Abstract
Objectives
Dysbiosis of intestinal microbiota has been implicated in ulcerative colitis (UC). Fucosyltransferase (FUT) 2 and FUT3 determine expression of histo-blood group antigens in the gut and may affect the intestinal microbiota. We investigated the association between FUT2 and FUT3 polymorphisms and UC in Chinese patients.
Methods
We genotyped FUT2 (rs281377, rs1047781 and rs601338) and FUT3 (rs28362459, rs3745635 and rs3894326) in 485 UC patients and 580 healthy controls using SNaPshot. We also evaluated expression of Lewis a and b antigens in the sigmoid colon of 7 UC patients and 7 patients with benign colonic polyps.
Results
The frequencies of mutant allele (A) and genotype (GA+AA) in FUT3 (rs3745635) were higher in UC patients than controls (P = 0.016, 95%CI: 1.339–1.699; P = 0.038, 95%CI: 1.330–1.742, respectively). Stratified analyses revealed that the frequencies of mutant allele (G) and genotype (TG+GG) of FUT3 (rs28362459) were significantly lower in patients with extensive colitis than those with distal colitis (P<0.001, 95%CI: 0.503–0.742; P = 0.001, 95%CI: 0.567–0.786, respectively). Similar conclusions were drawn for the mutant allele (A) and genotype (GA+AA) of FUT3 (rs3745635) in patients with extensive colitis compared to those with distal colitis (P = 0.006, 95%CI: 0.553–0.845; P = 0.011, 95%CI: 0.621–0.900, respectively). Although expression of Lewis b antigen in the sigmoid colon did not differ between UC patients and controls, Lewis a antigen expression was higher in the cryptic epithelium of both inflammatory and non-inflammatory sigmoid colon of UC patients than controls (P = 0.028).
Conclusions
Our findings indicated that polymorphisms in FUT3 and its intestinal expression might be associated with UC pathogenesis.
Introduction
Inflammatory bowel diseases (IBDs) are a group of chronic and non-specific intestinal inflammatory disorders including ulcerative colitis (UC) and Crohn’s disease (CD). Although the precise etiology of IBDs is not yet fully understood, abnormal host-microbial interactions have been implicated in the pathogenesis of IBD. Mucosal and fecal bacterial analyses have suggested that patients with IBD have less complex commensal bacteria population and higher numbers of mucosal-associated bacteria than healthy individuals [1–3]. Animal models have also indicated that the composition of intestinal microbiota influenced intestinal inflammation [4]. The composition of intestinal microbiota appears to be influenced by host genetics. For instance, in patients with IBD carrying the NOD2 and ATG16L1 risk alleles the intestinal microbiota contains lower levels of Faecalibacterium and higher levels of Escherichia [5].
The intestinal microbiota can be influenced by Fucosyltransferase (FUT) 2 and FUT3, which control presentation of histo-blood group antigens (HBGA) on the gastrointestinal mucosa and in bodily secretions. HBGA, including ABH antigens and Lewis antigens, act not only as binding sites for intestinal microbes such as Helicobacter pylori, Campylobacter jejuni, Norovirus and rotavirus [6–8], but also a carbon source for microbes including Escherichia coli [9]. FUT2 is located in chromosome 19q13, while FUT3 is mapped to chromosome 19p13, a genomic region containing putative susceptibility loci (IBD6) for IBD [10,11]. FUT2 influences presentation of ABH antigens in the gastrointestinal mucosa and their secretion. In people that express functional FUT2, termed secretors, expression of ABH antigens is widespread in the stomach and small intestine, but decreases progressively from the proximal to distal colon [12,13]. Homozygotes for loss-of-function alleles in FUT2 lack expression of ABH antigens in the gastrointestinal mucosa and bodily secretions and account for approximately 20% of the world’s population [14–16]. FUT2(rs601338, G428A) is the most common polymorphism in FUT2 Caucasian nonsecretors. In Chinese and Japanese populations, however, FUT2 (rs601338, G428A) is rare and instead the more common missense mutation FUT2 (rs1047781, A385T) is responsible for dramatically decreased expression of ABH antigens [15–17]. Additionally, FUT2 (rs281377, T357C) has been identified as a common silent mutation in Chinese non-secretors [18, 19].
FUT3 encodes α-(1,3/4)-fucosyltransferase required to synthesize Lewis a antigens, and mostly utilizes the H antigen, determined by FUT2, as a substrate to synthesize Lewis b antigen. Lewis b is mainly expressed in the proximal colon, but not expressed in the distal colon, whereas Lewis a is uniformly distributed throughout the colon [13]. Studies in Chinese populations have demonstrated that (rs28362459, T59G), (rs3745635, G508A) and (rs3894326, T1067A) were the most common polymorphic loci of FUT3 [18]. Furthermore, mutations of FUT3 (rs3745635) and (rs3894326) inactivate the enzyme [10, 20], while mutation of FUT3 (rs28362459) reduces the availability of α-(1,3/4)-fucosyltransferase [10, 19].
