Abstract
Pathology to vertebrate hosts has emerged repeatedly in the order Ophiostomatales. Occasional infections have been observed in Sporothrix mexicana at a low level of virulence, while the main pathogenic species cluster in a derived clade around S. schenckii s.str. In this paper, phylogeny and epidemiology of the members of this clade were investigated for 99 clinical and 36 environmental strains using four genetic loci, viz. rDNA ITS and partial CAL, TEF1, and TEF3; data are compared with amplified fragment length polymorphism (AFLP) genotyping. The four main species of the pathogenic clade were recognised. The species proved to show high degrees of endemicity, which enabled interpretation of literature data where live material or genetic information is lacking. The clade of four species comprised nine subclusters, which often had limited geographic distribution and were separate from each other in all partitions, suggesting low degrees of interbreeding between populations. In contrast, S. globosa exhibited consistent global distribution of identical AFLP types, suggesting another type of dispersal. Sporothrix brasiliensis is known to be involved in an expanding zoonosis and transmitted by cats, whereas S. globosa infections originated from putrid plant material, causing a sapronosis. Sporothrix schenckii s.str., the most variable species within the clade, also had a plant origin, with ecological similarities to that of S. globosa. A hypothesis was put forward that highly specific conditions in the plant material are required to promote the growth of Sporothrix. Fermented, self-heated plant debris may stimulate the thermodependent yeast-like invasive form of the fungus, which facilitates repeated infection of mammals.
Keywords: epidemiology, historical biogeography, phylogeny, sapronosis, Sporothrix, sporotrichosis, transmission routes, yeast conversion, zoonosis
INTRODUCTION
Sporotrichosis is a subcutaneous or cutaneous infection caused by traumatic inoculation of contaminated materials carrying inocula of Sporothrix species. Classically, the infection is known as rose ‘gardener’s disease’ (Engle et al. 2007) or ‘reed toxin’ (Song et al. 2013), as plants are often the source of the disease. The infection was first reported in 1898 in the USA by Benjamin R. Schenck (Schenck 1898). A second, similar case was described from Chicago two years thereafter (Hektoen & Perkins 1900) which led to the description of the pathogen as Sporothrix schenckii. During the century that followed (for a review, see Travassos & Lloyd 1980), the etiologic agent was supposed to be a single species that displayed a large diversity of virulence (de Lima et al. 2003), clinical features and routes of infection. Marimón et al. (2006, 2007), however, using molecular phylogenetic analyses showed that several sibling species were concerned. This was later confirmed by others applying additional gene regions (Criseo & Romeo 2010, Madrid et al. 2010, Rodrigues et al. 2014a, d). In retrospect, main groups recognized with multi-locus sequence data proved to correspond with phenotypic characters. Nowadays, the Sporothrix schenckii s.l. clade contains the clinically relevant species S. brasiliensis, S. globosa, and S. luriei in addition to S. schenckii sensu stricto (s.str.), while S. mexicana takes a remote phylogenetic position in the Ophiostoma-Sporothrix complex which is nested within the order Ophiostomatales (de Beer et al. 2003, de Beer & Wingfield 2013, Zhou et al. 2013).
Recently de Beer et al. (unpubl. data) delimited the genera Sporothrix and Ophiostoma on the basis of sequence data of four loci (LSU, ITS, CAL, BT2). Their classification matched with main ecological trends, i.e. Ophiostoma species were prevalently associated with bark beetles behind bark, whereas Sporothrix contained all major pathogens and/or occurred in plant debris or in soil. Some intermediate clades showed diverse ecologies and were phylogenetically ambiguous.
Traditionally, our understanding of the evolutionary history and phylogenetic relationships within Sporothrix has been fragmented due to the tendency of studying human pathogens and environmental species separately. Only a few studies attempted to integrate strains with dissimilar sources as well as geographic origins, moreover, sampling strategies are usually restricted to a short period of time (Dixon et al. 1991, Vismer & Hull 1997, Mesa-Arango et al. 2002, de Beer et al. 2003). Despite this segregation of information, focal outbreaks due to Sporothrix species are often connected to an environmental source, mainly involving traumatic inoculation of plant material into the cutaneous and subcutaneous tissues of subjects. Furthermore, assumptions of species distribution and ecological niche have been historically based on knowledge of morphological traits. However, common features used to recognize species, such as single-celled conidia disposed on clusters of denticles, are known to overlap among clinical and environmental Sporothrix species. As a result, cryptic entities were long-time overlooked throughout the taxonomic history of this genus. Today, multilocus sequencing provides a more reliable classification and enables in-depth studies of distribution and ecology.
When a species described with classical parameters is subdivided into a series of molecular siblings, and material for re-identification is not available, the older literature about this species becomes uninterpretable. This is a rather general consequence of drastic changes of taxonomic criteria. One of the first examples of such a new starting point was the case of Trichosporon, where the commonly used, physiologically defined species T. beigelii became obsolete after a molecular revision of the genus (Guého et al. 1992). In the case of sporotrichosis, abandoning existing literature would be highly inappropriate. Over the past century, large amounts of information have been collected on a worldwide scale. Barros et al. (2011) listed several hundreds of cases published during the last decade alone. In the current study 90 publications with case reports, case series and outbreaks, with a total of over 14 000 patients involved, are analysed. An early sporotrichosis epidemic that included over 200 cases during a 6-year period was reported from France by de Beurmann & Gougerot (1912). In the 1940s, the largest outbreak thus far took place in a gold mine in South Africa, involving over 3 000 miners developing the disease after having been infected via untreated wood contaminated by Sporothrix (Helm & Berman 1947). Other large epidemics were those related to plant materials like Sphagnum moss used in a nursery in the USA (Centers for Disease Control and Prevention 1988), rotten hay in Australia (Feeney et al. 2007), and reed and cornstalks in China (Wang & Sun 1982, Li et al. 1995, Song et al. 2013), with thousands of patients involved. The most recent outbreak of sporotrichosis was reported from south-east Brazil and was found to be caused by S. brasiliensis (Rodrigues et al. 2013b, 2014d). To understand the mechanisms behind the emergence of epidemics, outbreak data should be compared to historical information on Sporothrix infections. Re-interpretation of historical data in the light of modern molecular phylogeny is therefore compulsory.
The aim of the present study is to introduce a hypothetical system that enables to interpret and use at least part of the literature where sequence data are lacking. We collected pre-molecular papers, which contained interpretable case reports and geographical information. Available strains from each of these regions were sequenced and identified, and these data were compared to published materials. Frequencies of each of the identified species were compared with the assumption that their distributions in each region had largely remained unaltered. Additionally, clinical and environmental isolates deposited during the last century in the CBS culture collection (CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands) were re-identified with molecular techniques, which enables phylogenetic analysis of the human-pathogenic Sporothrix species in relation to other fungi that belong to the order Ophiostomatales.
MATERIAL AND METHODS
Fungal strains
A total of 205 strains were analysed, of which 109 were of clinical origin and 96 were environmental; all were maintained under the name ‘Sporothrix’ in the reference collection of the Centraalbureau voor Schimmelcultures Fungal Biodiversity Centre (CBS-KNAW), Utrecht, The Netherlands. Of these, 135 isolates comprising clinical (n = 99) and environmental (n = 36) strains belonged to the main pathogenic Sporothrix clade. Data on geographic origins and sources of isolation were collected and are listed in Table 1. All available type strains were included. Stock cultures were maintained on slants of 2 % malt extract agar (MEA) at 24 °C. Data on Ophiostoma species that are included in the current study were collected from GenBank, the accession numbers of used sequences are listed in Table 1.
Table 1.
Isolates of Sporothrix and Ophiostoma included in the study.
| Current/obsolete name | Strain | Country | Source | Genbank accession no. |
|||
|---|---|---|---|---|---|---|---|
| ITS | CAL | TEF1 | TEF3 | ||||
| S. brasiliensis | CBS 120339(T) | Brazil | Human | KP017087 | KP101421 | KP016994 | KP017184 |
| CBS 130106 | Brazil | Human | – | KP101422 | KP016985 | KP017204 | |
| CBS 130107 | Brazil | Human | – | KP101456 | KP016995 | KP017205 | |
| CBS 130109 | Brazil | Human | KP017088 | KP101455 | KP016996 | KP017138 | |
| CBS 130110 | Brazil | Human | KP017089 | KP101423 | KP016997 | KP017152 | |
| CBS 132985 | Brazil | Cat | – | KP101424 | KP016998 | KP017185 | |
| CBS 132987 | Brazil | Human | – | KP101425 | KP016999 | KP017194 | |
| CBS 132988 | Brazil | Human | – | KP101457 | KP016986 | KP017153 | |
| CBS 132989 | Brazil | Cat | – | KP101426 | KP016987 | KP017195 | |
| CBS 132990 | Brazil | Cat | – | KP101427 | KP016988 | KP017154 | |
| CBS 132991 | Brazil | Human | – | KP101428 | KP017000 | KP017155 | |
| CBS 132992 | Brazil | Human | – | KP101429 | KP017001 | KP017206 | |
| CBS 132993 | Brazil | Human | – | KP101430 | KP017002 | KP017186 | |
| CBS 132994 | Brazil | Dog | – | KP101431 | KP016989 | KP017156 | |
| CBS 132995 | Brazil | Cat | – | KP101432 | KP016990 | KP017157 | |
| CBS 132996 | Brazil | Cat | – | KP101433 | KP016991 | KP017158 | |
| CBS 132997 | Brazil | Cat | – | KP101449 | KP017003 | KP017139 | |
| CBS 132998 | Brazil | Cat | KP017090 | KP101434 | KP016992 | KP017159 | |
| CBS 132999 | Brazil | Cat | – | KP101453 | KP017004 | KP017187 | |
| CBS 133000 | Brazil | Cat | – | KP101450 | KP017005 | KP017140 | |
| CBS 133010 | Brazil | Cat | – | KP101440 | KP017013 | KP017145 | |
| CBS 133015 | Brazil | Cat | – | KP101445 | KP017018 | KP017149 | |
| CBS 133017 | Brazil | Cat | – | KP101458 | KP017020 | KP017150 | |
| CBS 133001 | Brazil | Cat | – | KP101451 | KP017005 | KP017141 | |
| CBS 133002 | Brazil | Cat | – | KP101452 | KP017005 | KP017142 | |
| CBS 133003 | Brazil | Cat | – | KP101454 | KP016982 | KP017188 | |
| CBS 133004 | Brazil | Dog | – | KP101435 | KP017008 | KP017189 | |
| CBS 133006 | Brazil | Cat | – | KP101436 | KP017009 | KP017143 | |
| CBS 133007 | Brazil | Cat | – | KP101437 | KP017010 | KP017190 | |
| CBS 133008 | Brazil | Cat | – | KP101438 | KP017011 | KP017144 | |
| CBS 133009 | Brazil | Cat | – | KP101439 | KP017012 | KP017191 | |
| CBS 133011 | Brazil | Cat | – | KP101441 | KP017014 | KP017146 | |
| CBS 133012 | Brazil | Cat | – | KP101442 | KP017015 | KP017147 | |
| CBS 133013 | Brazil | Cat | – | KP101443 | KP017016 | KP017192 | |
| CBS 133014 | Brazil | Cat | – | KP101444 | KP017017 | KP017148 | |
| CBS 133016 | Brazil | Cat | – | KP101446 | KP017019 | KP017193 | |
| CBS 133019 | Brazil | Cat | – | KP101447 | KP016993 | KP017196 | |
| CBS 133021 | Brazil | Cat | – | KP101448 | KP017021 | KP017151 | |
| S. globosa | CBS 120340(T) | Spain | Human | KP017084 | KP101459 | KP016949 | KP017165 |
| CBS 129717 | China | Human | – | KP101460 | KP016962 | KP017166 | |
| CBS 129718 | China | Human | – | KP101461 | KP016950 | KP017167 | |
| CBS 129719 | China | Human | KP017085 | KP101462 | KP016951 | KP017168 | |
| CBS 129720 | China | Human | – | KP101463 | KP016952 | KP017169 | |
| CBS 129721 | China | Human | – | KP101478 | KP016953 | KP017170 | |
| CBS 129722 | China | Human | – | KP101464 | KP016964 | KP017183 | |
| CBS 129723 | China | Human | – | KP101465 | KP016963 | KP017171 | |
| CBS 129724 | China | Human | – | KP101466 | KP016954 | KP017172 | |
| CBS 129725 | China | Human | – | KP101467 | KP016955 | KP017173 | |
| CBS 130104 | Spain | Human | – | KP101468 | KP016965 | KP017174 | |
| CBS 130105 | Spain | Human | – | KP101469 | KP016956 | KP017175 | |
| CBS 130115 | Spain | Human | – | KP101470 | KP016966 | KP017176 | |
| CBS 130116 | Spain | Human | – | KP101471 | KP016957 | KP017177 | |
| CBS 130117 | Japan | Human | – | KP101472 | KP016967 | KP017178 | |
| CBS 132923 | Brazil | Human | – | KP101473 | KP016958 | KP017179 | |
| CBS 132924 | Brazil | Human | KP017083 | KP101474 | KP016968 | KP017180 | |
| CBS 132925 | Brazil | Human | – | KP101475 | KP016959 | KP017203 | |
| CBS 292.55 | UK | Human | KP017086 | KP101476 | KP016960 | KP017181 | |
| CBS 340.35 | Japan | Human | – | KP101477 | KP016961 | KP017182 | |
| S. luriei | CBS 937.72(T) | South Africa | Human | AB128012 | AM747302 | KP016948 | KP017207 |
| S. mexicana | CBS 120341(T) | Mexico | Soil, rose tree | KP017072 | AM398393 | KP016931 | KP017230 |
