Abstract
Thirteen morphologically similar strains of barbatosphaeria- and tectonidula-like fungi were studied based on the comparison of cultural and morphological features of sexual and asexual morphs and phylogenetic analyses of five nuclear loci, i.e. internal transcribed spacer rDNA operon (ITS), large and small subunit nuclear ribosomal DNA, β-tubulin, and second largest subunit of RNA polymerase II. Phylogenetic results were supported by in-depth comparative analyses of common core secondary structure of ITS1 and ITS2 in all strains and the identification of non-conserved, co-evolving nucleotides that maintain base pairing in the RNA transcript. Barbatosphaeria is defined as a well-supported monophyletic clade comprising several lineages and is placed in the Sordariomycetes incertae sedis. The genus is expanded to encompass nine species with both septate and non-septate ascospores in clavate, stipitate asci with a non-amyloid apical annulus and non-stromatic ascomata with a long decumbent neck and carbonised wall often covered by pubescence. The asexual morphs are dematiaceous hyphomycetes with holoblastic conidiogenesis belonging to Ramichloridium and Sporothrix types. The morphologically similar Tectonidula, represented by the type species T. hippocrepida, grouped with members of Barbatosphaeria and is transferred to that genus. Four new species are introduced and three new combinations in Barbatosphaeria are proposed. A dichotomous key to species accepted in the genus is provided.
Keywords: phylogenetics, Ramichloridium, sequence analysis, spacer regions, Sporothrix, Tectonidula
INTRODUCTION
Barbatosphaeria is a non-stromatic perithecial ascomycete introduced for a single species Barbatosphaeria barbirostris and accommodated in the Sordariomycetes incertae sedis based on nuc28S rDNA sequences (Réblová 2007). The genus was erected for calosphaeria-like fungi characterised by dark ascomata growing between cortex and wood with long decumbent necks, often covered with red-brown pubescence; ellipsoid to oblong, hyaline, septate ascospores in unitunicate, clavate asci with a non-amyloid apical annulus. The asexual morphs are dematiaceous hyphomycetes with holoblastic denticulate conidiogenesis and resemble Ramichloridium and Sporothrix. Delimitation of Barbatosphaeria and its separation from the morphologically similar but distantly related members of the Calosphaeriales (Réblová et al. 2004, Vijaykrishna et al. 2004, Damm et al. 2008) entailed an understanding of life history, discovery of asexual morphs and revision of subtle morphological characters of asci and ascogenous hyphae.
Barbatosphaeria is closely related to the morphologically similar genus Tectonidula (Réblová & Štěpánek 2009), which differs from the former in the non-septate, allantoid to U- or horseshoe-shaped ascospores. Samuels & Candoussau (1996) hypothesised that the feature of allantoid ascospores is not common among perithecial ascomycetes and may contain to a certain extent phylogenetic information. In the phylogenetic analysis based on nuc28S rDNA sequences, both genera were grouped in one clade, but without significant statistical support (Réblová & Štěpánek 2009). At that time, both molecular and morphological data of the two type species seemed to support and warrant the distinction of two genera.
Several barbatosphaeria- and tectonidula-like fungi were collected in the years 2003–2013. They are characterised by an inconspicuous habitus, i.e. minute ascomata often arranged in circular or semi-circular groups under the periderm and usually collected incidentally when searching for other periderm-inhabiting fungi. Thirteen strains of these morphologically similar fungi were obtained in living culture. They can be divided into different groups based on sexual characters, primarily on morphology of the ascospores. They include isolates with septate and non-septate ascospores, ranging from ellipsoid, oblong, subcylindrical and curved to suballantoid, allantoid, U- to horseshoe-shaped or ¾ circular. These ascospore morpho-types represent a continuum between two extremes corresponding to the two type species of the previously recognised genera Barbatosphaeria and Tectonidula. Three recently collected strains match the description of Calosphaeria dryina, two belong to B. barbirostris and one to Tectonidula hippocrepida. The other seven strains represent new taxa and exhibit an undescribed range of ascospore morphology of members of Barbatosphaeria.