In recent years, several studies have linked several nucleotide polymorphisms in FUT2 to intestinal microbiota composition [21] and predisposition to CD [22, 23], celiac disease [24], type 1 diabetes [25] and primary sclerosing cholangitis [26], highlighting the essential role of host gene-microbiota interaction in autoimmune diseases. However, the conclusions drawn about the influence of FUT2 on UC, were not consistent. Additionally, few studies have investigated the influence of FUT3 polymorphisms on UC. In this study, we investigated the prevalence of FUT2 and FUT3 polymorphisms in a cohort of UC patients and healthy controls in Southeast China. We also evaluated intestinal expression of Lewis a and b antigens to further investigate the role of these genes in pathogenesis of UC.
Materials and Methods
Subjects
Between January 2004 and May 2015, 485 consecutive UC patients were recruited from The Second Affiliated Hospital of Wenzhou Medical University in Wenzhou city, Zhejiang province of Southeast China. UC was diagnosed according to colonoscopic, histo-pathological, radiologic and clinical findings, following the Lennard-Jones Criteria [27]. The severity of UC were evaluated by Truelove & Witt Activity Index [28]. In the corresponding period, a total of 580 age- and sex-matched healthy controls were enrolled at the Health Examination Center of The Second Affiliated Hospital of Wenzhou Medical University. Patients with autoimmune diseases, tumors, and IBD family history were excluded. Demographic data was collected (Table 1). The age and sex distribution did not differ significantly between these UC patients and controls. Of these UC patients, we obtained specimens of inflammatory lesions in the sigmoid colon during colonoscopy examination. Specimens of the adjacent non-inflammatory mucosa were also obtained in five of the 7 patients. In seven patients with benign colonic polyps, confirmed by colonoscopy examination and pathology, specimens of normal sigmoid colon mucosa were obtained during colonoscopy. The study was carried out in line with the Treaty of Helsinki and was approved by Ethics Committee of The Second Affiliated Hospital of Wenzhou Medical University. Written informed consent was obtained from all UC patients and controls.
Table 1. Demographic characteristics of UC patients and the controls.
| Characteristics | UC | Controls | P |
|---|---|---|---|
| Total number | 485 | 580 | |
| Sex (Female/Male) | 201/284 | 261/319 | 0.243 |
| Age (years) [mean (SD)] | 40.23(15.32) | 41.09(17.11) | 0.392 |
| Age of onset (years) [mean (SD)] | 39.05(14.47) | ||
| Smoking [n (%)] | 0.124 | ||
| Current or ex-smoker | 80(16.5) | 117(20.2) | |
| Never smoked | 405(83.5) | 463(79.8) | |
| Lesion location [n (%)] | |||
| Distal colitis | 311(64.1) | ||
| Extensive colitis | 174(35.9) | ||
| Severity of UC [n (%)] | |||
| Mild | 188(38.8) | ||
| Intermediate | 223(46.0) | ||
| Severe | 74(15.3) | ||
| Treatment [n (%)] | |||
| SASP/5-ASA | 408(84.1) | ||
| Prednisone | 118(24.3) | ||
| Antibiotics | 170(35.1) | ||
| Immunosuppressant | 9(1.9) | ||
| Infliximab | 0(0) | ||
| Colectomy | 2(0.4) |
UC, ulcerative colitis. SD, standard deviation. SASP, sulfasalazine. 5-ASA, 5-aminosalicylic acid.
Genomic DNA Extraction and Genotyping
Approximately 5 ml of peripheral blood was collected from each study subject in an EDTA tube. Genomic DNA was extracted from peripheral blood using the DNeasy Blood & Tissue kit (Qiagen GmbH), according to the manufacturer’s instructions. Genomic DNA was stored at 4°C for subsequent identification of genetic mutations.
We employed a Multiplex SNaPshot assay (Applied Biosystems, California, USA) to examine FUT2 and FUT3 genotypes. First, 10 ng of genomic DNA was added to a 10 μl PCR mixture containing 20 μmol dNTPs (Promega, Wisconsin, USA), 0.5 U FastStart Taq DNA polymerase (Roche, Basel, Switzerland), 1μl 10×PCR buffer with MgCl2 (15 mmol/L) (Roche, Basel, Switzerland) and amplification primers with a specific final concentration (Table 2). The thermal cycler conditions for multiplex PCR amplification were as follows: initial denaturation, 95°C for 5 min, amplification for 35 cycles at 94°C for 30 s, 65°C for 30 s and 72°C for 1 min, followed by a final elongation step at 72°C for 10 min. Subsequently, the PCR products were examined by electrophoresis on a 2.5% agarose gel. Second, we purified the PCR product using a mix of 1.5 U shrimp alkaline phosphatase (SAP) (New England Biolabs, Massachusetts, USA) and 2 U Exonuclease I (TAKARA, Dalian, China) at 37°C for 80 min, then 85°C for 15 min. Third, the multiplex SNaPshot sequencing reactions were carried out in a final volume of 7 μl containing 2 μl of purified multiple PCR products, 1 μl SNaPshot Multiplex Mix, 1 μl 5×Sequencing buffer (Applied Biosystems) and 3 μl SNaPshot sequencing primers (the final concentrations are listed in Table 2). The thermal cycler conditions were: initial denaturation at 96°C for 1 min followed by 25 cycles at 96°C for 10 s, 52°C for 5 s and 60°C for 30 s. Then depuration of product was performed with 1 U SAP at 37°C for 60 min and 75°C for 15 min. Finally, 1.5 μl of SNaPshot products were genotyped using the ABI 3730 Genetic Analyzer before they were mixed with 8 μl HiDiTM formamide and 0.5 μl GeneScan-120LIZ size standard (Applied Biosystems). Data were analyzed using GeneMapper 4.0 (Applied Biosystems). In order to guarantee quality of the study, about 3 percent of the samples were randomly selected and re-genotyped by direct sequencing. The regenotyping and SNaPshot results were in complete accordance with original results.