| CBS 120342 | Mexico | Carnation | KP017073 | AM398392 | KP016932 | KP017231 | |
| CBS 132927 | Brazil | Human | – | JF811340 | KP016933 | KP017232 | |
| CBS 132928 | Brazil | Human | – | JF811341 | KP016934 | KP017234 | |
| CBS 133192 | Italy | Dog | KP017075 | JX080721 | KP016935 | KP017233 | |
| S. schenckii | CBS 125601 | Colombia | Human | – | KP101395 | KP016973 | KP017120 |
| S. schenckii | CBS 117440 | South Africa | Human | KP017098 | KP101386 | KP017026 | KP017114 |
| CBS 117842 | South Africa | Human | – | KP101387 | KP017027 | KP017115 | |
| S. sp. / S. schenckii | CBS 115870 | South Africa | NK | – | – | KP016930 | KP017229 |
| CBS 130101 | Peru | Human | KP017095 | KP101390 | KP017022 | KP017197 | |
| CBS 130103 | Argentina | Human | – | KP101388 | KP017025 | KP017117 | |
| CBS 130111 | Colombia | Human | – | KP101401 | KP016974 | KP017123 | |
| CBS 130112 | Peru | Human | KP017096 | KP101391 | KP017023 | KP017132 | |
| CBS 130114 | Peru | Human | – | KP101392 | KP017024 | KP017133 | |
| CBS 130097 | Bolivia | Human | – | KP101396 | KP016969 | KP017137 | |
| CBS 130098 | Peru | Human | KP017091 | KP101397 | KP016979 | KP017121 | |
| CBS 130099 | Peru | Human | KP017092 | KP101398 | KP016980 | KP017122 | |
| CBS 132926 | Brazil | Human | – | KP101404 | KP016981 | KP017202 | |
| CBS 132961 | Brazil | Cat | – | KP101402 | KP016970 | KP017124 | |
| CBS 132962 | Brazil | Human | – | KP101403 | KP016975 | KP017125 | |
| CBS 132964 | Brazil | Human | – | KP101407 | KP017031 | KP017118 | |
| CBS 132966 | Brazil | Human | – | KP101400 | KP017032 | KP017126 | |
| CBS 132969 | Brazil | Human | – | KP101409 | KP017029 | KP017198 | |
| CBS 132970 | Brazil | Human | – | KP101405 | KP016976 | KP017127 | |
| CBS 132971 | Brazil | Human | – | KP101411 | KP016977 | KP017160 | |
| CBS 132972 | Brazil | Human | – | KP101412 | KP016978 | KP017199 | |
| CBS 132973 | Brazil | Human | – | KP101408 | KP017033 | KP017200 | |
| CBS 132974 | Brazil | Human | – | KP101410 | KP016971 | KP017128 | |
| CBS 132975 | Brazil | Human | – | KP101406 | KP017034 | KP017119 | |
| CBS 132976 | Japan | Human | – | KP101415 | KP017035 | KP017201 | |
| CBS 132977 | Mexico | Plant debris | – | KP101399 | KP017030 | KP017134 | |
| CBS 132981 | Brazil | Human | – | KP101414 | KP017036 | KP017129 | |
| CBS 132982 | Brazil | Human | – | KP101416 | KP016972 | KP017130 | |
| CBS 211.61 | South Africa | NK | KP017093 | KP101393 | KP016983 | KP017135 | |
| CBS 345.53 | The Netherlands | Human | – | KP101417 | KP017038 | KP017161 | |
| CBS 356.29 | Germany | NK | – | KP101413 | KP017037 | KP017131 | |
| CBS 359.36(T) | USA | NK | KP017100 | KP101420 | KP017041 | KP017163 | |
| CBS 444.67 | Mozambique | Human | KP017099 | KP101389 | KP017028 | KP017116 | |
| CBS 472.48 | NK | NK | KP017097 | KP101418 | KP017039 | KP017162 | |
| S. schenckii / S. sp. | CBS 498.86 | NK | NK | – | KP101419 | KP017040 | KP017164 |
| S. schenckii | CBS 938.72 | France | Human | KP017094 | KP101394 | KP016984 | KP017136 |
| S. abietinum | CBS 125.89 | Mexico | Abies vejarii | AF484453 | – | – | – |
| S. africanum | CBS 116566 | South Africa | Protea caffra | DQ316200 | – | – | – |
| S. aurorae | CBS 118837(T) | South Africa | Pinus elliottii | DQ396796 | – | – | – |
| S. brunneoviolacea | CBS 793.73 | Germany | Meadow soil | KP017069 | KP017106 | KP017061 | KP017112 |
| S. brunneoviolacea / S. inflata | CBS 101570 | USA | Endophyte in Vitis vinifera | KP017068 | KP017101 | KP017057 | KP017108 |
| S. brunneoviolacea | CBS 110895 | Austria | Root of Quercus petraea– | KP017104 | KP017062 | KP017109 | |
| CBS 110896 | Austria | Root of Quercus robur– | KP017102 | KP017058 | KP017110 | ||
| CBS 124561(T) | Spain | Soil | FN546959 | KP017103 | KP017059 | KP017113 | |
| CBS 124562 | Spain | Soil | FN546957 | – | – | – | |
| CBS 124564 | Spain | Soil | FN546958 | KP017105 | KP017060 | KP017111 | |
| S. curviconia | CBS 959.73(T) | NK | NK | – | – | KP017047 | KP017241 |
| S. sp. / S. curviconia | CBS 145.94 | NK | NK | KP017071 | – | – | – |
| S. sp. / S. curviconia | CBS 541.84 | Chile | Pinus radiata log | KC113234 | KP101483 | KP017046 | KP017239 |
| S. dentifundum | CBS 115790(T) | Hungary | Quercus wood | AY495434 | – | – | – |
| S. dimorphospora / S. inflata | CBS 553.74(T) | Canada | Soil | KP017082 | – | KP017052 | KP017209 |
| CBS 840.73 | Chile | Wood | – | – | KP017050 | KP017210 | |
| S. dimorphospora | CBS 125439 | USA | Soil | KP017080 | – | KP017048 | KP017208 |
| CBS 125440 | Spain | Soil | KP017081 | – | – | – | |
| CBS 125442 | Spain | Soil | FN546961 | – | KP017051 | KP017212 | |
| S. foliorum | CBS 326.37 | The Netherlands | Industrial strain | KP017067 | – | KP016929 | KP017240 |
| S. fusiforme | CBS 112912 | Azerbaijan | Populus nigra | AY280481 | – | – | – |
| S. gemellus | CBS 121959(T) | South Africa | Tarsonemus sp. fromProtea caffra | DQ821560 | – | – | – |
| S. inflata | CBS 239.68(T) | Germany | Soil, wheat field | AY495426 | – | KP017054 | KP017213 |
| CBS 794.73 | Sweden | Humus in Picea forest | KP017079 | – | KP017053 | KP017216 | |
| CBS 841.73 | Chile | Soil | AY495431 | – | KP017055 | KP017214 | |
| S. sp. / S. inflata | CBS 156.72 | The Netherlands | Greenhouse soil | – | – | KP017056 | KP017215 |
| S. lignivora | CBS 119147 | South Africa | Eucalyptus wood pole | KP017064 | KP017107 | KP017063 | KP017242 |
| CBS 119148(T) | South Africa | Eucalyptus wood | KP017065 | – | – | – | |
| CBS 119149 | South Africa | Eucalyptus wood pole | KP017066 | – | – | – | |
| S. lunatum | CBS 112927(T) | Austria | Carpinus betulus | AY280485 | – | – | – |
| S. pallida | CBS 111110 | Germany | Insect, Zootermopsisnevadensis | – | AM398382 | KP016937 | KP017217 |
| S. pallida / S. stylites | CBS 115868 | South Africa | Eucalyptus camaldulensis | EF127881 | – | KP016941 | KP017221 |
| CBS 115869 | South Africa | Wood utility pole | EF127884 | – | KP016942 | KP017222 | |
| CBS 115872 | South Africa | Wood pole | EF127882 | – | KP016938 | KP017220 | |
| S. pallida / S. humicola | CBS 118129(T) | South Africa | Soil | KP017076 | – | KP016939 | KP017223 |
| S. pallida / S. stylites | CBS 118848(T) | South Africa | Wood pole | KP017077 | – | KP016943 | KP017224 |
| S. pallida / S. nivea | CBS 131.56(T) | Japan | Stemonitis fusca | EF127880 | – | KP016944 | KP017227 |
| S. pallida | CBS 182.63 | The Netherlands | Soil | KC113233 | – | KP016940 | KP017228 |
| S. pallida / S. schenckii | CBS 201.53 | South Africa | Decaying grass | – | – | KP016945 | KP017225 |
| S. pallida / S. albicans | CBS 302.73(T) | UK | Soil | KP017078 | AM398396 | KP016946 | KP017226 |
| S. pallida / S. sp. | CBS 622.95 | NK | NK | – | – | KP016947 | KP017218 |
| S. pallida / S. sp. | CBS 623.95 | NK | NK | – | – | KP016936 | KP017219 |
| S. palmiculminatum | CBS 119590(T) | South Africa | Protea repens | DQ316191 | – | – | – |
| S. phasma | CBS 119721(T) | South Africa | Protea laurifolia | DQ316219 | – | – | – |
| S. protearum | CBS 116567 | South Africa | Protea caffra | DQ316203 | – | – | – |
| S. splendens | CBS 116569 | South Africa | Protea repens | DQ316215 | – | – | – |
| S. stenoceras | CBS 237.32(T) | Norway | Pine pulp | AF484462 | – | – | – |
| S. variecibatus | CBS 121960 | South Africa | Protea longifolia | DQ821569 | KP101479 | KP017042 | KP017235 |
| CBS 121961(T) | South Africa | Oodinychus sp. mite fromProtea repens | KP017070 | KP101481 | KP017043 | KP017236 | |
| CBS 121962 | South Africa | Eucalyptus sp. | DQ821567 | KP101482 | KP017044 | KP017237 | |
| CBS 123288 | NK | NK | – | KP101480 | KP017045 | KP017238 | |
| Ceratocystiopsis minuta | RJ705 | Poland | Picea abies | EU913697 | – | – | – |
| O. ainoae | CMW 1903 | Norway | Picea abies | HM031495 | – | – | – |
| O. angusticollis | CBS 186.86 | USA | Pinus banksiana | AY924383 | – | – | – |
| O. arduennense | MUCL 44866 | Belgium | Fagus sylvatica | AY573241 | – | – | – |
| O. bacillisporum | MUCL 45378 | Belgium | Fagus sylvatica | AY573258 | – | – | – |
| O. bicolor | CBS 492.77 | USA | Gallery of Ips sp. inPicea sp. | DQ268604 | – | – | – |
| O. bragantinum | CBS 430.92 | Brazil | Soil | FN546964 | – | – | – |
| CBS 474.91(T) | Brazil | Soil | FN546965 | – | – | – | |
| O. breviusculum | YCC-522, JCM 12501 | Japan | Single ascospore isolate from YCC-494 | AB200423 | – | – | – |
| O. canum | CBS 133.51 | Sweden | Pinus sylvestris | HM031489 | – | – | – |
| O. catonianum | C1084 | Italy | Pyrus | AF198243 | – | – | – |
| O. conicola | CBS 127.89 | Mexico | Cone with Conophthorus cembroides | AY924384 | – | – | – |
| O. coronatum | CBS 497.77 | NK | NK | AY924385 | – | – | – |
| O. denticiliatum | CMW 29493 | Norway | Scolytus ratzeburgi on Betula sp. | FJ804490 | – | – | – |
| O. fasciatum | UM 56 | Canada | Pseudotsuga menziesii | EU913720 | – | – | – |
| O. flexuosum | CBS 208.83 | Norway | Picea abies | AY924387 | – | – | – |
| O. floccosum | CBS 799.73 | Sweden | Soil | AF198231 | – | – | – |
| O. fumeum | CMW 26813 | South Africa | Eucalyptus cloeziana | HM051412 | – | – | – |
| O. fuscum | CMW 23196 | Finland | Pityogenes chalcographus on Picea abies | HM031504 | – | – | – |
| O. ips | CMW 7075 | NK | NK | AY546704 | – | – | – |
| O. japonicum | YCC-099 | NK | NK | GU134169 | – | – | – |
| O. karelicum | CMW 23099 | Russia | Scolytus ratzeburgi on Betula pendula | EU443762 | – | – | – |
| O. kryptum | DAOM229701 | NK | NK | AY304436 | – | – | – |
| O. minus | AU58.4 | Canada | Lodgepole pine lumber | AF234834 | – | – | – |
| CCMA12 | NK | NK | AY934511 | – | – | – | |
| O. montium | CMW13221 | NK | NK | AY546711 | – | – | – |
| O. multiannulatum | MUCL19062 | NK | NK | AY934512 | – | – | – |
| O. nigrocarpum | ATCC22391 | USA | Dendroctonus sp. | AF484474 | – | – | – |
| CBS 637.66(T) | USA | Abies sp. | AY280489 | – | – | – | |
| O. nikkoense | YCC-430 | Japan | NK | AB506674 | – | – | – |
| O. novoulmi | C510 | USA | Ulmus sp. | AF198236 | – | – | – |
| O. piceae | CBS108.21(T) | Germany | NK | AF198226 | – | – | – |
| O. piliferum | CBS 129.32 | The Netherlands | Scots pine | AF221070 | – | – | – |
| O. pluriannulatum | MUCL18372 | NK | NK | AY934517 | – | – | – |
| O. quercus | CMW2467 | France | Quercus sp. | AY466626 | – | – | – |
| O. rostrocoronatum | CBS 434.77(T) | USA | Woodpulp | AY194509 | – | – | – |
| O. saponiodorum | CMW29497 | Finland | Ips typographus onPicea abies | HM031507 | – | – | – |
| O. sejunctum | Ophi 1A | NK | NK | AY934519 | – | – | – |
| O. setosum | AU160-38 | NK | NK | AF128929 | – | – | – |
| O. subannulatum | CBS 188.86 | USA | Pinus | AY934522 | – | – | – |
| O. tapionis | CMW23266 | Finland | Hylastes brunneus onPinus sylvestris | HM031493 | – | – | – |
| O. tenellum | CBS 189.86 | USA | Pinus banksiana | AY934523 | – | – | – |
| O. tetropii | CBS 428.94 | Austria | Breeding system ofTetropium sp., Picea abies | AY934524 | – | – | – |
| O. triangulosporum | DSMZ4934 | NK | NK | AY934525 | – | – | – |
| Pesotum australiae | CMW6606 | Australia | Acacia mearnsii | EF408603 | – | – | – |
| Pesotum cupulatum | C1194 | USA | Pseudotsuga | AF198230 | – | – | – |
O = Ophiostoma, S = Sporothrix, NK = Not known, T = type culture.
DNA extraction
DNA was extracted following the Quick CTAB protocol. 1–10 mm3 fungal material was transferred to 2 mL screw-capped tubes filled with 490 μL CTAB-buffer 2× and 6–10 acid-washed glass beads. Ten μL proteinase K [10 mg/mL] (Sigma-Aldrich, St Louis, MO, USA) were added and mixed thoroughly for 10 min using a MoBio vortex (MoBio, Carlsbad, CA, USA). After that, 500 μL chloroform : isoamylalcohol (24 : 1) was added and shaken for 2 min followed by incubation for 60 min at 60 °C. Tubes were centrifuged for 10 min at 14 000 × g. The supernatant was collected in a new tube. To ∼400 μL DNA sample ∼270 μL of ice-cold iso-propanol (Sigma) was added and centrifuged again at 14 000 × g for 10 min and the upper layer was dissolved in 1 mL ice-cold ethanol 70 %. Tubes were centrifuged again at 14 000 × g for 2 min, air-dried and re-suspended in 50 μL TE-buffer (pH 8.0). Quality of genomic DNA was verified by running 2 μL DNA sample in a 0.8 % agarose gel. DNA was quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher, Wilmington, DE, USA). Samples were stored at −20 °C until further use.
DNA amplification and sequencing
Four gene regions were amplified for inclusion in the multi-locus sequence data analysis, i.e. rDNA internal transcribed spacer (ITS), and the partial genes calmodulin (CAL), translation elongation factor-1 (TEF1) and -3 (TEF3). Primers used for amplification and sequencing of CAL were CL1 and CL2a (O’Donnell et al. 2000). TEF primers were developed by B. Stielow (unpubl. data). PCR reactions were performed in a mixture containing 1.25 μL 10× PCR buffer, 6.7 μL ddH2O, 1 μL dNTP mix (2.5 mM), 0.25 μL of each primer (10 pmol), 0.06 μL Taq DNA polymerase (5 U/μL), 0.625 μL DMSO (Sigma), and 2.5 μL template DNA (100 ng/μL). PCR reactions were performed in a Hybaid Touchdown PCR machine (Hybaid, Middlesex, UK); the used annealing temperatures are listed in Table 2. PCR products were visualized by electrophoresis on a 1 % (w/v) agarose gel. Amplicons were purified using exoSAP-IT (Affymetrix, Santa Clara, CA, USA). The ABI Prism BigDye Terminator v. 3.1 (Applied Biosystems, Foster City, CA, USA) was applied according to the instructions provided by the manufacturer. Reactions were purified by using Sephadex G-50 ultrafine (GE Healthcare Bio-Sciences, Uppsala, Sweden) and sequencing was performed by using an ABI 3730xL automatic sequencer (Applied Biosystems).