It has proven difficult to evaluate these morphologically defined groups of strains based on conventional use of pairwise comparison of sequences with the divergence expressed in percentages. Such an approach does not reveal changes in nucleotides at the RNA structural level and their role in the RNA transcript. The study of variability within each group entailed a comparative analysis of 2D (secondary) structure of internal transcribed spacers ITS1 and ITS2 and application of the CBC species concept that has been used to delimit biological species (Coleman & Mai 1997, Coleman 2000). This concept is based on evolutionary processes at the RNA structural level that are responsible for maintaining the arrangement of base pairs in the RNA transcript and preserving the structure of the RNA helices, i.e. the compensatory base change (CBC) and hemi-compensatory base change (hCBC). On the other hand, any non-compensatory base change (non-CBC) may lead to a disruption of the stem structure (Leontis et al. 2002). The ITS1 and ITS2 loci represent rapidly evolving regions of the rDNA operon and display high sequence variability. Although they are non-coding, they include functionally constrained positions required for the folding of their transcripts that allow their own splicing and the correct processing of the rDNA genes (Côté et al. 2002). The ITS2 has been shown to retain a common core structure with several hallmarks in its RNA transcript that are evolutionarily constrained and universal among eukaryotes (Mai & Coleman 1997, Joseph et al. 1999, Coleman 2007). They also have proven useful for molecular taxonomic concepts (Coleman & Mai 1997).
We investigated the variability among 13 strains by comparing cultural and morphological characteristics of sexual and asexual morphs and phylogenetic analyses of five nuclear loci, i.e. the internal transcribed spacer rDNA operon (ITS), large and small subunit nuclear ribosomal DNA (nuc28S rDNA, nuc18S rDNA), β-tubulin, and second largest subunit of RNA polymerase II (rpb2). In addition, we performed in-depth comparative analyses of common core secondary structures of ITS1 and ITS2 of all strains and searched for non-conserved co-evolving nucleotides that maintain base pairing in the RNA transcript. All existing substitutions were mapped onto the predicted 2D models of ITS1 and ITS2 of B. barbirostris and evaluated.
MATERIALS AND METHODS
Herbarium material and fungal strains
Dry ascomata were rehydrated with water; material was examined with an Olympus SZX12 dissecting microscope and centrum material (including asci, ascospores and paraphyses) was mounted in Melzer’s reagent, 90 % lactic acid, lactophenol with cotton blue or aqueous cotton-blue (1 mg/mL). Hand sections of the ascomatal wall were studied in 3 % KOH. All measurements were made in Melzer’s reagent. Means ± standard deviations (SD) based on 20–25 measurements are given for dimensions of asci, ascospores, conidia and conidiogenous cells. Images were captured by differential interference (DIC) or phase contrast (PC) microscopy using an Olympus DP70 Camera operated by Imaging Software Cell on an Olympus BX51 compound microscope.
Multi-ascospore isolates were obtained from fresh material with the aid of a spore isolator (Meopta, Prague, Czech Republic). Ascospores and asci were spread on water agar, ascospores germinated within 48 h. Germinating ascospores were transferred and isolates were grown on potato-dextrose agar (PDA, Oxoid) and potato-carrot agar (PCA, Gams et al. 1998). Colonies were examined after 7, 21 and 30 d incubated at 25 °C in the dark. Cultures are maintained at CBS (CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands). Type material is deposited in PRM herbarium (National Museum in Prague). The Online auction colour chart (2004) was used as the colour standard.
DNA extraction, amplification and sequence alignment
Cultures used for DNA isolations were grown as previously described by Réblová et al. (2011) and DNA was extracted following the protocols of Lee & Taylor (1990). Procedures for amplifying and sequencing the ITS, nuc18S, nuc28S and rpb2 were performed as described in Réblová et al. (2011). A fragment of the 5’-end of the β-tubulin gene region was amplified and sequenced using primers Bt2a and Bt2b (Glass & Donaldson 1995) or benA1 (Geiser et al. 1998) and Bt2b. Sequences were edited using Sequencher v. 5.0 software (Gene Codes Corp., Ann Arbor, MI, USA).
GenBank accession numbers for newly sequenced taxa and other homologous sequences of members of the Sordariomycetes retrieved from GenBank are listed in Table 1. Sequences were manually aligned in BioEdit v. 7.0.9.0 (Hall 1999). 2D models of the ITS1 and ITS2 obtained for individual members of Barbatosphaeria were used to determine the positions of homologous nucleotides in the ITS. The nuc18S and nuc28S alignments were enhanced by utilizing the homologous 2D structure of Saccharomyces cerevisiae (Gutell 1993, Gutell et al. 1993) in order to improve the decisions on homologous characters and introduction of gaps. These procedures and alignment of the protein-coding genes were performed as described in Réblová & Réblová (2013).