Table 2. Amplification and extension primers of FUT2 and FUT3.
| SNP | Amplification Primer (5′→3′) | Size (bp) | Conc.(μM) | Extension primer (5′→3′) | Size (bp) | Conc. (μM) |
|---|---|---|---|---|---|---|
| FUT2 rs281377 | F:TCAACATCAAAGGCACTGGGACC R:TGGCGGAGGTGGTGGTAGAA | 338 | 2.5 | 18T-TGGCAGAACTACCACCTGAA | 38 | 1 |
| FUT2 rs1047781 | F:TCAACATCAAAGGCACTGGGACC R:TGGCGGAGGTGGTGGTAGAA | 338 | 2.5 | 38T-TGGAGGAGGAATACCGCCAC | 58 | 1 |
| FUT2 rs601338 | F:TCAACATCAAAGGCACTGGGACC R:TGGCGGAGGTGGTGGTAGAA | 338 | 2.5 | CACCGGCTACCCCTGCTCCT | 20 | 1 |
| FUT3 rs28362459 | F:AAGAAACACACAGCCACCAGCAG R:AATGACCCTCACTCCTCTCTCCTCT | 137 | 1 | 31T-GCGCCGCTGTCTGGCCGCAC | 51 | 1 |
| FUT3 rs3745635 | F:GGCTGAGTCCGGCTTCCAGTT R:TATCATGTCCAACCCTAAGTCACGC | 280 | 1 | 5T-CCGACATCTTCACGCCCTAC | 25 | 1 |
| FUT3 rs3894326 | F:AGGTCCCTAGCAGGCAAGTCTTC R:CTGGGCACTGGATTTCTGCAAGG | 254 | 1 | 21T-CAGGTACCAGACGGTGCGCAGCA | 44 | 1 |
Bold characters refer to the 5’ poly-thymidine tail.
Immunohistochemistry
For immunohistochemical analysis, formalin-fixed paraffin-embedded tissue sections were de-paraffinized with xylene and rehydrated with ethanol solutions and distilled water. Antigen retrieval was performed by heating the sections in EDTA antigen retrieval buffer (pH 9.0) at 99°C for 25 min. Then the slides were washed in PBS (pH 7.4) three times for 5 min. After blocking in endogenous peroxidase with 3% hydrogen peroxide solution at room temperature for 25 min, the slides were again washed in PBS (pH 7.4) three times for 5 min. To block unspecific antibody binding, sections were incubated with 3% BSA (Solarbio, Beijing, China) for 30 min. Sections were then incubated overnight at 4°C in PBS (pH 7.4) containing primary antibodies, blood group Lewis a and b antibodies (Catalog number: sc-51512 and sc-51513, Santa Cruz Biotech, Texas, U.S.A.) at 1:20 in PBS (pH 7.4). Sections were washed three times in PBS then incubated with polyclonal goat anti-mouse immunoglobulins/HRP (Dako, Glostrup, Denmark) at room temperature for 50 min. The the slides were rinsed in PBS three times for 5 min and HRP was added. Rabbit/Mouse (DAB+). Harris staining was performed to highlight the nucleus of cells.
Lewis antigen expression was evaluated quantitatively using the Image Pro Plus 6.0 analysis system (Media Cybernetics, Silver Spring, MD) by calculating the mean density, which was integrated optical density (IOD) divided by area of interest [29]. Three 200X fields under microscope of each slide were randomly selected and the mean density was obtained for further statistic analysis.
Statistical Analysis
Hardy-Weinberg equilibrium for genotypes was evaluated by chi-square test both in UC patients and the controls. Either chi-square or Fisher’s exact test was applied to compare categorical variables such as alleles and genotypes. Unconditional logistic regression analysis was employed to investigate the allelic and genotypic distributions in UC patients, stratified by their clinical characteristics. Covariants included age, gender, smoking, lesion location and severity of UC. Linkage disequilibrium (LD) was estimated and visualized by Haploview 4.2. Haplotypes were reconstructed and haplotype frequencies were calculated by Phase 2.1 software [30]. Mann-Whitney U-test was applied to compare expression of Lewis antigens. All statistical data were input into SPSS 17.0 software (SPSS for Windows version 17.0, Chicago, IL, USA). A two-tailed P value less than 0.05 was considered significant.