Table 2.
Success rates of sequencing for each gene.
| Contigs (%) | S. brasiliensis (n = 44) | S. schenckii (n = 43) | S. globosa (n = 22) | Environmental species (n = 96) |
|---|---|---|---|---|
| ITS (%) | 81.8 | 100 | 100 | 70.8 |
| CAL (%) | 95.5 | 100 | 90.9 | 62.5 |
| TEF1 (%) | 95.5 | 79.1 | 100 | 96.9 |
| TEF3 (%) | 97.7 | 97.7 | 100 | 100 |
Phylogenetic analyses
Consensus sequences were assembled by using SeqMan package of Lasergene software v. 8.1 (DNAStar, Madison, WI, USA) and alignments were made in BioEdit v. 7.0.5.2 software (Hall 1999). The genetically diverse ITS sequences were aligned by using MUSCLE program (www.ebi.ac.uk/Tools/msa/muscle), while sequences of the CAL, TEF1 and TEF3 sequences were aligned by using the server version of the MAFFT program v. 7.0 (www.ebi.ac.uk/Tools/msa/mafft/) (Katoh & Standley 2013). Retrieved alignments were manually checked to avoid mis-paired bases. All sequences determined in this study were deposited in GenBank and the accession numbers are listed in Table 1.
The best-fit evolutionary model was determined by application of ModelTest v. 0.1.1. Bayesian analysis was performed with MrBayes v. 3.1.2 (Ronquist et al. 2012). Four MCMC chains were run simultaneously for 1 × 107 generations. Bootstrapped Maximum Likelihood analysis was performed by using RAxML-VI-HPC v. 7.0.3 (Stamatakis et al. 2008) as implemented on the Cipres portal (www.phylo.org/) with non-parametric bootstrapping using 1 000 replicates.
Amplified fragment length polymorphism genotyping
The Sporothrix isolates were subjected to amplified fragment length polymorphism (AFLP) genotyping using a previously described procedure (Chowdhary et al. 2013). However, for the amplification of the DNA fragments the selective cytosine residue of the EcoRI primer was replaced by an adenine residue (5’-Flu-GACTGCGTACCAATTCAA-3’), while the MseI primer remained the same with one selective residue (5’-GATGAGTCCTGACTAAG-3’). After amplification, amplicons were 50× diluted using ddH2O; 1 μL of the diluted amplicon was then added to a mixture of 8.9 μL ddH2O and 0.1 μL LIZ600 (Applied Biosystems) followed by a heating step for 1 min at 96 °C followed by cooling down to 4 °C. Fragment analysis was carried out using an ABI3500xL Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions. Raw data were then inspected visually after importation into BioNumerics v. 6.6 (Applied Maths, St. Martens-Latem, Belgium) and analysed by UPGMA clustering using the Pearson correlation coefficient.
Meta-analysis
We analysed the existing medical and veterinary literature on human and veterinary cases of sporotrichosis from the first publication 1898 till present. A search was initiated using the PubMed database for which the MeSH terms ‘Sporothrix’ and ‘sporotrichosis’, yielded in total 705 results. Reports on treatment, immunology, antifungals and virulence factors, as well as book chapters and reports that also include other diseases were neglected. The focus was then placed on cases and case series from 1940 up to now; case reports with insufficient data were discarded. Over 14 000 cases published in 90 reports were collected; the selection covered countries all over the world. The search also included ∼2 827 cases published in Chinese language. Numbers are approximate because some cases had been used in repeated publications; we tried to exclude duplicates when individual cases were numbered. Cases were listed geographically on the basis of identifiable entities, such as Europe, China, or Brazil. The statistical method for Table 5 is the χ2 test.
RESULTS
Judging from literature data, the most endemic regions are China (3 299 cases), South Africa (3 154 cases), and Brazil (5 814 cases). Less frequently, sporotrichosis occurs in Japan, Australia, India, and the remaining Americas outside the eastern part of South America. The disease is less prevalent in Europe, except for the unique outbreak involving 200 cases occurring in France over a period of six years at the beginning of last century (Beurmann & Gougerot 1912).
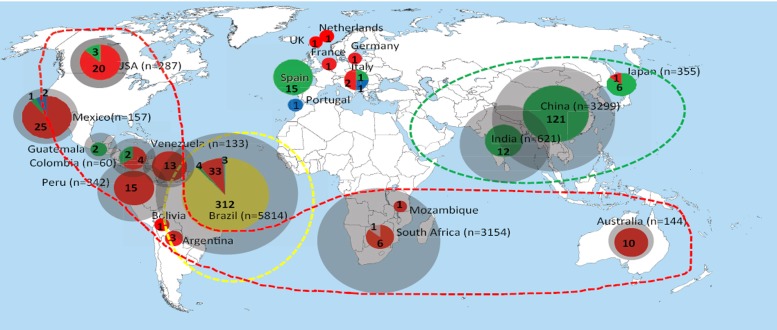
Outside the recent Brazilian epidemic, nearly all cases and case series were published to be caused by Sporothrix schenckii s.l.; the subdivision of this taxon into four molecular siblings occurred only in 2006 (Marimón et al. 2006). We aimed to recognise the individual siblings retrospectively by comparing contemporary distributions of molecular species with historical biogeography abstracted from published data. A comparison of the number of published cases (Fig. 1; grey circles) and the number of sequenced strains in the same area (Fig. 1; coloured) is given. Distributions of molecular species as percentages of the total numbers of sequenced cases in the same defined area are given in Table 3 and Fig. 1. In most of the defined areas a single molecular species is preponderant (> 80 %). Main calculated endemic areas with their prevalent species are as follows: Asia S. globosa (99.3 %), Australia and southern Africa S. schenckii (94 %), south-eastern South America S. brasiliensis (88 %), western part of South America and Central and North America S. schenckii (89 %). The percentages indicate statistical probabilities that the prevalent endemic species was concerned in historical publications without sequence data. In European countries the low number of cases hinders to ascertain predominant species.
Fig. 1.

Geographic distribution of sporotrichosis caused by S. brasiliensis, S. schenckii, and S. globosa according to case reports published over 70 years, compared with sequenced isolates and with expression of statistical probabilities that the prevalent endemic species was concerned in historical publications without sequence data. Samples were categorised as sequenced and non-sequenced specimens. The sizes of circumferences are roughly proportional to the numbers of cases / strains included. Numbers reported within the pies denote the number of strains examined. Main endemic areas indicated by dotted lines.
Table 3.
Estimated distributions of molecular species on the basis of percentages of sequenced strains compared to the total number of published cases in the respective area.
| Continent / region | Country | Reported cases | Sequenced isolates | S. brasiliensis % | S. globosa % | S. schenckii % | S. mexicana % | S. luriei |
|---|---|---|---|---|---|---|---|---|
| Asia | China | 3299 | 121 | 121: 100 % | ||||
| India | 621 | 12 | 12: 100 % | |||||
| Japan | 355 | 7 | 6: 86 % | 1: 14 % | ||||
| Australia | Australia | 144 | 10 | 10: 100 % | ||||
| Africa | South Africa | 3154 | 7 | 6: 86 % | 1 | |||
| Mozambique | 1 | 1: 100 % | ||||||
| Western and southern parts of South America, Central and North America | Peru | 342 | 15 | 15: 100 % | ||||
| Argentina | 3 | 3: 100% | ||||||
| Bolivia | 1 | 1: 100 % | ||||||
| Mexico | 157 | 28 | 1: 4 % | 25: 89 % | 2: 7 % | |||
| Venezuela | 133 | 13 | 13: 100 % | |||||
| Guatemala | 55 | 2 | 2: 100 % | |||||
| Columbia | 60 | 6 | 2: 33 % | 4: 67 % | ||||
| USA | 287 | 23 | 3: 13 % | 20: 87 % | ||||
| Eastern South America | Brazil | 5814 | 352 | 312: 88.4 % | 4: 1.1 % | 33: 9.3 % | 3: 0.8 % | |
| Europe | 6 | 24 | 17: 71 % | 5: 21 % | 2: 8 % | |||
| Total | 625 | 312 | 168 | 137 | 7 | 1 |
In order to have a complete overview of molecular species occurring in humans and animals and their potential routes of transmission, we sequenced all strains deposited in the CBS collection over the last hundred years under the name ‘Sporothrix’. Using standard primers, sequencing efficiency proved to differ slightly between species. Success rates of sequencing for each gene are listed in Table 2. In clinical strains, the largest percentages of poor sequences were encountered in TEF1 and ITS in S. schenckii and S. brasiliensis, viz. 20.9 % and 18.2 %, respectively. Strains of environmental species outside the S. schenckii clade generally generated good results with TEF1 and TEF3, but a somewhat higher percentages of failure were obtained with ITS and CAL (Table 2).
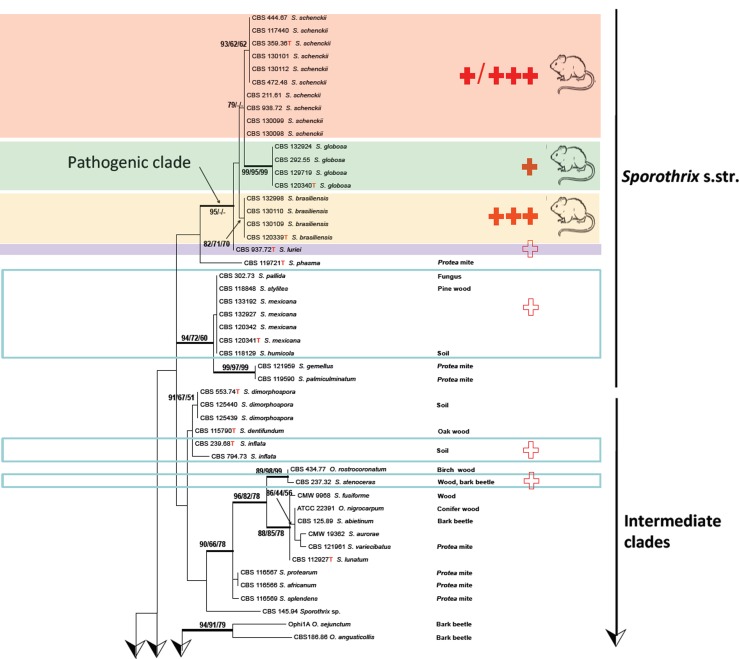
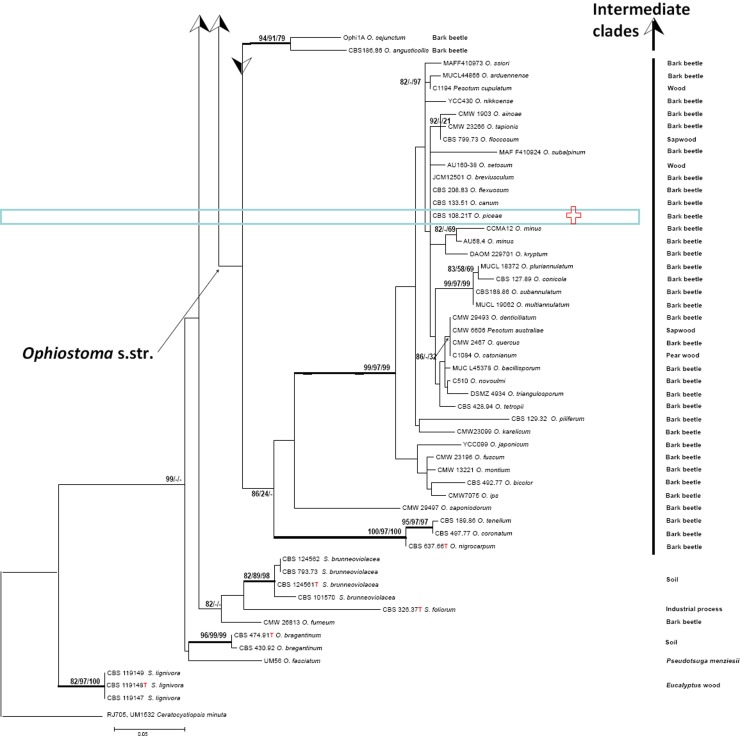
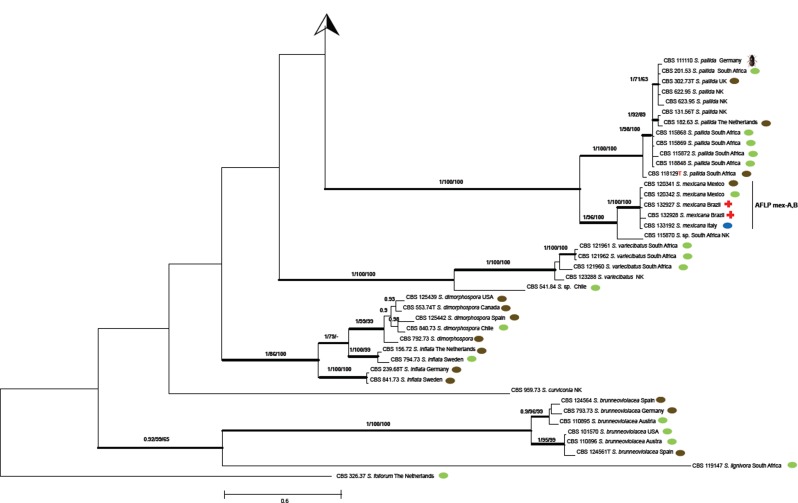
ITS sequences could be aligned confidently over the entire order Ophiostomatales; a general tree is presented in Fig. 2, using Ceratocystiopsis minuta RJ705 as outgroup. The complete alignment included 101 sequences for ITS, 37 generated in this study and 64 retrieved from GenBank. ITS sequences produced an 807 bp-long alignment (327 for ITS1, 191 for ITS2), and included 334 invariable characters, 246 variable parsimony-informative sites (34 %), and 72 singletons. Several highly confident clades could be recognized, one of which (Fig. 2) consisted of Ophiostoma species associated with bark beetles (bootstrap support 86 %). Outside this clade several more bark beetle-associated species were noted, at significant distance and separated by Ophiostoma species with other habitats. Several intermediary lineages were found in soil and in Protea infructescences, intermingled with occasional wood-inhabiting taxa (Fig. 2). The ultimate clade contained four potentially human-pathogenic Sporothrix species. Sporothrix stenoceras represented a separate clade, with 99 % statistical support and distinct from the clades containing pathogenic species (Marimón Clades I–III and VI) and a saprophytic clade (Marimón Clade IV) (Marimón et al. 2006).
Fig. 2.
Phylogenetic relationships inferred from PhyML based on ITS sequences of 101 strains belonging to Sporothrix and Ophiostoma. The numbers close to the branches represent indices of support (ML/NJ/MP) based on 1 000 bootstrap replications. Branches with bootstrap support value higher than 80 % are indicated in bold.