Table 1.
A list of fungi, isolate information and new sequences determined for this study and those retrieved from GeneBank. The asterisk (*) denotes ex-type strains of Barbatosphaeria. GenBank accession numbers in bold were generated for this study.
| Classification | Taxon | Source | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|
| ITS | β-tubulin | nuc28S | nuc18S | rpb2 | |||
| Sordariomycetes | |||||||
| genera incertae sedis | Barbatosphaeria arboricola | CBS 114120 | KM492885 | KM492872 | KM492861 | KM492848 | KM492900 |
| CBS 127689* | KM492887 | KM492874 | KM492862 | KM492849 | KM492901 | ||
| CBS 127690 | KM492886 | KM492873 | HQ878592 | HQ878596 | HQ878602 | ||
| Barbatosphaeria barbirostris | CBS 121149 | KM492889 | KM492876 | EF577059 | KM492851 | KM492903 | |
| M.R. 3767 | KM492888 | KM492875 | KM492863 | KM492850 | KM492902 | ||
| Barbatosphaeria dryina | CBS 127691 | KM492890 | KM492877 | KM492864 | KM492852 | KM492904 | |
| CBS 137796 | KM492891 | KM492878 | KM492865 | KM492853 | KM492905 | ||
| CBS 137798 | KM492892 | KM492879 | KM492866 | KM492854 | – | ||
| Barbatosphaeria fimbriata | M.R. 3694 | KM492893 | KM492880 | KM492867 | KM492855 | KM492906 | |
| Barbatosphaeria hippocrepida | ICMP 17630* | KM492894 | KM492881 | FJ617557 | HQ878599 | HQ878608 | |
| Barbatosphaeria neglecta | CBS 127693* | KM492895 | KM492882 | KM492868 | KM492856 | – | |
| Barbatosphaeria sp | M.R. 3730 | KM492897 | KM492884 | KM492870 | KM492858 | KM492908 | |
| Barbatosphaeria varioseptata | CBS 137797* | KM492896 | KM492883 | KM492869 | KM492857 | KM492907 | |
| Ceratostomella pyrenaica | CBS 117116 | – | – | DQ076323 | DQ076324 | – | |
| Cryptadelphia groenendalensis | DAOM 231136 | – | – | AY281104 | – | – | |
| Lentomitella cirrhosa | ICMP 15131 | – | – | AY761085 | AY761089 | KM492911 | |
| Lentomitella crinigera | CBS 113655 | – | – | AY761086 | – | – | |
| Lentomitella pallibrunnea | SMH 2534 | – | – | EU527993 | – | – | |
| Lentomitella tropica | SMH 3225 | – | – | EU527999 | – | – | |
| Natantiella ligneola | CBS 123410 | KM492899 | – | FJ617555 | KM492859 | KM492909 | |
| CBS 123470 | KM492898 | – | FJ617556 | HQ878598 | HQ878605 | ||
| Rhamphoria delicatula | CBS 132724 | – | – | FJ617561 | JX066711 | JX066702 | |
| Rhodoveronaea varioseptata | CBS 123473 | – | – | FJ617560 | JX066710 | JX066700 | |
| Xylomelasma sordida | CBS 131683 | – | – | KM492871 | KM492860 | KM492910 | |
| Annulatascaceae | Annulatascus velatisporus | A70-18 | – | – | AY316354 | – | – |
| Annulusmagnus triseptatus | CBS 131483, CBS 128831 | – | – | GQ996540 | JQ429242 | JQ429258 | |
| Ascitendus austriascus | CBS 131685 | – | – | GQ996539 | GQ996542 | JQ429257 | |
| Boliniales | Camarops microspora | CBS 649.92 | – | – | AY083821 | DQ471036 | DQ470937 |
| Cornipulvina ellipsoides | SMH 1378 | – | – | DQ231441 | – | – | |
| Calosphaeriales | Calosphaeria pulchella | CBS 115999 | – | – | AY761075 | AY761071 | GU180661 |
| Jattaea algeriensis | STE-U 6399, CBS 120871 | – | – | EU367457 | EU367462 | HQ878603 | |
| Togniniella microspora | CBS 113648 | – | – | AY761076 | AY761073 | GU180660 | |
| Chaetosphaeriales | Chaetosphaeria ciliata | ICMP 18253 | – | – | GU180637 | GU180614 | GU180659 |
| Chaetosphaeria curvispora | ICMP 18255 | – | – | GU180636 | AY502933 | GU180655 | |
| Coniochaetales | Coniochaeta discoidea | SANK 12878, CBS 158.80 | – | – | AY346297 | AJ875179 | AY780191 |