Results
Comparison of alleles and genotypes of FUT2 and FUT3 between UC patients and controls
In this study, the genotype distributions of FUT2 and FUT3 were shown to be in good agreement with Hardy-Weinberg equilibrium both in UC patients and the controls (all P>0.05). The allele and genotype frequencies of SNPs in FUT2 did not differ significantly between UC patients and the controls (all P>0.05). However, the frequencies of mutant allele (A) and genotype (GA+AA) in FUT3 (rs3745635) were higher in UC patients than the controls (P = 0.016, 95%CI: 1.339–1.699; P = 0.038, 95%CI: 1.330–1.742, respectively). In addition, the most common homozygous mutation of FUT2 (rs601338), which led to nonsecretors in Caucasians, was not observed in our study. Several healthy controls (17.3%) were homozygous for FUT2 (rs1047781 T), thus nonsecretors (Table 3, Dataset in S1 Dataset).
Table 3. Allelic and genotypic distributions of FUT2 and FUT3 between UC patients and the controls.
| Genotype/allele | Controls(580) n(%) | UC (485)n(%) | OR (95%CI) | P |
|---|---|---|---|---|
| FUT2 rs281377 | ||||
| TT | 441(76.0) | 356(73.4) | ||
| TC | 126(21.7) | 114(23.5) | ||
| CC | 13(2.2) | 15(3.1) | ||
| TC+CC | 139(24.0) | 129(26.6) | 0.871(1.150–1.517) | 0.324 |
| allele C | 152(13.1) | 144(14.8) | 0.904(1.156–1.478) | 0.247 |
| FUT2 rs1047781 | ||||
| AA | 160(27.6) | 155(32.0) | ||
| AT | 314(54.1) | 247(50.9) | ||
| TT | 106(18.3) | 83(17.1) | ||
| AT+TT | 420(72.4) | 330(68.0) | 0.623(0.811–1.056) | 0.119 |
| allele T | 526(45.3) | 413(42.6) | 0.753(0.894–1.061) | 0.200 |
| FUT2 rs601338 | ||||
| GG | 575(99.1) | 479(98.8) | ||
| GA | 5(0.9) | 6(1.2) | ||
| AA | 0(0) | 0(0) | ||
| GA+AA | 5(0.9) | 6(1.2) | 0.437(1.441–4.749) | 0.547 |
| allele A | 5(0.4) | 6(0.6) | 0.437(1.438–4.726) | 0.548 |
| FUT3 rs28362459 | ||||
| TT | 332(57.2) | 281(57.9) | ||
| TG | 219(37.8) | 174(35.9) | ||
| GG | 29(5.0) | 30(6.2) | ||
| TG+GG | 248(42.8) | 204(42.1) | 0.761(0.972–1.241) | 0.819 |
| allele G | 277(23.9) | 234(24.1) | 0.830(1.013–1.237) | 0.895 |
| FUT3 rs3745635 | ||||
| GG | 435(75.0) | 336(69.3) | ||
| GA | 136(23.4) | 133(27.4) | ||
| AA | 9(1.6) | 16(3.3) | ||
| GA+AA | 145(25.0) | 149(30.7) | 1.016(1.330–1.742) | 0.038 |
| allele A | 154(13.3) | 165(17.0) | 1.055(1.339–1.699) | 0.016 |
| FUT3 rs3894326 | ||||
| TT | 471(81.2) | 406(83.7) | ||
| TA | 105(18.1) | 76(15.7) | ||
| AA | 4(0.7) | 3(0.6) | ||
| TA+AA | 109(18.8) | 79(16.3) | 0.611(0.841–1.156) | 0.286 |
| allele A | 113(9.7) | 82(8.5) | 0.635(0.856–1.153) | 0.305 |
Haplotype analysis of FUT2 and FUT3
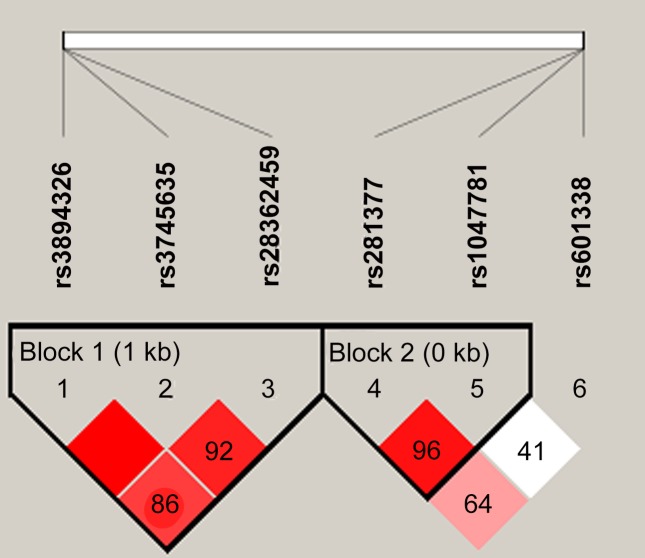
Two LD blocks were presented as follows: block rs281377-rs1047781 in FUT2 (D' = 0.96, r2 = 0.11) and block rs3894326-rs3745635-rs28362459 in FUT3 [rs3894326/rs3745635 (D' = 1, r2 = 0.01), rs3894326/rs28362459 (D' = 0.86, r2 = 0.23), rs3745635/rs28362459 (D' = 0.92, r2 = 0.47)] (Fig 1). Furthermore, we conducted a haplotype analysis in all study subjects to investigate whether any specific haplotype would confer risk of or protection from UC. No haplotypes differed significantly between UC patients and the controls (all P>0.05, Table 4).
Fig 1. Linkage disequilibrium patterns of FUT2 and FUT3 in Chinese Han populations.