A multilocus tree (Fig. 3) excluding the major part of Ophiostoma was based on 135 selected isolates including 99 clinical strains and 36 representative environmental strains of species having all three genes for multilocus studies. Lengths of generated amplicons were 792 bp, 526 bp, and 255 bp for CAL, TEF1, and TEF3, respectively. Of the 1 573 nucleotides sequenced, 942 (59.9 %) were constant, 464 (29.5 %) were parsimony-informative, and 146 (9.3 %) were variably parsimony non-informative sites. Over the entire dataset, the lowest number of variable sites was 92 (17.5 %) in the TEF1 fragment, and the highest was 432 (54.5 %) in the CAL fragment; extended data per gene are provided in Table 4.
Fig. 3.
Phylogenetic relationship inferred from Bayesian statistics based on concatenated CAL, TEF1 and TEF3 sequences of 135 strains of Sporothrix species. Bootstrap and posterior probabilities values were added to respective branches (BI/ML/NJ). Branches with bootstrap support values higher than 80 % are indicated in bold.
Table 4.
Variability of loci used in 136 Sporothrix isolates examined.
| Locus | No. of bp sequenced | % variable sites | % parsimony-informative sites | % singleton sites |
|---|---|---|---|---|
| CAL | 792 | 54.5 | 42.8 | 11.7 |
| TEF1 | 526 | 17.5 | 13.5 | 4.0 |
| TEF3 | 255 | 33.7 | 21.2 | 12.5 |
| Combined genes | 1573 | 38.8 | 29.5 | 9.28 |
Phylogenetic trees were analysed using independent and combined datasets. All 135 sequences generated in this study, except for one CAL which was retrieved from GenBank. Gene trees of CAL, TEF1, and TEF3 presented a higher discriminatory power and similar topologies to the ITS tree. Sequences could be aligned confidently over the entire dataset. The combined tree based on CAL, TEF1, and TEF3 data of 135 strains is given in Fig. 3, basically comprising the upper part (Sporothrix s.str.) of the ITS tree (Fig. 2) and using Sporothrix foliorum, CBS 326.37 as outgroup. The best-fit model of evolution was estimated to be HKY+GAMMA. Fig. 3 shows the majority-rule consensus trees, deduced by Bayesian inferences sampled by MCMC and were selected to demonstrate the tree topology.
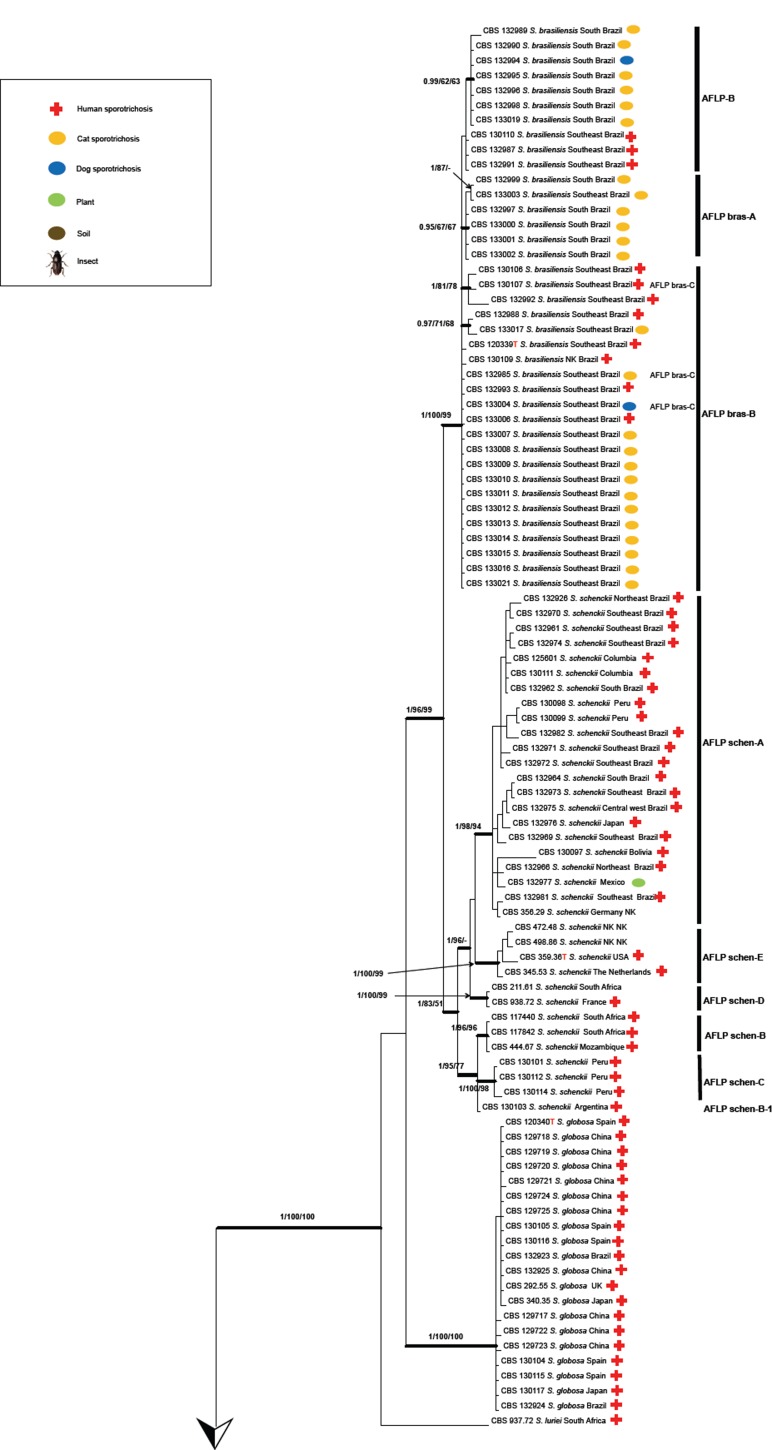
The combined tree of 135 isolates was subdivided into several main groups (Fig. 3), five of which were also observed in previous studies (Marimón et al. 2006, 2007, Madrid et al. 2009, Rodrigues et al. 2013b, 2014d). Each described species could be distinguished at high bootstrap values. Five subclades were discernible within S. schenckii, which almost entirely matched with AFLP groups A–E (below; Fig. 4) and were mostly geographically restricted (Fig. 5). CBS 130103 was the single strain from Argentina clustering in AFLP group B from South Africa with small deviations in AFLP profile (Fig. 4) as well as in sequence data (Fig. 3). In S. brasiliensis several small sets of strains had bootstrap support, but groups were too similar to allow meaningful distinction. Sporothrix globosa was homogeneous. Sporothrix mexicana was found to be nested in an environmental clade comprising saprobic species (Fig. 2; Marimón Clade IV).
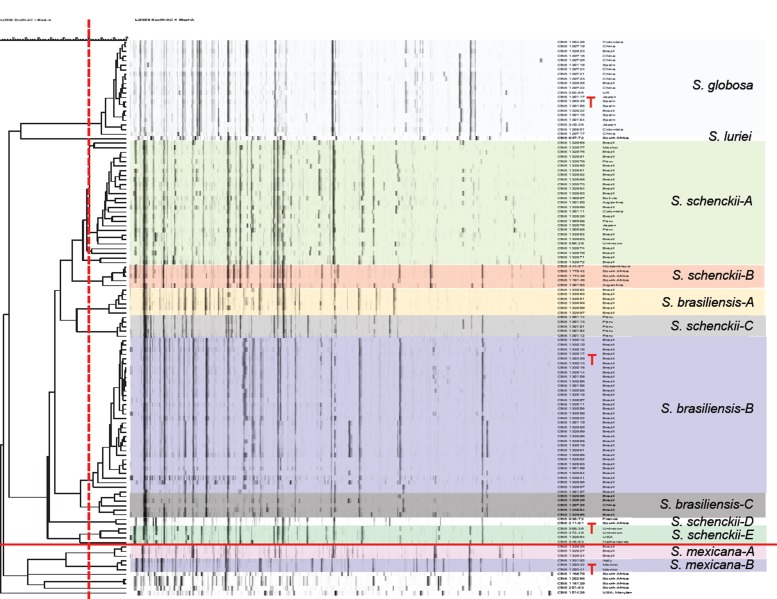
Fig. 4.
Amplified fragment length polymorphism (AFLP) profiles of 122 strains of Sporothrix. Clustering of AFLP banding pattern of isolates of Sporothrix was done by UPGMA. Red vertical bars represents cut-off for distinction of clusters. Strains of S. mexicana and below are phylogenetically unrelated.
Fig. 5.
Minimum spanning tree of AFLP data showing the relationships among 135 Sporothrix isolates, showing prevalent endemism of subclusters. Each dot corresponds to a unique genotype.
AFLP data of 116 strains of the potentially human- and animal-pathogenic species (Pathogenic clade in Fig. 2) are listed in Fig. 4. At a cut-off level of 70 % 13 groups could be recognised. All strains of S. globosa clustered in a single group. Sporothrix brasiliensis consisted of three groups, and S. schenckii of five groups (AFLP A–E; Fig. 4), the members of which matched with the groups found with multilocus sequence data (Fig. 3). In AFLP, Sporothrix mexicana contained two groups that could not be seen in the combined tree, and the single available strain of S. luriei deviated from all remaining species. In each of the groups of S. brasiliensis and S. schenckii, strains were clustered according to their geographic origin at low geographic distances, but numerous strains of S. globosa with identical profiles were repeatedly found to originate from different continents (Fig. 4).
Table 3 shows the number of sequences analysed, relative to the geography of isolation of the respective strain. In most cases, one species is predominant in each defined area. On the basis of this predominance, the probability of attribution of a given strain from that area was calculated. Using this probability, historical case reports without sequence data could be attributed to either S. brasiliensis, S. globosa, or S. schenckii; cases reported as S. luriei and S. mexicana were too rare for this calculation and were left blank in Table 3. Probability of Chinese strains as S. globosa, for example, was 100 %, whereas in Europe none of the species was predominant and thus such calculation was impossible. Sporotrichosis is rare in Europe, and etiologic agents may have been imported, which would explain the relatively high species diversity in this continent.
Table 5 provides an overview of cases of sporotrichosis published in the world literature with an accent on case series and including the great majority of cases published to date. Sporotrichosis classically was reported to occur in temperate and subtropical climates with a relatively high humidity. From Table 5 it appears that this holds true for S. schenckii. Hyperendemic areas are Brazil, Peru, Uruguay, Venezuela, and South Africa. In the hyperendemic area of north-east China and Japan, where S. globosa is prevalent, the climate is relatively cold (p = 10-6). In Asian countries, where S. globosa is prevalent, a preponderance of female patients is noted (p = 10-6). In Australia and South Africa more males are involved, partly associated with outdoor work, such as was the case with the miner epidemic in Witwatersrand by S. schenckii. In American countries male : female ratios are variable; no significant deviation from an equal ratio was noted in S. brasiliensis (Table 5).
Table 5.
Overview of published cases and case series of sporotrichosis with possible identification on the basis of regional sequence data.