| Coniochaeta ostrea | CBS 507.70 | – | – | DQ470959 | DQ471007 | DQ470909 | |
| Coronophorales | Bertia moriformis | SMH 3344, SMH 4320 | – | – | AY695261 | – | AY780151 |
| Chaetosphaerella phaeostroma | SMH 4585 | – | – | AY346274 | – | AY780172 | |
| Diaporthales | Diaporthe phaseolorum | FAU 458, NRRL 13736 | – | – | U47830 | L36985 | AY641036 |
| Gnomonia gnomon | CBS 199.53 | – | – | AF408361 | DQ471019 | DQ470922 | |
| Valsa ambiens | AR 3516 | – | – | AF362564 | DQ862056 | DQ862025 | |
| Glomerellales | Glomerella cingulata | MCA 2498, FAU 513 | – | – | DQ286199 | M55640 | DQ858455 |
| Kylindria peruamazonensis | CBS 838.91 | – | – | GU180638 | GU180609 | GU180656 | |
| Monilochaetes infuscans | CBS 379.77 | – | – | GU180645 | GU180619 | GU180658 | |
| Reticulascus clavatus | CBS 125296 | – | – | GU180643 | GU180622 | – | |
| Hypocreales | Pseudonectria rousseliana | AR 2716, CBS 114049 | – | – | U17416 | AF543767 | DQ522459 |
| Trichoderma viride | GJS 89-127, IFFI 13001 | – | – | AY489726 | AF525230 | EU252006 | |
| Virgatospora echinofibrosa | CBS 110115 | – | – | AY489724 | AY489692 | EF692516 | |
| Magnaporthales | Gaeumannomyces graminis | AR 3401, M 57 | – | – | AF362557 | JF414874 | – |
| Magnaporthe grisea | Ina168, 70-15 | – | – | AB026819 | DQ493955 | – | |
| Muraeriata africana | GKM 1084 | – | – | EU527995 | – | – | |
| Melanosporales | Melanospora tiffanii | ATCC 15515 | – | – | AY015630 | AY01561 | AY015637 |
| Melanospora zamiae | ATCC 12340, CBS 421.87 | – | – | AY046579 | AY046578 | DQ368634 | |
| Microascales | Corollospora maritima | AFTOL 5011, JK 4834 | – | – | FJ176901 | U46871 | DQ368632 |
| Lignincola laevis | AFTOL 737, A169-1D | – | – | U46890 | AF050487 | DQ836886 | |
| Microascus trigonosporus | BS 218.31, ATCC52470 | – | – | DQ470958 | DQ471006 | AF107792 | |
| Petriella setifera | CCFC 226737, CBS 385.87, | – | – | AY281100 | U43908 | DQ368640 | |
| CBS 110344 | |||||||
| Ophiostomatales | Ophiostoma piliferum | DAOM 226737, CBS 129.32, | – | – | AY281094 | AJ243295 | DQ470905 |
| CBS 158.74 | |||||||
| Ophiostoma stenoceras | CBS 139.51 | – | – | DQ836904 | DQ836897 | DQ836891 | |
| Papulosaceae | Brunneosporella aquatica | HKUCC 3708 | – | – | AF132326 | – | – |
| Fluminicola coronata | HKUCC 3717 | – | – | AF132332 | – | – | |
| Papulosa amerospora | JK 5547F | – | – | DQ470950 | DQ470998 | DQ470901 | |
| Sordariales | Gelasinospora tetrasperma | CBS 178.33 | – | – | DQ470980 | DQ471032 | DQ470932 |
| Lasiosphaeria ovina | SMH 1538, CBS 958.72 | – | – | AF064643 | AY083799 | AY600292 | |
| Sordaria fimicola | SMH 4106, MUCL 937, CBS 723.96 | – | – | AY780079 | X69851 | DQ368647 | |
| Xylariales | Anthostomella torosa | JK 5678E | – | – | DQ836902 | DQ836895 | DQ836885 |
| Graphostroma platystoma | CBS 270.87 | – | – | DQ836906 | DQ836900 | DQ836893 | |
| Xylaria hypoxylon | AFTOL 51 | – | – | AY544648 | AY544692 | DQ470878 | |
| Leotiomycetes | |||||||
| Helotiales | Leotia lubrica | AFTOL 1, isolate unknown for nuc18S | – | – | AY544644 | L37536 | DQ470876 |
| Microglossum rufum | AFTOL 1292 | – | – | DQ470981 | DQ471033 | DQ470933 | |
Two sequence alignments were constructed: 1) ITS combined with β-tubulin; and 2) nuc18S and nuc28S combined with rpb2. Individual alignments were concatenated to combine sequences for the bi- and multilocus alignments. Alignments are deposited in TreeBASE (Study no. 16315).