Numbers indicated the percentage of D' between 2 SNPs and in the dark area which have no digital represents D' = 1. Dark color indicated strong connection. Block 1 illustrated linkage disequilibrium in the 3 SNP in FUT3, and block 2 showed that rs281377 was in linkage disequilibrium with rs1047781 in FUT2.
Table 4. Haplotype frequencies of FUT2 and FUT3 in patients with UC and the controls.
| Groups | FUT2 (rs281377-rs1047781) | FUT3 (rs3894326- rs3745635-rs28362459) | ||||
|---|---|---|---|---|---|---|
| TT | TA | CA | TGT | TAG | AGG | |
| UC | 0.425 | 0.427 | 0.148 | 0.733 | 0.158 | 0.071 |
| Controls | 0.450 | 0.419 | 0.127 | 0.752 | 0.129 | 0.093 |
No positive results were obtained for each haplotype of FUT2 and FUT3 between UC patients and the controls (all P >0.05). The haplotype frequencies lower than 0.030 were not presented in the table.
Association between FUT2 and FUT3 polymorphisms and clinical characteristics of patients with UC
Unconditional logistic regression analysis was used to evaluate the associations between FUT2 and FUT3 polymorphisms and clinical characteristics of UC patients. Covariants included age, gender, smoking, lesion location and severity of UC. The frequencies of mutant allele (G) and genotype (TG+GG) of FUT3 (rs28362459) were significantly lower in patients with extensive colitis than in those with distal colitis (P<0.001, 95%CI: 0.503–0.742; P = 0.001, 95%CI: 0.567–0.786, respectively). The frequencies of mutant allele (A) and genotype (GA+AA) of FUT3 (rs3745635) in patients with extensive colitis were significantly lower than in patients with distal colitis (P = 0.006, 95%CI: 0.553–0.845; P = 0.011, 95%CI: 0.621–0.900, respectively) (Table 5). However, none of the six SNPs of FUT2 and FUT3 were significantly associated with UC severity in this sample (all P>0.05).
Table 5. Association of FUT2 and FUT3 polymorphisms with clinical characteristics of UC patients.
| Genotype/Allele | Distal colitis(n = 311)(%) | Extensive colitis(n = 174)(%) | OR(95%CI) | P |
|---|---|---|---|---|
| FUT2 rs281377 | ||||
| TT | 231(74.3) | 125(71.8) | ||
| TC+CC | 80(25.7) | 49(28.2) | 0.746(1.132–1.717) | 0.560 |
| T allele | 532(85.5) | 294(84.5) | ||
| C allele | 90(14.5) | 54(15.5) | 0.753(1.086–1.566) | 0.660 |
| FUT2 rs1047781 | ||||
| AA | 90(28.9) | 65(37.4) | ||
| AT+TT | 221(71.1) | 109(62.6) | 0.461(0.683–1.012) | 0.057 |
| A allele | 346(55.6) | 211(60.6) | ||
| T allele | 276(44.4) | 137(39.4) | 0.623(0.814–1.063) | 0.131 |
| FUT2 rs601338 | ||||
| GG | 308(99.0) | 171(98.3) | ||
| GA+AA | 3(1.0) | 3(1.7) | 0.360(1.801–9.022) | 0.468 |
| G allele | 619(99.5) | 345(99.1) | ||
| A allele | 3(0.5) | 3(0.9) | 0.360(1.794–8.938) | 0.469 |
| FUT3 rs28362459 | ||||
| TT | 162(52.1) | 119(68.4) | ||
| TG+GG | 149(47.9) | 55(31.6) | 0.340(0.503–0.742) | 0.000 |
| T allele | 450(72.3) | 286(82.2) | ||
| G allele | 172(27.7) | 62(17.8) | 0.409(0.567–0.786) | 0.001 |
| FUT3 rs3745635 | ||||
| GG | 202(65.0) | 134(77.0) | ||
| GA+AA | 109(35.0) | 40(23.0) | 0.362(0.553–0.845) | 0.006 |
| G allele | 502(80.7) | 303(87.1) | ||
| A allele | 120(19.3) | 45(12.9) | 0.429(0.621–0.900) | 0.011 |
| FUT3 rs3894326 | ||||
| TT | 253(81.4) | 153(87.9) | ||
| TA+AA | 58(18.6) | 21(12.1) | 0.350(0.599–1.025) | 0.060 |
| T allele | 562(90.4) | 326(93.7) | ||
| A allele | 60(9.6) | 22(6.3) | 0.381(0.632–1.050) | 0.074 |
The covariants also included sex, age of onset, smoking, severity of the disease. Nevertheless, these covariants did not affect the allelic and genotypic distributions of FUT2 and FUT3 in UC patients and the controls.