| Species | Year | Country | n | Host | M/F pre-ponderance | Climate | Occupation | Probable transmission | Clinical form (D, F, LC, SYS, other) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| S. brasiliensis | 1989 | Brazil | 5 | Human | Warm, humid | Cat owner, veterinarian | Cat | LC | Larsson et al. 1989 | |
| 1998 | Brazil | 1 | Human | M | Warm, humid | D | Al-Tawfiq & Wools 1998 | |||
| 2001 | Brazil | 66 | Human | F | Warm, humid | Cat | F, LC | Barros et al. 2001 | ||
| 2003 | Brazil | 24 | Human | F | Warm, humid | Housewife | Cat | D | Barros et al. 2003 | |
| 2004 | Brazil | 178 | Human | F | Warm, humid | Housewife, student | Cat | D, F, LC | Barros et al. 2004 | |
| 2005 | Brazil | 304 | Human | M | Warm, humid | Farmer, teacher, student | F, LC | da Rosa et al. 2005 | ||
| 2005 | Brazil | 2 | Human | F | Warm, humid | Cat | Conjunctiva, F | Schubach et al. 2005 | ||
| 2006 | Brazil | 2 | Human | M | Warm, humid | Arthritis | Appenzeller et al. 2006 | |||
| 2008 | Brazil | 81 | Child | F | Warm, humid | Cat | LC | Barros et al. 2008a | ||
| 2008 | Brazil | 94 | Human | F | Warm, humid | Housewife | Cat | D, F, LC, SYS | arros et al. 2008b | |
| 2008 | Brazil | 759 | Human | F | Warm, humid | Car | Schubach et al. 2008 | |||
| 2010 | Brazil | 10 | Human | M | Warm, humid | Farmer | Armadillo hunting | F, LC | Alves et al. 2010 | |
| 2011 | Brazil | 5 | Human | F | Warm, humid | Cat | LC | Costa et al. 2011 | ||
| 2012 | Brazil | 1848 | Human | F | Warm, humid | Cat | Silva et al. 2012 | |||
| 2012 | Brazil | 21 | Human (HIV) | M | Warm, humid | D, F, LC | Freitas et al. 2012 | |||
| 2012 | Brazil | 92 | Cat | Warm, humid | D, F, SYS | Madrid et al. 2012 | ||||
| 11 | Dog | M | Warm, humid | F, D, LC | ||||||
| 2013 | Brazil | 4 | Human | F | Warm, humid | Cat | F with dacryocystitis | Freitas et al. 2013 | ||
| 2014 | Brazil | 3804 | Cat | Warm, humid | Pereira et al. 2014 | |||||
| S. globosa | 1982 | China | 273 | Human | Cool | Worker in paper factory | Decaying reed | F, LC | Wang & Sun 1982 | |
| 1986 | China | 232 | Human | F | Warm, humid | Wu 1986 | ||||
| 1997 | China | 142 | Human | Cool | F, LC | Jin et al. 1997 | ||||
| 1998 | China | 400 | Human | F | Cool | Farmer | Reed | F, LC | Song et al. 1998 | |
| 1999 | China | 237 | Human | Warm, humid | Ran et al. 1999 | |||||
| 2005 | China | 224 | Human | Cool | F, LC | Yang et al. 2005 | ||||
| 2007 | China | 48 | Human | Cool | F, LC | Gao et al. 2007 | ||||
| 2008 | China | 447 | Human | Cool | F, LC | Zhang & Lin 2008 | ||||
| 2008 | China | 226 | Human | Cool | F, LC | Fu et al. 2008 | ||||
| 2011 | China | 585 | Human | Cool | F, LC | Li et al. 2011 | ||||
| 2011 | China | 15 | Infant | F | Cool | F, LC | Song et al. 2011 | |||
| 2013 | China | 457 | Human | F | Cool | D, F, LC | Song et al. 2013 | |||
| 1992 | India | 1 | Human | F | Cool | Pulmonary | Padhye et al. 1992 | |||
| 1994 | India | 12 | Human | F | Humid | Farmer | LC | Chakrabarti et al. 1994 | ||
| 1998 | India | 17 | Human | M | Cool | Khaitan et al. 1998 | ||||
| 1999 | India | 25 | Human | F | Cool, humid | Horticulture, forest, farming | Ghosh et al. 1999 | |||
| 2007 | India | 21 | Human | F | Cool, humid | F, LC | Mehta et al. 2007 | |||
| 2008 | India | 9 | Human | F | Cool | Agarwal et al. 2008 | ||||
| 2009 | India | 1 | Human | M | Cool | Medical attendant | Cat | LC | Yegneswaran et al. 2009 | |
| 2011 | India | 224 | Human | F | Cool, humid | Plant | F, LC | Bhutia et al. 2011 | ||
| 2012 | India | 1 | Human | F | Cool | F | Tilak et al. 2012 | |||
| 2012 | India | 305 | Human | F | Cool, humid | F, LC | Verma et al. 2012 | |||
| 1986 | Japan | 200 | Human | F | Cool | Farmer | Plant | F, LC | Itoh et al. 1986 | |
| 2009 | Japan | 155 | Human | 1/1 | Cool | F, LC | Takenaka et al. 2009 | |||
| 1990 | Malaysia | 5 | Human | M | Warm, humid | Vet student, cat owner | Cat | LC | Zamri-Saad et al. 1990 | |
| 2012 | Malaysia | 19 | Human | F | Warm, humid | D, F, LC | Tang et al. 2012 | |||
| 2005 | Laos | 1 | Human | F | Warm, humid | Farmer | Wood | LC | Newton et al. 2005 | |
| S. mexicana | 2011 | Portugal | 1 | Human | M | Warm, dry | D | Dias et al.2011 | ||
| S. schenckii | 1940 | South Africa | 3000 | Human | Warm, humid | Untreated mine wood | Helm & Berman 1947 | |||
| 1997 | South Africa | 154 | Human | M | Cool, dry | Recreational, farmer | F, LC | Vismer & Hull 1997 | ||
| 2010 | Brazil | 120 | Dog | Warm, humid | Barros et al. 2010 | |||||
| 1965 | USA | 148 | Human | M | Temperate | Nursery | Plant thorn | Alessio et al. 1965 | ||
| 1977 | USA | 1 | Human | F | Temperate | Lab technician | Yeast of Sporothrix | F | Thompson & Kaplan 1977 | |
| 1978 | USA | 17 | Human | M | Temperate | Forestry worker | Sphagnum moss | Pulmonary | Powell et al. 1978 | |
| 1987 | USA | 1 | Human | M | Temperate | SYS | Gullberg et al. 1987 | |||
| 1991 | USA | 84 | Human | M | Temperate | Forestry worker | Sphagnum moss | Dixon et al. 1991 | ||
| 1992 | USA | 1 | Human | F | Temperate | Lab technician | Yeast of Sporothrix | F | Cooper et al. 1992 | |
| 1997 | USA | 9 | Human | M | Temperate | Tree nursery | Sphagnum moss | LC | Hajjeh et al. 1997 | |
| 1997 | USA | 5 | Human | M | Temperate | Hay bale | Dooley et al. 1997 | |||
| 2002 | USA | 1 | Human | M | Temperate | Fire ant | LC | Miller & Keeling 2002 | ||
| 2003 | USA | 1 | Infant | F | Temperate | F, larynx | Khabie et al. 2003 | |||
| 2007 | USA | 1 | Human(HIV) | M | Temperate | D | Vilela et al. 2007 | |||
| 2007 | USA | 1 | Dog | F | Temperate | F | Whittemore & Webb 2007 | |||
| 2009 | USA | 14 | Cat | Temperate | F, D, LC | Crothers et al. 2009 | ||||
| 4 | Dog | Temperate | F, D, LC | |||||||
| 4 | Horse | Temperate | F, LC | |||||||
| 1 | Donkey | Temperate | LC | |||||||
| 2009 | USA | 1 | Infant | Temperate | F | Tlougan et al. 2009 | ||||
| 2011 | USA | 1 | Human | F | Temperate | Cat | F | Rees & Swartzberg 2011 | ||
| 1998 | Australia | 16 | Human | M | Warm, dry | Mouldy hay | F, LC | Conias & Wilson 1998 | ||
| 2007 | Australia | 11 | Human | M | Warm, dry | Hay | Feeney et al. 2007 | |||
| 2012 | Australia | 31 | Human | M | Humid | Plant debris | Sivagnanam et al. 2012 | |||
| 2013 | Australia | 86 | Human (lower immune) | Warm, dry | Pulmonary | Aung et al. 2013 | ||||
| 1994 | Mexico | 4 | Human | Rust-stained tent | Campos et al. 1994 | |||||
| 2001 | Mexico | 50 | Human | F | Farmer, housewife | LC | Espinosa-Texis et al. 2001 | |||
| 2006 | Mexico | 55 | Human | M | F, LC | Macotela-Ruiz & Nochebuena-Ramos 2006 | ||||
| 2007 | Mexico | 25 | Child | Plant, soil, squirrel, cat, rat | D, F, LC | Bonifaz et al. 2007 | ||||
| 2007 | Mexico | 13 | Human | M | LC | Arenas et al. 2007 | ||||
| 1943 | Uruguay | 46 | Human | M | Cool, humid | Mackinnon 1943 | ||||
| 1969 | Uruguay | 157 | Human | M | Warm | Armadillo | Mackinnon et al. 1969 | |||
| 2004 | Uruguay | 42 | Human | M | Warm | Armadillo | F, LC | Civila et al. 2004 | ||
| 2000 | Peru | 238 | Human | M | Cool, dry | Farmer, student, housewife, infant | D, F, LC | Pappas et al. 2000 | ||
| 2011 | Peru | 20 | Human | M | Cool, dry | F | Ramírez-Soto et al. 2011 | |||
| 2010 | Columbia | 60 | Human | M | Warm, humid | Farmer | F, LC | Rubio et al. 2010 | ||
| 2013 | Venezuela | 87 | Human | M | Warm, humid | Farmer, student, housewife | Plant, insect, metal | F, LC | Mata-Essayag et al. 2013 |
NC = No comparison; NK = Not known; D = disseminated; F = fixed; LC = lymphocutaneous; SYS = systemic; S = Sporothrix.
Globally the most common clinical form is lymphocutaneous (LC) sporotrichosis, but in China the fixed cutaneous form is prevalent (p = 0.037). The mode of transmission of sporotrichosis often remains unclear. Trauma was mentioned in many cases, but was difficult to define in some cases because small traumata are easily neglected by patients. In some cases absence of trauma was explicitly mentioned (Table 5) and thus subcutaneous inoculation does not need to be apparent for the onset of sporotrichosis. Contact with decaying plant material was frequently noted in S. globosa and S. schenckii, whereas S. brasiliensis was significantly more associated with cat-transmission (p = 10-6). Other animal sources, such as fish, squirrel, armadillo, rat, dog, or insect, were mentioned in S. schenckii and S. brasiliensis, but not in S. globosa (Table 5).
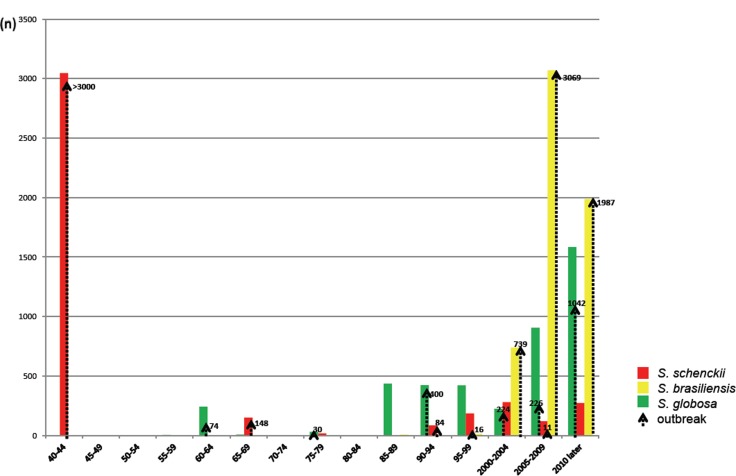
Many of the reports listed in Table 5 were case series from a common substrate during a fixed period. These outbreaks outnumber case series from various sources (Table 5). The total number of cases series and outbreaks reported in the literature comprises > 14 000 cases, while the number of reports of single cases is relatively low; this low number is only partially influenced by the fact that single cases with insufficient data were discarded from our meta-analysis. The outbreak-character of sporotrichosis is further demonstrated in Fig. 6, where case series during intervals of 5 years are listed. Several large outbreaks, such as by S. brasiliensis in south-east Brazil and S. globosa in Jilin, north-east China, are still ongoing. Fig. 6 does not list the case series of 200 infections in France during the years 1904–1911 (Beurmann & Gougerot 1912).
Fig. 6.
Timeline of epidemics and case series caused by three main pathogenic Sporothrix species since 1940. Vertical bars represent gross number of cases, vertical arrows denote case series and sapronoses or zoonoses.
DISCUSSION
For the present study, four gene regions were analysed: the rDNA ITS domain, the partial calmodulin (CAL) gene, and two regions in the translation elongation factor (TEF1 and TEF3). Molecular taxonomy of Sporothrix is particularly based on CAL (Marimón et al. 2006), although ITS performs equally well in distinguishing the main species, as demonstrated by Zhou et al. (2013) and Rodrigues et al. (2014b, d) and confirmed with a larger dataset in the present study. The tree topology of three combined genes CAL, TEF1, and TEF3 proved to be similar to that of the ITS tree. The most variable gene is CAL (variable sites 54.5 %) followed by TEF3 (33.7 %) and TEF1 (17.5 %). A similar range of variability was found with the number of parsimony-informative sites (CAL → TEF3 → TEF1). Diagnostics with CAL is optimal because intraspecific variability is small compared to barcoding gaps between species, yielding a highly resolved phylogenetic tree (data not shown). The genes analysed tend to differ in PCR performance, although ITS may present a higher number of negatives than usual in fungi (Table 2) (Schoch et al. 2012).
The genus Sporothrix is embedded in the order Ophiostomatales. The core genus is Ophiostoma, which is classically known to comprise fungi that live in association with bark beetles. De Beer et al. (unpubl. data) delimited the two genera, maintaining nearly all arthropod-associated species in Ophiostoma. Ecologies of the 32 accepted Sporothrix species were quite diverse. Virulence to mammals is nearly exclusively found in a small group of species around S. schenckii. The ITS tree combining these groups demonstrate that phylogenetic distances are moderate and comply with classification in a single order; ITS was alignable with reasonable confidence over the entire dataset. The maximum ITS distance (measured by similarity in BioNumerics) from S. brasiliensis CBS 133019 to S. foliorum CBS 326.37 was 15 %. The span over diversity within the clade with pathogenic species (S. brasiliensis CBS 133019 to S. mexicana CBS 120342) was 4.3 % for ITS and 8 % for three genes analysed. We randomly sequenced all strains deposited in the CBS collection over a century and found that only 3 isolates might represent undescribed species (Table 1). It is questionable whether S. inflata and S. dimorphospora are different species. Within the habitats analysed sampling has apparently been sufficient to cover extant biodiversity.
The occurrence of two pronounced types of ecology within the Ophiostomatales, viz. human pathogenicity and bark beetle association, is remarkable. In the ITS tree (Fig. 2) the species living inside bark beetle galleries constitute a well-supported clade, matching Ophiostoma (de Beer et al. 2003). Some of the species in Fig. 2 have been isolated from sapwood and a role of the bark beetle has then usually not been proven. Outside this clade explicit bark beetle-association is uncommon. Arthropod dispersal remains common in other clades, however, as exemplified by clades with species from mites in Protea infructescences. As the intermediate species in the phylogenetic trees (Fig. 2) exhibit other types of ecology, there is no obvious link between bark beetle-association and human pathogenicity.
The remaining groups compose a polytomy from an unresolved backbone with bootstrap values below 80 % (Fig. 2). Comparing the clades over the entire tree, we observe a consistent decrease of bark beetle association outside Ophiostoma, concomitantly with an increasing vertebrate infectivity. Inside Ophiostoma, a single case of human infection in a leukemic patient was described in Ophiostoma piceae (Bommer et al. 2009). Outside the pathogenic clade (Fig. 3), two cases were reported by S. stenoceras (Mariat et al. 1968), while S. pallida infections are represented by sporadic cases related to impairment of the immune system, e.g. in transplant recipients (Morrison et al. 2013). In all these species, human pathology is highly exceptional (white crosses in Fig. 2), except for several S. mexicana cases (Dias et al. 2011, Rodrigues et al. 2013a) where human infection is relatively common, but mild. Sporothrix luriei is a very rare species, with a single proven case from South Africa (Ajello & Kaplan 1969) and two cases with unproved culture from Italy and India, respectively (Alberici et al. 1989, Padhye et al. 1992). The species is reported to be highly virulent (Fernández-Silva et al. 2012). The remaining species of the pathogenic clade (Guarro et al. 1999), S. schenckii, S. globosa and S. brasiliensis occur in epidemic proportions with several thousands of cases each. Virulence has been tested in animal inoculations (Arrillaga-Moncrieff et al. 2009, Castro et al. 2013, Fernandes et al. 2013) using mice as model animal. Sporothrix brasiliensis presented more fungal burden, dissemination capacity and massive infiltration in infected tissues compared to S. schenckii and S. globosa. Highest virulence was observed in S. brasiliensis which correlates with high degree of pathogenicity in felines (Rodrigues et al. 2013b) and humans (Silva-Vergara et al. 2012). A connection among disease severity, humoral response and protein secretion revealed a common immunogenic protein of 60 kDa recognised by antisera in all virulent isolates of Sporothrix (Fernandes et al. 2013). Further, this molecule was also shown to be a component of the cell wall of S. brasiliensis and S. schenckii (Castro et al. 2013) and appears to be relevant during infection.
Sporothrix brasiliensis shows low degrees of variability and has been suggested to be clonal (Rodrigues et al. 2013b, 2014d). The low variability of this taxon is also supported by low chromosomal polymorphisms (Sasaki et al. 2014) and homogeneous susceptibility profiles to antifungal agents (Rodrigues et al. 2014c). In our dataset (Fig. 3) a supported clade matched with AFLP group A and mainly contained strains from southern Brazil; strains of the top clade had the preponderant AFLP type B. The occurrence of separate genotypes among strains indicated that the Brazilian S. brasiliensis epidemic has at least two distinct sources as was proposed earlier by Rodrigues et al. (2013b) and confirmed here by AFLP data.
Sporothrix globosa strains analysed were strictly identical. Isolates studied by Yu et al. (2013) were divided into two highly supported subclades (S. globosa I and S. globosa II). Group I comprised the majority of Chinese clinical isolates, three Chinese environmental isolates from reed, corn stalks, and soil, the type strain of S. globosa, and some isolates from the USA, India, Japan, Brazil, and the UK. Sporothrix globosa II included a small part of Chinese clinical isolates and a single isolate from Italy. More material is needed to establish whether S. globosa is preponderantly clonal.
As noticed earlier (Marimón et al. 2006, 2007), S. schenckii is the likely ancestral, most variable species within the pathogenic clade. This genetic variability is reflected in significant differences in genome size and chromosome profiles generated by pulsed-field gel electrophoresis that usually display 4 to 7 chromosomal bands, ranging from 2.0 to 7.0 Mb (Sasaki et al. 2014). In our dataset, five supported subgroups were visible in all partitions (Fig. 3), which were also recognisable in AFLP data (Fig. 4). This suggests that the groups are separate lineages with limited gene flow. Nearly all lineages A–E of S. schenckii have restricted geographic distributions (Fig. 5). Four of the clusters are concurrent with groups distinguished by Marimón et al. (2006, 2007).
