1) and processed using XDS (Kabsch, 2010). Structures were determined by molecular replacement using PHASER (McCoy, 2007) as implemented in the PHENIX suite (Adams et al., 2010). Electron density map inspection and model building was performed using COOT (Emsley et al., 2010). Model refinement was performed using PHENIX (Adams et al., 2010), PDB-REDO (Joosten et al., 2014), and REFMAC5 (Murshudov et al., 2011).
Supplementary Material
Acknowledgments
We would like to thank S. Eustermann and D. Neuhaus for providing purified sortase enzyme and the sequence of the DNA dumbbell used in this study, J. Benovic and M. Lemmon for manuscript feedback, W. Englander for helpful discussions, the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interactions Facility at Thomas Jefferson University (P30CA56036), the SIBYLS 12.3.1 beamline and staff of the Advanced Light Source (Lawrence Berkeley Laboratory), and support from the NIH with postdoctoral fellowship awards (GM108360 to JMD and CA174221 to JDS) and a research grant (GM087282 to JMP and BEB).
Footnotes
ACCESSION NUMBERS
The coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank with the following accession codes: 5ds3 (PARP-1 ΔHD CAT) and 5dsy (PARP-2 ΔHD CAT).
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online.
AUTHOR CONTRIBUTIONS
JMD performed the HXMS experiments and analyzed the data. MFL performed protein purification and mutagenesis, biochemical assays, and crystal structure determination. JED, CDC, and KRK contributed to HXMS experiments and analysis. AAR performed protein purification and mutagenesis, biochemical assays, and crystal structure determination. MM and JDS performed protein purification and mutagenesis. JMD, MFL, BEB, JMP designed the experiments, analyzed the data, and wrote the manuscript. BEB and JMP directed the research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–92. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci U S A. 2014;111:10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- Chapman JD, Gagne JP, Poirier GG, Goodlett DR. Mapping PARP-1 Auto-ADP-ribosylation Sites by Liquid Chromatography-Tandem Mass Spectrometry. J Proteome Res. 2013;12:1868–1880. doi: 10.1021/pr301219h. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–68. [PMC free article] [PubMed] [Google Scholar]
- D’Silva I, Pelletier JD, Lagueux J, D’Amours D, Chaudhry MA, Weinfeld M, Lees-Miller SP, Poirier GG. Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim Biophys Acta. 1999;1430:119–126. doi: 10.1016/s0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- DeNizio JE, Elsasser SJ, Black BE. DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res. 2014;42:4318–4331. doi: 10.1093/nar/gku090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Videler H, Yang JC, Cole PT, Gruszka D, Veprintsev D, Neuhaus D. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J Mol Biol. 2011;407:149–170. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Wu W-F, Langelier M-F, Yang J-C, Easton LE, Riccio AA, Pascal JM, Neuhaus D. Structural basis for DNA single-strand break detection and signaling by human PARP-1. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–96. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, Wheeler D, Redpath P, Li J, Romero G, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8:1819–1831. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquerel E, Sobol RW. ARTD1 (PARP1) activation and NAD in DNA repair and cell death. DNA Repair (Amst) 2014;23:27–32. doi: 10.1016/j.dnarep.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné JP, Ethier C, Defoy D, Bourassa S, Langelier MF, Riccio AA, Pascal JM, Moon KM, Foster LJ, Ning Z, Figeys D, Droit A, Poirier GG. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair. 2015 doi: 10.1016/j.dnarep.2015.02.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Haikarainen T, Narwal M, Joensuu P, Lehtio L. Evaluation and Structural Basis for the Inhibition of Tankyrases by PARP Inhibitors. ACS Med Chem Lett. 2013;5:18–22. doi: 10.1021/ml400292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle AN, Resing KA, Goldsmith EJ, Ahn NG. Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc Natl Acad Sci U S A. 2001;98:956–961. doi: 10.1073/pnas.98.