Expression of Lewis a and b antigens in the sigmoid colon
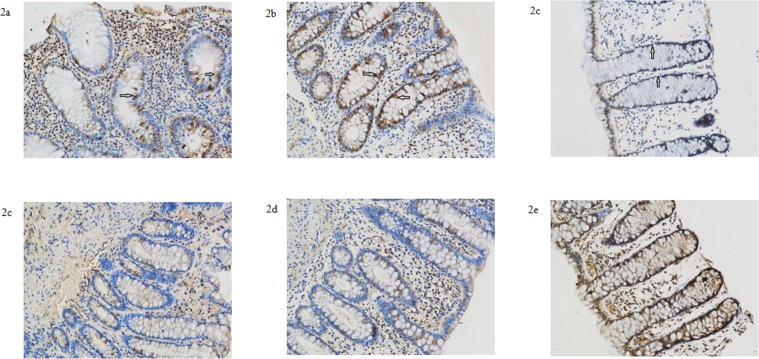
We used immunohistochemical staining techniques to investigate expression of Lewis a and b antigens in the sigmoid colon of 7 UC patients and 7 patients with benign colonic polyps. Lewis a and b were expressed in the sigmoid colon of all studied samples, regardless of the FUT2 or FUT3 genotype (Fig 2, Table 6). In the normal mucosa of patients with benign colonic polyps, Lewis a was mostly expressed in the epithelium, while expression of Lewis b was observed in the cells and extracellular matrix. Immunohistochemical staining of Lewis a indicated increased expression in the cryptic epithelium of inflammatory lesions in UC patients than the normal mucosa of patients with benign colonic polyps (P = 0.028), although there was no difference of Lewis a expression in surface epithelium between the two groups. Notably, the extent of Lewis a staining did not differ significantly between inflammatory lesions and adjacent non-inflammatory mucosa of UC patients. In addition, the expression of Lewis b did not differ significantly among the inflammatory lesions, adjacent non-inflammatory mucosa of UC patients and normal control mucosa.
Fig 2. Representative Sigmoid Colon specimens of UC patients and patients with benign colonic polyps.
Expression of Lewis a antigen (a-c) and Lewis b antigen (d-f) in the sigmiod specimens of inflammatory and adjacent non-inflammatory tissue of UC patients and normal controls. Samples a, b, d, e were derived from patient 5, described in Table 6. Samples c and f were derived from normal tissue of patients with benign colonic polyps. Immunohistochemical staining indicated increased expression of Lewis a antigen in the cryptic epithelium in inflammatory lesions from UC patients and normal mucosa from patients with benign colonic polyps (see arrows in a-c). Expression in the epithelium did not differ dramatically between UC patients and patients with benign colonic polyps. Expression of Lewis b antigen did not differ dramatically between the three groups (d–f).
Table 6. Demographic characteristics of UC patients for immunohistochemistry study.
| Case | Sex | Age | Smo-king | Lesion location | Disease severity | FUT2 Genotype | FUT3 Genotype |
|---|---|---|---|---|---|---|---|
| 1 | Male | 56 | yes | Distal | Mild | FUT2 rs1047781 ATFUT2 rs281377 CT | FUT3 rs28362459 GTFUT3 rs3894326 AT |
| 2 | Female | 45 | no | Distal | Severe | FUT2 rs1047781 TT | Wild type |
| 3 | Female | 22 | no | Extensive | Mild | Wild type | FUT3 rs28362459 GT |
| 4 | Male | 42 | no | Distal | Mild | Wild type | Wild type |
| 5 | Male | 47 | yes | Distal | Intermediate | Wild type | Wild type |
| 6 | Female | 42 | no | Distal | Intermediate | Wild type | Wild type |
| 7 | Female | 51 | no | Extensive | Intermediate | Wild type | Wild type |
One UC patient who carried homozygous mutation of FUT2 (rs1047781) expressed same levels of Lewis a and b antigens. This might indicate that FUT2 genotypes have a minimal effect on expression of Lewis a and b antigens. We also noticed that expression of Lewis a and b antigen was decreased in one UC patient carrying mutant genotypes of FUT3 (rs28362459 GT, rs3894326 AT) and FUT2 (1047781 AT, rs281377 CT).
Discussion
In this study, we investigated the frequency of mutations in FUT2 and FUT3 in Chinese patients with UC and healthy controls. In this population we found that FUT2 polymorphisms were not significantly associated with UC. However, the frequencies of mutant allele (A) and genotype (GA+AA) of FUT3 (rs3745635) were higher in UC patients than in the controls. Furthermore, stratified analyses also indicated that the mutant alleles and genotypes of FUT3 (rs28362459 and rs3745635) were more common in patients with distal colitis than those with extensive colitis. These findings suggested that mutations in FUT3 not only are associated with UC, but also might affect the location of lesions.
To our knowledge, the role of FUT3 in UC has not previously been reported. However, recent studies have linked several mutations in FUT3 to the risk of colon cancer [31], peptic ulcer, atrophic gastritis [32,33], Norovirus infection [34], type 2 diabetes [35] and coronary heart disease [36]. Moreover, the products of FUT3, Lewis a and b antigen, are involved in host-microbial interactions [6–9].
Next, we used immunohistochemistry to evaluate expression of Lewis a and b antigens in the sigmoid colon of UC patients and patients with benign colonic polyps. In our study, Lewis a and b were expressed in the sigmoid colon of all studied samples, including one UC patient (case 2) who carried homozygous mutation of FUT2 rs1047781. This might indicate that the FUT2 genotypes have a minimal effect on expression of Lewis a and b antigens.