As can be concluded from combined AFLP and MLST data, remarkable differences between species are noted. Sporotrichosis in Brazil dates back to 1907, when the disease was first diagnosed in naturally infected mice (Lutz & Splendore 1907). Sporothrix brasiliensis strains with closely similar AFLP profiles have very limited distribution. Despite the large expansion of populations during the Rio de Janeiro zoonotic epidemic, strains with identical genotypes are found at small geographic distances (Rodrigues et al. 2014d). The species thus seems to have a slow vector of dispersal. Several authors (Barros et al. 2004, Madrid et al. 2012, Rodrigues et al. 2013b) have demonstrated a role of felines in transmission of S. brasiliensis. Cats are relatively sedentary, and thus the zoonosis is expected to expand geographically at a slow pace. Sporothrix globosa also shows low degrees of variation, but a significant difference with S. brasiliensis is observed in that identical multilocus genotypes are repeatedly found at very large geographic distances. For example, molecularly identical strains CBS 125438, CBS 129719, and CBS 132923 originated from Colombia, China, and Brazil, respectively, while also strains from UK, Japan, Spain, Brazil, and China had identical genotypes. Obviously a rapid vector of dispersal is at work in S. globosa. Given the large distances between identical strains, airborne distribution seems likely. The absence of S. globosa from Africa and Australia then remains puzzling, but this perhaps can be explained by sampling effects. Notably, S. globosa infections are derived from plant debris (Wang & Sun 1982, Li et al. 1995, Song et al. 2013) and is classically known as ‘reed toxin’ (Song et al. 2013). Cats have never been observed as sources of infection in endemic areas of S. globosa. Conversely, plants have never been observed as sources of infection by S. brasiliensis.
In this respect, S. schenckii seems intermediate. Classically the infection is known as ‘rose gardener’s disease’, suggesting a plant source of infection and traumatic inoculation (Rodrigues et al. 2014d). In our dataset, CBS 132977 in group A originated from plant material in Mexico, while the remaining strains of that group were of clinical origin. In the literature a connection between S. schenckii and plants has been made many times. Dixon et al. (1991) described a sapronosis of 84 cases, studying 21 clinical isolates which proved to be identical to strains from Sphagnum moss by RFLP. A similar report was that of Hajjeh et al. (1997) in an outbreak of sporotrichosis from Sphagnum moss among nine out of 65 nursery workers. Feeney et al. (2007) described an outbreak of sporotrichosis from hay in Australia, where S. schenckii was shown to be the etiologic agent by ITS sequencing. The large epidemic from South Africa in the forties of the previous century, with more than 3 000 cases, was proven to have untreated mining wood as source of infection, and disappeared after the wood had been impregnated with creosote (Helm & Berman 1947).
From the above it is obvious that historical outbreak data are needed to understand the behaviour of individual Sporothrix species. Data of Feeney et al. (2007) and Yu et al. (2013) could be verified as S. schenckii and S. globosa, respectively by GenBank submissions, but in many cases neither molecular data nor strains were available for study. Given the geographic structuring of almost all Sporothrix populations, we used geographically defined sets of sequenced strains (Fig. 1) to deduce the most probable identity of historical strains in the same region. Ratios of numbers of strains sequenced per region, compared to the number of reported cases from the same region, are illustrated in Fig. 1. For example, all strains from China sequenced thus far, i.e. 112 strains from Yu et al. (2013) and Tan et al. (2013), plus nine from the present study, were identified as S. globosa. Thus, the historical probability in China to be S. globosa is 100 %, while in the USA the strains are expected to be S. schenckii with a probability of 87 %. Data are summarised in Table 3, where we took 80 % as cut-off below which percentage historical data could not be interpreted.
Summarising published cases since 1940 (Table 5), a remarkable phenomenon becomes apparent. Most of the published cases concerned case series or outbreaks, either with plant origins (sapronoses) or feline origins (zoonoses). Fig. 6 shows all cases since 1940 in a histogram accumulatively in 5-year intervals. The great majority of cases were part of an outbreak, the numbers of cases per series varying from 5 to 3 069. The smallest outbreak concerns patients being infected from the same heap of hay stored in an old house (Dooley et al. 1997). Although neglected by the present study, individual cases are uncommon, even in older literature when only few cases had been published. The oldest outbreak is that in France during the period 1906–1911, which started 8 yr after the first description of Sporothrix by Hektoen & Perkins in 1900. Since then, sporotrichosis has remained rare in Europe, and part of the etiologic agents may have been imported, which would explain the relatively high species diversity in this continent.
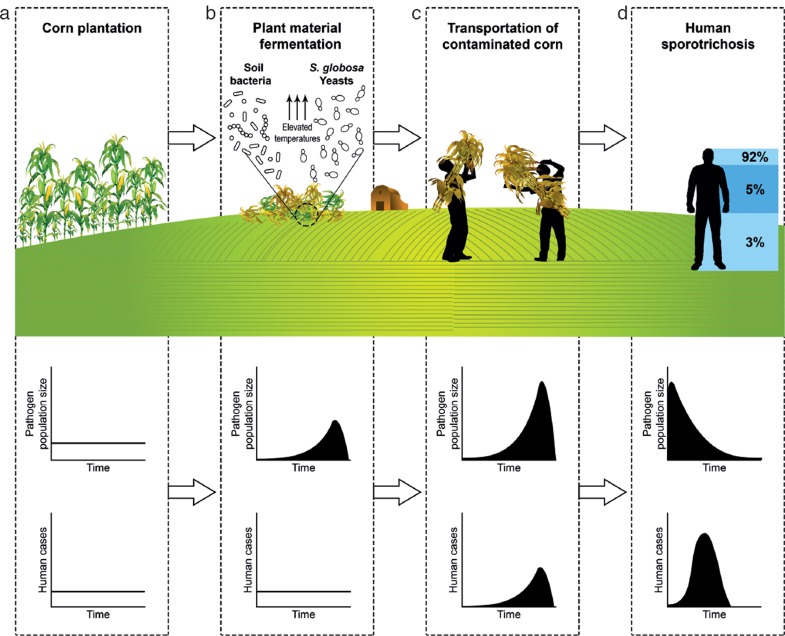
How can we explain this outbreak behaviour observed in all Sporothrix species irrespective of their different modes of transmission? The plant-borne species (S. schenckii and S. globosa) are found on decaying plant material or wood. In each plant-borne species, large differences are observed in type of plant material. In S. schenckii, for example, this was mining wood, rose thorns, and Sphagnum moss. In Sporothrix species with plant material as source of infection we have to assume that not the host plant species, but the condition of the plant material is significant. This condition has to be highly special. Notably, Sphagnum moss is used worldwide in large quantities, but only a few outbreaks of sporotrichosis have been described. As another example, decaying hay is ubiquitous material, and thus infections with regular intervals over time would be expected, but we consistently observe occasional infections from a common source, i.e. outbreaks. Therefore we hypothesise that Sporothrix species are not plant pathogens, but require particular conditions in decaying plant material, which are reached only occasionally. We postulate that a particular state of decay and fermentation of the plant material promotes excessive growth of Sporothrix. High temperature and humidity, associated with metabolic changes (induction of respiratory system) and oxidative stress due decay and fermentation may shift the morphology, favouring the invasive yeast growth form (Klein & Tebbets 2007). This hypothesis is illustrated in Fig. 7. In a small-scale study in The Netherlands (Y. Zhang, unpubl. data) we were unable to detect Sporothrix in growing corn plants. The species is hypothesised to grow exponentially in corn debris (Fig. 7b) serving as a potential inoculum for corn harvesters; the prevalent clinical type in north-east China is facial (Xia et al. 2009). At disappearance of the infectious material, the human sapronosis will die out with some delay (Fig. 7d), matching with the observation that most cases in China’s Jilin Province become apparent during winter (Song et al. 2013).
Fig. 7.
Schematic overview of the hypothesis for the dissemination of the plant-born pathogen Sporothrix globosa, applied to the large human epidemics occurring in corn crops in north-east China. a. The species has not been found as an endophyte. b. Specific conditions in decaying plant material, such as fermentation, may stimulate excessive growth of the thermodependent yeasts, which facilitates infections in mammals. c. Onset of facial infections during transportation of corn debris. d. Delayed development of human infection during decline of growth in plant debris.
Occasionally, Sporothrix infections have been described that transmitted by animals very different from cats, which may be warm-blooded vertebrates but also arthropods: bites by squirrels, bats, fire ants, and spiders have been recorded (Moaven et al. 1999, Miller & Keeling 2002, Alves et al. 2010, Rodrigues et al. 2014a). The fungus was also isolated from plant material in armadillo burrows (Mackinnon et al. 1969, Rodrigues et al. 2014a). The conditions of plant decay in the burrow may be suitable for the development of Sporothrix, and subsequent dispersal by the armadillo may be expected.
Thus, despite the preponderance of cat vectors, the animal host species may vary, just as the plant host species did. This leads us to a hypothesis of wild animals occasionally providing conditions similar to those in fermented plant material. Cats take up propagules from the soil and easily transmit them to their mouth by licking. Conditions in animal saliva at the feline body temperature (normal range 37.7–39.1 °C) might be a stimulating factor for the production of the Sporothrix yeast phase. Cat saliva has a pH of 7.5–8.0, which is similar to that of self-heating bulk corn debris (around 8.0) and optimal for the mould-to-yeast conversion. With a hypothesis of conditional similarities between fermenting plant material and animal digestive tracts, the unique host shift of Sporothrix from plant to animal becomes understandable.
In south-east Brazil, transmission occurs nearly always by cats. Cat saliva is a stable environment, and despite the presence of antibodies – which generally have a low impact on fungal infections – repeated colonisation by Sporothrix once it has adapted to these conditions may be expected. The large outbreak in this area (da Rosa et al. 2005, Schubach et al. 2004, 2008, Silva et al. 2012, Pereira et al. 2014) suggests that the number of cases increases relative to the number of patients and cat vectors. Several peculiarities of cats may facilitate the dispersal of the fungus in the environment within limited endemic areas. Firstly, they are the most common pet animals with close contact to humans. Secondly, given the hypothesized origin of Sporothrix in cat saliva and its transmission to claws during licking, cat mobility and clawing enable them to take up and transmit the fungus, either each other during play or fight with house cats or stray cats, or transmit the fungus to human hosts via scratches or bites (Madrid et al. 2012).
Cat-transmitted cases also occur in S. schenckii, but at a much lower frequency (Rodrigues et al. 2013b), suggesting that this is not an exclusive relationship between S. brasiliensis and the feline host. In 1952, a first cat-transmitted case was reported from the USA. Until 1988, cat-associated cases and reports of transmission to human were sporadic. Four Malaysian veterinary students and one cat owner developed lesions of sporotrichosis after being bitten or scratched by cats with apparent fight wounds (Zamri-Saad et al. 1990). Crothers et al. (2009) described a series of 14 cases in cats, six of which were disseminated. The few outbreaks involving cats during this period invariably was limited to people in close contact with cats (Zamri-Saad et al. 1990, Cooper et al. 1992). This condition changed considerably since around 1998 in Brazil, when the incidence of cat and cat-transmitted cases increased dramatically to more than 5 000 human and feline cases in an expanding area around Rio de Janeiro and São Paulo (Barros et al. 2004, 2008b, Schubach et al. 2008, Pereira et al. 2014).
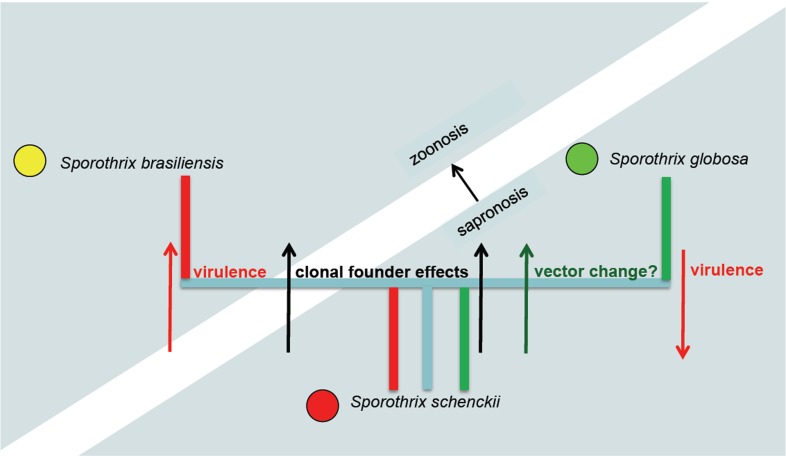
Sporothrix schenckii has been supposed to be the ancestral species on phylogenetic grounds, mainly because of its high degree of variability in all markers, its wide distribution, and hypothetical presence of sexuality as judged from a balanced mating type distribution (Teixeira et al. In press). The species seems preponderantly plant-transmitted. The share of cat-transmitted cases is much lower than in S. brasiliensis, and much higher than in S. globosa, where it is zero. In humans, disseminated cases occur almost only in immunocompromised patients, while cats seem to be relatively susceptible to infection. Host-shifts from plant material to animals are likely to have occurred already within S. schenckii, which diminishes the value of genomic comparisons between species. The main (preponderantly) clonal offshoots S. brasiliensis and S. globosa seem to have adapted successfully to their respective new habitats. This evolutionary hypothesis is summarised in Fig. 8. The ancestral species S. schenckii contains divergent genotypes with different behaviour. The clonal offshoot S. brasiliensis on average has increased virulence and is cat-transmitted; thus a shift from sapronoses to zoonoses takes place. The clonal offshoot S. globosa has lower virulence and has maintained sapronotic behaviour, but its vector of distribution seems to have changed.
Fig. 8.
Diagram of hypothetical processes of evolution from highly diverse ancestral species S. schenckii and clonal offshoots S. globosa and S. brasiliensis. Due to differential selection processes, the derived species differ significantly in virulence, transmission and type of epidemic caused.
Sporothrix mexicana is phylogenetically distant to the main pathogenic clade of S. schenckii, S. globosa, and S. brasiliensis, and thus is unlikely to have played a role in the evolutionary host shift described above. Information on the few clinical strains available from the 1950s and 1970s decade is very limited (Rodrigues et al. 2013a). Dias et al. (2011) described one disseminated case without history of trauma from Portugal in an immunocompetent patient.
A certain degree of gender predominance was discernible which differed between species (Table 5). Considering all publications with > 1 case, we noticed that in S. globosa in 11 case series female hosts were predominant, vs zero times male hosts. This is in agreement with Verma et al. (2012) who reported a female preponderance in the epidemiology of sporotrichosis in Himachal Pradesh, a small hill state in north-west India, usually related to agriculture practice. In S. schenckii these figures were opposite: one female vs eight males. In S. brasiliensis female / male ratios were more or less equal (7/9). Gender ratios have been reported to differ, mainly related to urban migration. In Brazil, many housewives stay at home and tend to be in charge of caring cats, while males have a larger chance to be infected during outdoor activities (Schubach et al. 2008). In rural areas of north-east China, most males go to cities to find jobs, leaving females, elderly and children at home, having increased chance of infection. Sporothrix schenckii infection classically seems to be connected with male agricultural activities (Rodrigues et al. 2014d). As most publications are unclear about gender ratios, more epidemiological study is necessary.