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill DM, Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J Biol Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- Jagtap PG, Southan GJ, Baloglu E, Ram S, Mabley JG, Marton A, Salzman A, Szabo C. The discovery and synthesis of novel adenosine substituted 2,3-dihydro-1H-isoindol-1-ones: potent inhibitors of poly(ADP-ribose) polymerase-1 (PARP-1) Bioorg Med Chem Lett. 2004;14:81–85. doi: 10.1016/j.bmcl.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–20. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto H, Fujii Y, Hirano S, Yokota Y, Taketo A. Genetic and biochemical properties of streptococcal NAD-glycohydrolase inhibitor. J Biol Chem. 2006;281:9181–9189. doi: 10.1074/jbc.M506879200. [DOI] [PubMed] [Google Scholar]
- Langelier MF, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr Opin Struct Biol. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem. 2011a;286:10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Servent KM, Pascal JM. Purification of human PARP-1 and PARP-1 domains from Escherichia coli for structural and biochemical analysis. Methods Mol Biol. 2011b;780:209–226. doi: 10.1007/978-1-61779-270-0_13. [DOI] [PubMed] [Google Scholar]
- Langelier MF, Riccio AA, Pascal JM. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42:7762–7775. doi: 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Servent KM, Rogers EE, Pascal JM. A Third Zinc-binding Domain of Human Poly(ADP-ribose) Polymerase-1 Coordinates DNA-dependent Enzyme Activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl MA, Pinkner JS, Anderson PJ, Hultgren SJ, Caparon MG. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog. 2005;1:e35. doi: 10.1371/journal.ppat.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos A, Gryllos I, Hakansson A, Srivastava A, Kokkotou E, Wessels MR. Enhancement of streptolysin O activity and intrinsic cytotoxic effects of the group A streptococcal toxin, NAD-glycohydrolase. J Biol Chem. 2006;281:8216–8223. doi: 10.1074/jbc.M511674200. [DOI] [PubMed] [Google Scholar]
- Miranda EA, Dantzer F, O’Farrell M, de Murcia G, de Murcia JM. Characterisation of a gain-of-function mutant of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1995;212:317–325. doi: 10.1006/bbrc.1995.1972. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narwal M, Venkannagari H, Lehtio L. Structural basis of selective inhibition of human tankyrases. J Med Chem. 2012;55:1360–1367. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
- Pion E, Bombarda E, Stiegler P, Ullmann GM, Mely Y, de Murcia G, Gerard D. Poly(ADP-ribose) polymerase-1 dimerizes at a 5′ recessed DNA end in vitro: a fluorescence study. Biochemistry. 2003;42:12409–12417. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- Robu M, Shah RG, Petitclerc N, Brind’Amour J, Kandan-Kulangara F, Shah GM. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci U S A. 2013;110:1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KW, Kim DS, Kraus WL. New Facets in the Regulation of Gene Expression by ADP-Ribosylation and Poly(ADP-ribose) Polymerases. Chem Rev. 2015;115:2453–81. doi: 10.1021/cr5004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J, Dunn CA, Singh N, Veith S, Hasan-Olive MM, et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014;20:840–55. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Ghosh J, Elam JS, Pinkner JS, Hultgren SJ, Caparon MG, Ellenberger T. Structural basis of Streptococcus pyogenes immunity to its NAD+ glycohydrolase toxin. Structure. 2011;19:192–202. doi: 10.1016/j.str.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Front Oncol. 2013;3:301. doi: 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JD, Tholey RM, Langelier MF, Planck JL, Schiewer MJ, Lal S, Bildzukewicz NA, Yeo CJ, Knudsen KE, Brody JR, et al. Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res. 2014;74:31–37. doi: 10.1158/0008-5472.CAN-13-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of Human Poly(ADP-ribose) Polymerase-1 Is Important for Enzyme Activity and Contains a Novel Zinc-Ribbon Motif. Biochemistry. 2008;47:5804–5813. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- Tsuge H, Nagahama M, Oda M, Iwamoto S, Utsunomiya H, Marquez VE, Katunuma N, Nishizawa M, Sakurai J. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc Natl Acad Sci U S A. 2008;105:7399–404. doi: 10.1073/pnas.0801215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.