Notably, expression of Lewis a and b antigen remained unchanged in one UC patient (case 3) with mutant FUT3 (rs28362459) genotype GT, while decreased in one UC patient (case 1) carrying mutant genotypes of FUT3 (rs28362459 GT, rs3894326 AT) and FUT2 (1047781 AT, rs281377 CT). These observations suggested that heterozygous mutation of rs3745635 reduced the function of α-(1,3/4)-fucosyltransferase more significantly than rs28362459. These observations may suggest a cumulative effect of FUT3 mutations on its expression.
In comparison to healthy control samples, expression of Lewis a was higher in the cryptic epithelium in both inflammatory lesions and non-inflammatory tissue from UC patients, although no difference was found in surface epithelium between these groups. This finding suggested the potential role of Lewis a antigen in the pathogenesis of UC. Notably, the finding that expression of Lewis a antigen was also increased in non-inflammatory tissue might indicate that Lewis a antigen expression was upregulated before inflammation occurred. However, the sample size involved in the genotype-phenotype study is too small to draw any precise conclusions. Studies involving a larger number of patients and genotypes will be required to illustrate the genotype-phenotype effect of the two genes.
Lewis antigens have previously been implicated in intestinal microbiota composition [6, 21, 34]. The intestinal microbiota of individuals with negative Lewis blood groups were recently reported to contain a less rich and diverse range of bacteria than those with Lewis a phenotype [21]. In addition, fucose released from Lewis antigens was reported to play a key role in the pathogenicity and metabolism of enterohaemorrhagic Escherichia coli (EHEC) [9].
Some studies have previously explored the association between FUT2 polymorphisms and IBD. McGovern et al. found that in a Caucasian population non-secretion, caused by a FUT2 (rs601338) polymorphism, was associated with CD, but not UC. However, in a Finnish population the wild GG genotype of FUT2 (rs601338) was associated with enhanced risk of UC [24]. And in a Chinese Han population mutations in FUT2 (rs281377 and rs601338) predisposed patients to UC, while FUT2 (rs1047781 and rs601338) polymorphisms were associated with UC in Uyghur population from northwest China [37]. In this study we found no association between FUT2 polymorphisms and UC. Although our finding may indicate the absence of association between UC and FUT2 polymorphisms in Chinese population, the conclusions are limited by the relatively small sample size analyzed in this study. Given the multifactorial nature of UC, many rare genetic variants may contribute to its development, and a wide range of rare variants, or the potential dependency of multiple variants, is not easily identified in these relatively small sample sizes.
Our results, however, suggested that mutations in FUT3 and its intestinal expression might be associated with UC susceptibility in Chinese patients. Studies involving larger cohort of UC patients will be required to validate the role of FUT3 in UC.
Supporting Information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study project was supported by grants from Natural Science Foundation of Zhejiang Province (Grant number: LY14H030012, http://www.zjnsf.gov.cn/; recipient: YJ) and Zhejiang Provincial Health Bureau (Grant number: 2014KYB157 and 2012KYA132, http://www.zjwst.gov.cn/; recipient: YJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mondot S, Kang S, Furet JF, Aguirre de Carcer D, McSweeney C, Morrison M, et al. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis. 2011; 17: 185–192. 10.1002/ibd.21436 [DOI] [PubMed] [Google Scholar]
- 2.Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009; 44: 180–186. 10.1080/00365520802433231 [DOI] [PubMed] [Google Scholar]
- 3.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010; 139: 1844–1854. 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- 4.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998; 66: 5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011; 17: 179–184. 10.1002/ibd.21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shirato H, Ogawa S, Ito H, Sato T, Kameyama A, Narimatsu H, et al. Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding. J Virol. 2008; 82: 10756–10767. 10.1128/JVI.00802-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen [Fucα(1→2)Galβ(1→4)GlcNAc], and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003; 278: 14112–14120. [DOI] [PubMed] [Google Scholar]
- 8.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, et al. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012; 485: 256–259. 10.1038/nature10996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012; 492: 113–117. 10.1038/nature11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mollicone R, Reguigne I, Kelly RJ, Fletcher A, Watt J, Chatfield S, et al. Molecular basis for Lewis alpha(1,3/1,4)-fucosyltransferase gene deficiency (FUT3) found in Lewis-negative Indonesian pedigrees. J Biol Chem. 1994; 269: 20987–20994. [PubMed] [Google Scholar]
- 11.Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000; 66: 1863–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujitani N, Liu Y, Toda S, Shirouzu K, Okamura T, Kimura H. Expression of H type 1 antigen of ABO histo-blood group in normal colon and aberrant expressions of H type 2 and H type 3/4 antigens in colon cancer. Glycoconj J. 2000; 17: 331–338. [DOI] [PubMed] [Google Scholar]