Conclusions
Our data and the review of the existing literature have shown that Sporothrix is unique in the fungal kingdom by its prevalent occurrence in the form of outbreaks, and that these outbreaks differ fundamentally from each other. In S. brasiliensis a huge zoonosis is taking place today, while the contemporary outbreak of similar dimensions in China is a sapronosis. In the ancestral species S. schenckii most outbreaks are sapronoses, but small zoonoses with cats as prime susceptible hosts have also been observed. It is significant to public health to consider these distinctions. Sapronoses, providing very special conditions promoting fungal growth, basically can be controlled by removal of the plant biomass allowing this contamination. In contrast, the zoonoses of cats compose a much more diffuse source of infection, which is more difficult to control. In addition, the ancestral species S. schenckii contains a mixture of strains that are susceptible to antifungals widely used in cutaneous infections, e.g. terbinafin, ketoconazole, and itraconazole, and strains with decreased susceptibility (Stopiglia et al. 2014). A large difference is noted particularly with azoles between the highly susceptible species S. brasiliensis and the resistant species S. globosa (Marimón et al. 2008). Selection and clonal expansion of resistant strains during epidemics may increase the significance of sporotrichosis as a human disease.
Acknowledgments
The research presented in this paper was supported by the KNAW - FES project ‘Barcoding the CBS Collections’, by KNAW China Desk Project 11CDP009, and the project was co-funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant No. (1-965-34-HiCi) which the authors gratefully acknowledge. Bert Gerrits van den Ende is thanked for technical assistance.
References
- Agarwal S, Gopal K, Umesh , et al. 2008. Sporotrichosis in Uttarakhand (India): a report of nine cases. International Journal of Dermatology 47: 367–371. [DOI] [PubMed] [Google Scholar]
- Ajello L, Kaplan W. 1969. A new variant of Sporothrix schenckii. Mycoses 12: 633–644. [DOI] [PubMed] [Google Scholar]
- Al-Tawfiq JA, Wools KK. 1998. Disseminated sporotrichosis and Sporothrix schenckii fungemia as the initial presentation of human immunodeficiency virus infection. Clinical Infectious Diseases 26: 1403–1406. [DOI] [PubMed] [Google Scholar]
- Alberici F, Paties CT, Lombardi G, et al. 1989. Sporothrix schenckii var. luriei as the cause of sporotrichosis in Italy. European Journal of Epidemiology 5: 173–177. [DOI] [PubMed] [Google Scholar]
- Alessio DJ d’, Leavens LJ, Strumpf GB, et al. 1965. An outbreak of Sporotrichosis in Vermont associated with Sphagnum moss as the source of infection. New England Journal of Medicine 272: 1054–1058. [DOI] [PubMed] [Google Scholar]
- Alves SH, Boettcher CS, Oliveira DC, et al. 2010. Sporothrix schenckii associated with armadillo hunting in southern Brazil: epidemiological and antifungal susceptibility profiles. Revista Sociedade Brasiliera de Medicina Tropical 43: 523–525. [DOI] [PubMed] [Google Scholar]
- Appenzeller S, Appenzeller S, Amaral TN, et al. 2006. Sporothrix schenckii infection presented as monoarthritis: report of two cases and review of the literature. Clinical Rheumatology 25: 926–928. [DOI] [PubMed] [Google Scholar]
- Arenas R, Miller D, Campos-Macias P. 2007. Epidemiological data and molecular characterization (mtDNA) of Sporothrix schenckii in 13 cases from Mexico. International Journal of Dermatology 46: 177–179. [DOI] [PubMed] [Google Scholar]
- Arrillaga-Moncrieff I, Capilla J, Mayayo E, et al. 2009. Different virulence levels of the species of Sporothrix in a murine model. Clinical Microbiology and Infection 15: 651–655. [DOI] [PubMed] [Google Scholar]
- Aung AK, Teh BM, McGrath C, et al. 2013. Pulmonary sporotrichosis: case series and systematic analysis of literature on clinico-radiological patterns and management outcomes. Medical Mycology 51: 534–544. [DOI] [PubMed] [Google Scholar]
- Bachmeyer C, Buot G, Binet O, et al. 2006. Fixed cutaneous sporotrichosis: an unusual diagnosis in West Europe. Clinical Experimental Dermatology 31: 479–481. [DOI] [PubMed] [Google Scholar]
- Barros MB, Almeida Paes R de, Schubach AO. 2011. Sporothrix schenckii and sporotrichosis. Clinical Microbiology Reviews 24: 633–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MB, Costa DL, Schubach TM, et al. 2008a. Endemic of zoonotic sporotrichosis: profile of cases in children. Pediatric Infectious Disease Journal 27: 246–250. [DOI] [PubMed] [Google Scholar]
- Barros MB, Schubach AO, Galhardo MC, et al. 2003. Sporotrichosis with wide-spread cutaneous lesions: report of 24 cases related to transmission by domestic cats in Rio de Janeiro, Brazil. International Journal of Dermatology 42: 677–681. [DOI] [PubMed] [Google Scholar]
- Barros MB, Schubach AO, Schubach TM, et al. 2008b. An epidemic of sporotrichosis in Rio de Janeiro, Brazil: epidemiological aspects of a series of cases. Epidemiology and Infection 136: 1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MB, Schubach OA, Valle AC do,et al. 2004. Cat-transmitted sporotrichosis epidemic in Rio de Janeiro, Brazil: description of a series of cases. Clinical Infectious Diseases 38: 529–535. [DOI] [PubMed] [Google Scholar]
- Barros MB, Schubach TM, Galhardo MC, et al. 2001. Sporotrichosis: an emergent zoonosis in Rio de Janeiro. Memórias do Instituto Oswaldo Cruz 96: 777–779. [DOI] [PubMed] [Google Scholar]
- Barros MB, Schubach TP, Coll JO, et al. 2010. Sporotrichosis: development and challenges of an epidemic. Revista Panamericana Salud Pública 27: 455–460. [In Portuguese.] [PubMed] [Google Scholar]
- Beer ZW de, Harrington TC, Vismer HF, et al. 2003. Phylogeny of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 95: 434–441. [PubMed] [Google Scholar]
- Beer ZW de, Wingfield MJ. 2013. Emerging lineages in the Ophiostomatales. In: The Ophiostomatoid fungi: expanding frontiers. CBS Biodiversity Series 12. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- Beurmann L, Gougerot H. 1912. Les Sporotrichose. Librairie Felix Alcan, Paris, France. [Google Scholar]
- Bhutia PY, Gurung S, Yegneswaran PP, et al. 2011. A case series and review of sporotrichosis in Sikkim. The Journal of Infections in Developing Countries 5: 603–608. [DOI] [PubMed] [Google Scholar]
- Bommer M, Hütter ML, Stilgenbauer S, et al. 2009. Fatal Ophiostoma piceae infection in a patient with acute lymphoblastic leukaemia. Journal of Medical Microbiology 58: 381–385. [DOI] [PubMed] [Google Scholar]
- Bonifaz A, Saúl A, Paredes-Solis V, et al. 2007. Sporotrichosis in childhood: clinical and therapeutic experience in 25 patients. Pediatric Dermatology 24: 369–372. [DOI] [PubMed] [Google Scholar]
- Cafarchia C, Sasanelli M, Lia RP, et al. 2007. Lymphocutaneous and nasal sporotrichosis in a dog from southern Italy: case report. Mycopathologia 163: 75–79. [DOI] [PubMed] [Google Scholar]
- Campos P, Arenas R, Coronado H. 1994. Epidemic cutaneous sporotrichosis. International Journal of Dermatology 33: 38–41. [DOI] [PubMed] [Google Scholar]
- Castro RA, Kubitschek-Barreira PH, Teixeira PA, et al. 2013. Differences in cell morphometry, cell wall topography and gp70 expression correlate with the virulence of Sporothrix brasiliensis clinical isolates. PLoS ONE 8: e75656.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. 1988. Multistate outbreak of sporotrichosis in seedling handlers. Morbidity and Mortality Weekly Report 37: 652–653. [PubMed] [Google Scholar]
- Chakrabarti A, Roy SK, Dhar S, et al.1994. Sporotrichosis in north-west India. Indian Journal of Medical Research 100: 62–65. [PubMed] [Google Scholar]
- Chowdhary A, Sharma C, Duggal S, et al. 2013. New clonal strain of Candida auris, Delhi, India. Emerging Infectious Diseases 19: 1670–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civila ES, Bonasse J, Conti-Díaz IA, et al. 2004. Importance of the direct fresh examination in the diagnosis of cutaneous sporotrichosis. International Journal of Dermatology 43: 808–810. [DOI] [PubMed] [Google Scholar]
- Conias S, Wilson P. 1998. Epidemic cutaneous sporotrichosis: report of 16 cases in Queensland due to mouldy hay. Australasian Journal of Dermatology 39: 34–37. [DOI] [PubMed] [Google Scholar]
- Cooper CR, Dixon DM, Salkin IF. 1992. Laboratory-acquired sporotrichosis. Journal of Medical and Veterinary Mycology 30: 169–171. [DOI] [PubMed] [Google Scholar]
- Costa RO, Bernardes-Engemann AR, Azulay-Abulafia L, et al. 2011. Sporotrichosis in pregnancy: case reports of 5 patients in a zoonotic epidemic in Rio de Janeiro, Brazil. Anais Brasilieras de Dermatologia 86: 995–998. [DOI] [PubMed] [Google Scholar]
- Criseo G, Malara G, Romeo O, et al. 2008. Lymphocutaneous sporotrichosis in an immunocompetent patient: a case report from extreme southern Italy. Mycopathologia 166: 159–162. [DOI] [PubMed] [Google Scholar]
- Criseo G, Romeo O. 2010. Ribosomal DNA sequencing and phylogenetic analysis of environmental Sporothrix schenckii strains: comparison with clinical isolates. Mycopathologia 169: 351–358. [DOI] [PubMed] [Google Scholar]
- Crothers SL, White SD, Ihrke PJ, et al. 2009. Sporotrichosis: a retrospective evaluation of 23 cases seen in northern California (1987–2007). Veterinary Dermatology 20: 249–259. [DOI] [PubMed] [Google Scholar]
- Dias NM, Oliveira MM, Santos C, et al. 2011. Sporotrichosis caused by Sporothrix mexicana, Portugal. Emerging Infectious Diseases 17: 1975–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DM, Salkin IF, Duncan RA, et al. 1991. Isolation and characterization of Sporothrix schenckii from clinical and environmental sources associated with the largest U.S. epidemic of sporotrichosis. Journal of Clinical Microbiology 29: 1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley DP, Bostic PS, Beckius ML. 1997. Spook house sporotrichosis. A point-source outbreak of sporotrichosis associated with hay bale props in a Halloween haunted-house. Archives of Internal Medicine 157: 1885–1887. [DOI] [PubMed] [Google Scholar]
- Engle J, Desir J, Bernstein JM. 2007. A rose by any other name. Skinmed 6: 139–141. [DOI] [PubMed] [Google Scholar]
- Espinosa-Texis A, Hernández-Hernández F, Lavalle P, et al. 2001. Study of 50 patients with sporotrichosis. Clinical and laboratory assessment. Gaceta Médicina de México 137: 111–116. [PubMed] [Google Scholar]
- Feeney KT, Arthur IH, Whittle AJ, et al. 2007. Outbreak of sporotrichosis, Western Australia. Emerging Infectious Diseases 13: 1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes GF, Santos PO dos, Rodrigues AM, et al. 2013. Characterization of virulence profile, protein secretion and immunogenicity of different Sporothrix schenckii sensu stricto isolates compared with S. globosa and S. brasiliensis species. Virulence 4: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Silva F, Capilla J, Mayayo E, et al. 2012. Virulence of Sporothrix luriei in a murine model of disseminated infection. Mycopathologia 173: 245–249. [DOI] [PubMed] [Google Scholar]
- Freitas DF, Lima IAR, Curi CL, et al. 2013. Acute dacryocystitis: another clinical manifestation of sporotrichosis. Memórias do Instituto Oswaldo Cruz 109: 162–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas DF, Siqueira-Hoagland B de, ,Valle AC do, et al. 2012. Sporotrichosis in HIV-infected patients: report of 21 cases of endemic sporotrichosis in Rio de Janeiro, Brazil. Medical Mycology 50: 170–178. [DOI] [PubMed] [Google Scholar]
- Fu AH, Yu Y, Wang LP. 2008. Clinical data analysis of 226 cases sporotrichosis. Journal of Pathogen Biology 8: 637. [In Chinese.] [Google Scholar]
- Gao F, Wang Q, Li TN. 2007. Clinical analysis of 48 cases sporotrichosis of Shenyang region.Chinese Journal of Mycology 3: 146–147. [In Chinese.] [Google Scholar]
- Ghosh A, Chakrabarti A, Sharma VK, et al. 1999. Sporotrichosis in Himachal Pradesh (north India). Transactions of the Royal Society of Tropical Medicine and Hygiene 93: 41–45. [DOI] [PubMed] [Google Scholar]
- Guarro J, Gené J, Stchigel AM. 1999. Developments in fungal taxonomy. Clinical Microbiology Reviews 12: 454–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guého E, Smith MT, Hoog GS de, et al. 1992. Contributions to a revision of the genus Trichosporon. Antonie van Leeuwenhoek 61: 289–316. [DOI] [PubMed] [Google Scholar]
- Gullberg RM, Quintanilla A, Levin ML, et al. 1987. Sporotrichosis: recurrent cutaneous, articular, and central nervous system infection in a renal transplant recipient. Reviews of Infectious Diseases 9: 369–375. [DOI] [PubMed] [Google Scholar]
- Hajjeh R, McDonnell S, Reef S, et al. 1997. Outbreak of sporotrichosis among tree nursery workers. Journal of Infectious Diseases 176: 499–504. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hektoen L, Perkins CF. 1900. Refractory subcutaneous abscess caused by Sporothrix schenckii: a new pathogenic fungus. Journal of Experimental Medicine 5: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Berman C. 1947. The clinical, therapeutic and epidemiological features of the sporotrichosis infection on the mines.In Sporotrichosis infection on mines of the Witwatersrand. Proceedings of the Transvaal Mine Medical Officers’ Association, December 1944, Johannesburg: 59–67. [Google Scholar]
- Itoh M, Okamoto S, Kariya H. 1986. Survey of 200 cases of sporotrichosis. Dermatologica 172: 209–213. [DOI] [PubMed] [Google Scholar]
- Jin XZ, Jiang RH, Li FQ, et al. 1997. Analysis of 142 cases of sporotrichosis. Chinese Journal of Dermatology 5: 331. [In Chinese.] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabie N, Boyce TG, Roberts GD, et al. 2003. Laryngeal sporotrichosis causing stridor in a young child. International Journal of Pediatric Otorhinolaryngology 67: 819–823. [DOI] [PubMed] [Google Scholar]
- Khaitan BK, Lakhanpal S, Banerjee U, et al. 1998. Sporotrichosis in an unusual location − dermatologically and geographically. Indian Journal of Pathology and Microbiology 41: 461–463. [PubMed] [Google Scholar]
- Klein BS, Tebbets B. 2007. Dimorphism and virulence in fungi. Current Opinions in Microbiology 10: 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson CE, Gonçalves M, de A, Araujo VC, et al. 1989. Feline sporotrichosis: clinical and zoonotic aspects. Revista do Instituto de Medicina Tropical Sao Paulo 31: 351–358. [In Portuguese.] [DOI] [PubMed] [Google Scholar]