- 13.Yuan M, Itzkowitz SH, Palekar A, Shamsuddin AM, Phelps PC, Trump BF, et al. Distribution of blood group antigens A, B, H, Lewisa, and Lewisb in human normal, fetal, and malignant colonic tissue. Cancer Res. 1985; 45: 4499–4511. [PubMed] [Google Scholar]
- 14.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995; 270: 4640–4649. [DOI] [PubMed] [Google Scholar]
- 15.Yip SP, Lai SK, Wong ML. Systematic sequence analysis of the human fucosyltransferase 2 (FUT2) gene identifies novel sequence variations and alleles. Transfusion. 2007; 47: 1369–1380. [DOI] [PubMed] [Google Scholar]
- 16.Kudo T, Iwasaki H, Nishihara S, Shinya N, Ando T, Narimatsu I, et al. Molecular genetic analysis of the human Lewis histo-blood group system. II. Secretor gene inactivation by a novel single missense mutation A385T in Japanese nonsecretor individuals. J Biol Chem. 1996; 271: 9830–9837. [DOI] [PubMed] [Google Scholar]
- 17.Liu YH, Koda Y, Soejima M, Pang H, Wang BJ, Kim DS, et al. The fusion gene at the ABO-secretor locus (FUT2): absence in Chinese populations. J Hum Genet. 1999; 44: 181–184. [DOI] [PubMed] [Google Scholar]
- 18.Liu YH, Koda Y, Soejima M, Pang H, Wang B, Kimura H. Lewis (FUT3) genotypes in two different Chinese populations. J Forensic Sci. 1999; 44: 82–86. [PubMed] [Google Scholar]
- 19.Koda Y, Kimura H, Mekada E. Analysis of Lewis fucosyltransferase genes from the human gastric mucosa of Lewis-positive and -negative individuals. Blood. 1993; 82: 2915–2919. [PubMed] [Google Scholar]
- 20.Nishihara S, Narimatsu H, Iwasaki H, Yazawa S, Akamatsu S, Ando T, et al. Molecular genetic analysis of the human Lewis histo-blood group system. J Biol Chem. 1994; 269: 29271–29278. [PubMed] [Google Scholar]
- 21.Wacklin P, Tuimala J, Nikkilä J, Tims Sebastian, Mäkivuokko H, Alakulppi N, et al. Faecal Microbiota Composition in Adults Is Associated with the FUT2 Gene Determining the Secretor Status. PloS one. 2014; 9: e94863 10.1371/journal.pone.0094863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forni D, Cleynen I, Ferrante M, Cassinotti A, Cagliani R1, Ardizzone S, et al. ABO histo-blood group might modulate predisposition to Crohn's disease and affect disease behavior. Journal of Crohn's and Colitis. 2014; 8: 489–494. 10.1016/j.crohns.2013.10.014 [DOI] [PubMed] [Google Scholar]
- 23.McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010; 19: 3468–3476. 10.1093/hmg/ddq248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parmar AS, Alakulppi N, Paavola-Sakki P, Kurppa K, Halme L, Färkkilä M, et al. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens. 2012; 80: 488–493. 10.1111/tan.12016 [DOI] [PubMed] [Google Scholar]
- 25.Smyth DJ, Cooper JD, Howson JM, Clarke P, Downes K, Mistry T, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011; 60: 3081–3084. 10.2337/db11-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folseraas T, Melum E, Rausch P, Juran BD, Ellinghaus E, Shiryaev A, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012; 57: 366–375. 10.1016/j.jhep.2012.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989; 170: 2–6. [DOI] [PubMed] [Google Scholar]
- 28.Truelove SC, Witts LJ. Cortisone in ulcerative colitis; final report on a therapeutic trial. Br Med J. 1955; 2: 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun S, Chen G, Xu M, Qiao Y, Zheng S. Differentiation and migration of bone marrow mesenchymal stem cells transplanted through the spleen in rats with portal hypertension. PLoS one. 2013; 8: e83523 10.1371/journal.pone.0083523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from populationgenotype data. Am J Hum Genet. 2003; 73: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duell EJ, Bonet C, Muñoz X, Lujan-Barroso L, Weiderpass E, Boutron-Ruault MC, et al. Variation at ABO his-blood group and FUT loci and diffuse and intestinal gastric cancer risk in a European population. Int J Cancer. 2015; 136: 880–893. 10.1002/ijc.29034 [DOI] [PubMed] [Google Scholar]
- 32.Magalhaes A, Reis C A. Helicobacter pylori adhesion to gastric epithelial cells is mediated by glycan receptors. Brazilian Journal of Medical and Biological Research. 2010; 43:611–668. [DOI] [PubMed] [Google Scholar]
- 33.Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002; 297: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlsson B, Kindberg E, Buesa J, Rydell GE, Lidón MF, Montava R, et al. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII. 4 Norovirus infection. PLoS One. 2009; 4: e5593 10.1371/journal.pone.0005593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petit J M, Morvan Y, Mansuy-Collignon S, Viviani V, Vaillant G, Matejka G, et al. Hypertriglyceridaemia and Lewis (AB-) phenotype in non-insulin-dependent diabetic patients. Diabetes & metabolism. 1997; 23: 202–204. [PubMed] [Google Scholar]
- 36.Ellison R C, Zhang Y, Myers R H, Swanson JL, Higgins M, Eckfeldt J, et al. Lewis blood group phenotype as an independent risk factor for coronary heart disease (the NHLBI Family Heart Study). The American journal of cardiology. 1999; 83:345–348. [DOI] [PubMed] [Google Scholar]
- 37.Aheman A, Luo HS, Gao F. Association of fucosyltransferase 2 gene variants with ulcerative colitis in Han and Uyghur patients in China. World J Gastroenterol. 2012; 18: 4758–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.