- Li HY, Song J, Zhang Y, et al. 1995. Epidemiological survey of sporotrichosis in Zhaodong, Heilongjiang. Chinese Journal of Dermatology 28: 401–402. [In Chinese.] [Google Scholar]
- Li SS, Liu HS, Zheng H, et al. 2011. Clinical analysis of 585 cases of cutaneous sporotrichosis. Chinese Journal of Dermatology 44: 161–164. [In Chinese.] [Google Scholar]
- Lima RF de, Schäffer GV, Moraes Borba C de. 2003. Variants of Sporothrix schenckii with attenuated virulence for mice. Microbes and Infection 5: 933–938. [DOI] [PubMed] [Google Scholar]
- Lutz A, Splendore A. 1907. Sobre uma micose observada em homens e ratos: Contribuição para o conhecimento das assim chamadas esporotricoses. Revista Médica de São Paulo 21: 443–450. [Google Scholar]
- Mackinnon JE. 1943. The dependence on the weather of the incidency of sporotrichosis. Mycopathologia 4: 367–374. [Google Scholar]
- Mackinnon JE, Conti-Díaz IV, Gezuele E, et al. 1969. Isolation of Sporothrix schenckii from nature and considerations on its pathogenicity and ecology. Sabouraudia 7: 38–45. [PubMed] [Google Scholar]
- Macotela-Ruiz E, Nochebuena-Ramos E. 2006. Esporotricosis en algunas comunidades rurales de la sierra norte de Puebla: informe de 55 casos (Septiembre 1995–Diciembre 2005). Gaceta Médica de México 142: 377–380. [PubMed] [Google Scholar]
- Madrid H, Cano J, Gené J, et al. 2009. Sporothrix globosa, a pathogenic fungus with widespread geographical distribution. Revista Iberoamericana de Micologia 26: 218–222. [DOI] [PubMed] [Google Scholar]
- Madrid H, Gené J, Cano J, et al. 2010. Sporothrix brunneoviolacea and Sporothrix dimorphospora, two new members of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 102: 1193–1203. [DOI] [PubMed] [Google Scholar]
- Madrid IM, Mattei AS, Fernandes CG, et al. 2012. Epidemiological findings and laboratory evaluation of sporotrichosis: A description of 103 cases in cats and dogs in southern Brazil. Mycopathologia 173: 265–273. [DOI] [PubMed] [Google Scholar]
- Magand F, Perrot JL, Cambazard F, et al. 2009. Autochthonous cutaneous sporotrichosis in France. Annales de Dermatologie et Véneréologie 136: 273–275. [In French.] [DOI] [PubMed] [Google Scholar]
- Mariat F, Escudié A, Gaxotte P. 1968. Isolement de souches de Ceratocystis sp. À forme conidienne Sporotrichum, de cuirs chevelus humains et de poils de rats. Comparaison avec l’espèce pathogène Sporotrichum schenckii. Comptes Rendus Hebdomaires des Séances de l’Académie de Sience 267: 974–976. [PubMed] [Google Scholar]
- Marimón R, Cano J, Gené J, et al. 2007. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. Journal of Clinical Microbiology 45: 3198–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimón R, Gené J, Cano J, et al. 2006. Molecular phylogeny of Sporothrix schenckii. Journal of Clinical Microbiology 44: 3251–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimón R, Serena C, Gené J, et al. 2008. In vitro antifungal susceptibilities of five species of Sporothrix. Antimicrobial Agents and Chemotherapy 52: 732–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Essayag S, Delgado A, Colella MT, et al. 2013. Epidemiology of sporotrichosis in Venezuela. Internal Journal of Dermatology 52: 974–980. [DOI] [PubMed] [Google Scholar]
- Mayorga R, Cáceres A, Toriello C, et al. 1978. An endemic area of sporotrichosis in Guatemala. Sabouraudia 16: 185–198. [In French.] [PubMed] [Google Scholar]
- Mehta KI, Sharma NL, Kanga AK, et al. 2007. Isolation of Sporothrix schenckii from the environmental sources of cutaneous sporotrichosis patients in Himachal Pradesh, India: results of a pilot study. Mycoses 50: 496–501. [DOI] [PubMed] [Google Scholar]
- Mesa-Arango AC, Rocío Reyes-Montes M del, , Pérez-Mejía A, et al. 2002. Phenotyping and genotyping of Sporothrix schenckii isolates according to geographic origin and clinical form of sporotrichosis. Journal of Clinical Microbiology 40: 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SD, Keeling JH. 2002. Ant sting sporotrichosis. Cutis 69: 439–442. [PubMed] [Google Scholar]
- Moaven LD, Altman SA, Newnham AR. 1999. Sporotrichosis mimicking necrotising arachnidism. Medical Journal of Australia 171: 685–686. [DOI] [PubMed] [Google Scholar]
- Morrison AS, Lockhart SR, Bromley JG, et al. 2013. An environmental Sporothrix as a cause of corneal ulcer. Medical Mycology Case Reports 2: 88–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton PN, Chung WH, Phetsouvanh R, et al. 2005. Sporotrichosis, plain of jars, Lao People’s Democratic Republic. Emerging Infectious Diseases 11: 1496–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg H, Aoki T, et al. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- Padhye AA, Kaufman L, Durry E, et al. 1992. Fatal pulmonary sporotrichosis caused by Sporothrix schenckii var. luriei in India. Journal of Clinical Microbiology 30: 2492–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas PG, Tellez I, Deep AE, et al. 2000. Sporotrichosis in Peru: description of an area of hyperendemicity. Clinical Infectious Diseases 30: 65–70. [DOI] [PubMed] [Google Scholar]
- Pereira SA, Gremião IDF, Kitada AAB, et al. 2014. The epidemiological scenario of feline sporotrichosis in Rio de Janeiro, State of Rio de Janeiro, Brazil. Revista da Sociedade Brasileira de Medicina Tropical. 47: 392–393. [DOI] [PubMed] [Google Scholar]
- Powell KE, Taylor A, Phillips BJ, et al. 1978. Cutaneous sporotrichosis in forestry workers. Epidemic due to contaminated Sphagnum moss. Journal of the American Medical Association 240: 232–235. [PubMed] [Google Scholar]
- Ramírez-Soto MC, Andagua-Castro J, Lizárraga-Trujillo J, et al. 2011. Sporotrichosis in patients attending a reference center in Abancay, Peru. Revista Peruana Medicina Experimental y Salud Publica 28: 508–512. [In Spanish.] [DOI] [PubMed] [Google Scholar]
- Ran YP, Liu DC, Li ZY, et al. 1999. 10,982 cases of fungal skin disease from 1981 to 1997. Chinese Journal of Dermatology 5: 336. [In Chinese.] [Google Scholar]
- Rees RK, Swartzberg JE. 2011. Feline-transmitted sporotrichosis: A case study from California. Dermatol Online Journal 17: 2. [PubMed] [Google Scholar]
- Rodrigues AM, Bagagli E, Camargo ZP de, et al. 2014a Sporothrix schenckii sensu stricto isolated from soil in an armadillo’s burrow. Mycopathologia 177: 199–206. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, Hoog GS de,, Camargo ZP de.2013a. Emergence of pathogenicity in the Sporothrix schenckii complex. Medical Mycology 51: 405–412. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, Hoog GS de, Camargo ZPde. 2014b. Genotyping species of the Sporothrix schenckii complex by PCR-RFLP of calmodulin. Diagnostic Microbiology and Infectious Diseases 78: 383–387. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, Hoog GS de, Cássia Pires D de, et al. 2014c. Genetic diversity and antifungal susceptibility profiles in causative agents of sporotrichosis. BMC Infectious Diseases 14: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AM, Hoog GS de, Zhang Y, et al. 2014d. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerg Microbes Infect 3: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AM, Melo Teixeira M de, Hoog GS de, et al. 2013b. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Neglected Tropical Diseases 7, 6: e2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Mark P, van der, et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa AC da, Scroferneker ML, Vettorato R, et al. 2005. Epidemiology of sporotrichosis: a study of 304 cases in Brazil. Journal of the American Academy Dermatology 52: 451–459. [DOI] [PubMed] [Google Scholar]
- Rubio G, Sanchez G, Porras L, et al. 2010. Esporotricosis: prevalencia, perfil clinico y epidemiologico en un centro de referencia en Colombia. Revista Iberoamericana de Micologia 27: 75–79. [DOI] [PubMed] [Google Scholar]
- Sasaki AA, Fernandes GF, Rodrigues AM, et al. 2014. Chromosomal polymorphism in the Sporothrix schenckii complex. PLoSONE 9: e86819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck BR. 1898. On refractory subcutaneous abscess caused by a fungus possibly related to the Sporotricha. Bulletin of the Johns Hopkins Hospital 9: 286–290. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universalDNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubach A, Barros MB, Schubach TM, et al. 2005. Primary conjunctival sporotrichosis: two cases from a zoonotic epidemic in Rio de Janeiro, Brazil. Cornea 24: 491–493. [DOI] [PubMed] [Google Scholar]
- Schubach A, Barros MB, Wanke B. 2008. Epidemic sporotrichosis. Current Opinions in Infectious Diseases 21: 129–133. [DOI] [PubMed] [Google Scholar]
- Schubach TM, Schubach A, Okamoto T, et al. 2004. Evaluation of an epidemic of sporotrichosis in cats: 347 cases (1998–2001). Journal of the American Veterinary Medical Association 224: 1623–1629. [DOI] [PubMed] [Google Scholar]
- Silva MB, Costa MM, Torres CC, et al. 2012. Urban sporotrichosis: a neglected epidemic in Rio de Janeiro, Brazil. Cadernos Saúde Pública 28: 1867–1880. [In Portuguese.] [DOI] [PubMed] [Google Scholar]
- Silva-Vergara ML, Camargo ZP de, Silva PF, et al. 2012. Disseminated Sporothrix brasiliensis infection with endocardial and ocular involvement in an HIV-infected patient. American Journal of Tropical Medicine and Hygiene 86: 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam S, Bannan AM, Chen SC, et al. 2012. Sporotrichosis (Sporothrix schenckii infection) in the New South Wales mid-north coast, 2000–2010. Medical Journal of Australia 196: 588–590. [DOI] [PubMed] [Google Scholar]
- Song J, Li HY, Yang XH. 1998. 108 cases children sporotrichosis analysis report. The Chinese Journal of Dermatovenereology 4: 230. [In Chinese.] [Google Scholar]
- Song Y, Li SS, Zhong SX, et al. 2013. Report of 457 sporotrichosis cases from Jilin province, northeast China, a serious endemic region. Journal of the European Academy of Dermatology and Venereology 27: 313–318. [DOI] [PubMed] [Google Scholar]
- Song Y, Yao L, Zhong SX, et al. 2011. Infant sporotrichosis in northeast China: a report of 15 cases. International Journal of Dermatology 50: 522–529. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web Servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stopiglia CDO, Magagnin CM, Castrillón MR, et al. 2014. Antifungal susceptibilities and identification of species of the Sporothrix schenckii complex isolated in Brazil. Medical Mycology 52: 56–64. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Sato S, Nishimoto K. 2009. Survey of 155 sporotrichosis cases examined in Nagasaki Prefecture from 1951 to 2007. Nippon Ishinkin Gakkai Zasshi 50: 101–108. [DOI] [PubMed] [Google Scholar]
- Tan JW, Liu W, Wan Z, et al. 2013. Reclassification of 33 clinical strains of Sporothrix from northern China based on phenotypic and molecular characters. Mycosystema 32: 161–167. [In Chinese.] [Google Scholar]
- Tang MM, Tang JJ, Gill P, et al. 2012. Cutaneous sporotrichosis: a six-year review of 19 cases in a tertiary referral center in Malaysia. International Journal of Dermatology 51: 702–708. [DOI] [PubMed] [Google Scholar]
- Teixeira M, Rodrigues AM, Tsui CKM, et al. In press. Asexual propagation of a virulent clone complex in human and feline outbreak of sporotrichosis. Eukaryotic Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DW, Kaplan W. 1977. Laboratory-acquired sporotrichosis. Sabouraudia 15: 167–170. [DOI] [PubMed] [Google Scholar]
- Tilak R, Kumari V, Bansal M, et al. 2012. Lymphocutaneous sporotrichosis in an adolescent girl presenting as mycetoma. The International Journal of Low Extremity Wounds 11: 184–186. [DOI] [PubMed] [Google Scholar]
- Tlougan BE, Podjasek JO, Patel SP, et al. 2009. Neonatal sporotrichosis. Pediatric Dermatology 26: 563–565. [DOI] [PubMed] [Google Scholar]
- Travassos LR, Lloyd KO. 1980. Sporothrix schenckii and related species of Ceratocystis. Microbiological Reviews 44: 683–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Verma GK, Singh G, et al. 2012. Sporotrichosis in sub-himalayan India. PLoS Neglected Tropical Diseases 6: e1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela R, Souza GF, Fernandes Cota G, et al. 2007. Cutaneous and meningeal sporotrichosis in a HIV patient. Revista Iberoamericana de Micologia 24: 161–163. [DOI] [PubMed] [Google Scholar]
- Vismer HF, Hull PR. 1997. Prevalence, epidemiology and geographical distribution of Sporothrix schenckii infections in Gauteng, South Africa.Mycopathologia 137: 137–143. [DOI] [PubMed] [Google Scholar]
- Wang ZC, Sun BF. 1982. Investigation report of incidence of sporotrichosis in Jilin province. Journal of Jilin University (Medical Edition) 3: 82–83. [In Chinese.] [Google Scholar]
- Whittemore JC, Webb CB. 2007. Successful treatment of nasal sporotrichosis in a dog. Canadian Veterinary Journal 48: 411–414. [PMC free article] [PubMed] [Google Scholar]
- Wu SX. 1986. Analysis report of 131 cases of sporotrichosis in elderly people and children, Chinese Journal of Dermatology 19, 4: 199–200. [In Chinese.] [Google Scholar]
- Xia JX, Mu Y, Pan SS, et al. 2009. Clinical analysis of 10 cases of nasal fixed sporotrichosis. Chinese Journal of Mycology 4: 353–354. [In Chinese.] [Google Scholar]
- Yang FZ, Li YZ, Yao JY. 2005. Clinical analysis of 224 cases of cutaneous sporotrichosis. Chinese Journal of Leprosy Skin Disease 21: 59–60. [In Chinese.] [Google Scholar]
- Yegneswaran PP, Sripathi H, Bairy I, et al. 2009. Zoonotic sporotrichosis of lymphocutaneous type in a man acquired from a domesticated feline source: report of a first case in southern Karnataka, India. International Journal of Dermatology 48: 1198–1200. [DOI] [PubMed] [Google Scholar]
- Yu X, Wan Z, Zhang Z, et al. 2013. Phenotypic and molecular identification of Sporothrix isolates of clinical origin in Northeast China. Mycopathologia 176: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamri-Saad M, Salmiyah TS, Jasni S, et al. 1990. Feline sporotrichosis: an increasingly important zoonotic disease in Malaysia. Veterinary Record 127: 480. [PubMed] [Google Scholar]
- Zhang JD, Lin JP. 2008. Clinical analysis of 316 cases of cutaneous sporotrichosis. Chinese Journal of Mycology 3: 207–210. [In Chinese.] [Google Scholar]
- Zhou X, Rodrigues AM, Feng P, et al. 2013. GlobalITS diversity in the Sporothrix schenckii complex. Fungal Diversity 60: 1–13. [Google Scholar]