Phylogenetic analyses
Phylogenetic relationships were resolved based on analyses of ITS, β-tubulin, nuc18S, nuc28S, and rpb2 sequences of 13 isolates representing eight species of Barbatosphaeria. We analysed the first 2/3 of the 5’ half of the nuc28S (corresponding to the first 1 197 nucleotides of S. cerevisiae), the almost entire nuc18S, and the 5–7 segments of the rpb2. Bases 1–137 of the nuc18S, 1–76 of the nuc28S, 1–60 of the rpb2 alignments were excluded from analyses because of the incompleteness of the 5’-end of the majority of the available sequences. The coding regions (exons) 4, 5 and partly 6 of the β-tubulin in total length of 262 nucleotides were analysed, non-coding regions were excluded. All characters of the ITS sequences were included. Two outgroups were used for the two phylogenies. Two members of the Leotiomycetes, Leotia lubrica and Microglossum rufum, were used to root the multilocus phylogeny. Two strains of Natantiella ligneola were used to root the ITS-β-tubulin phylogeny.
Each combined dataset was partitioned into several subsets of nucleotide sites. The ITS-β-tubulin analysis was partitioned into ITS; the three exons of β-tubulin were analysed together and partitioned into first, second and third codon positions. The three-gene dataset was partitioned into nuc28S, nuc18S, and first, second and third codon positions of rpb2.
The program MrModeltest2 v. 2.3 (Nylander 2008) was used to infer the appropriate substitution model that would best fit the model of DNA evolution for each sequence dataset and each partition of the combined datasets. Maximum likelihood (ML) and Bayesian inference (BI) analyses were used to estimate phylogenetic relationships. ML analysis was performed with RAxML-HPC v. 7.0.3 (Stamatakis et al. 2005, Stamatakis 2006) with a GTRCAT model of evolution. Nodal support was determined by non-parametric bootstrapping (BS) with 1 000 replicates.
The BI analysis was performed in a likelihood framework as implemented in MrBayes v. 3.0b4 software package to reconstruct phylogenetic trees (Huelsenbeck & Ronquist 2001). For the ITS dataset we used SYM+I+G substitution model. For the β-tubulin dataset we used GTR+I substitution model for the first and second codon position, and GTR model for the third codon position. For the combined nuc18S, nuc28S, and rpb2 dataset we used for each partition the GTR+I+G substitution model. Two Bayesian searches were performed using the default parameters. Analyses were run for 10 M generations, with trees sampled every 1 000 generations. Tracer v. 1.6.0. (Rambaut et al. 2013) was used to confirm convergence of trees and burn-in. The first 50 000 trees, which represented the burn-in phase of the analysis, were discarded. The remaining trees were used for calculating posterior probabilities (PP) of recovered branches (Larget & Simon 1999).
Prediction of 2D structure models of ITS1 and ITS2
Predicting the 2D structure of the ITS is essential for constructing a reliable multiple sequence alignment to compare nucleotides at homologous positions (in helices and loops) while searching for non-conserved co-evolving nucleotides that maintain base pairing. Consensus 2D structure models for the ITS1 and ITS2 were built using the PPfold program v. 3.0 (Sukosd et al. 2012), which uses an explicit evolutionary model and a probabilistic model of structures, and relies on multiple sequence alignment of related RNA sequences. In the case of ITS1 we obtained a rough structure with distinguished helices H1 and H3. Final 2D models created for all members of Barbatosphaeria were further improved using the Mfold program (Zuker 2003) and then adjusted manually if necessary, based on comparison of homologous positions in the alignment. In the case of ITS2 the rough structure built using the PPfold program contained only H1 and a tip of H3. This was probably because the input multiple sequence alignment containing thirteen sequences possessed little evolutionary signal. Therefore, we used the ITS2 Database III (Koetschan et al. 2010) to build 2D structures for members of this genus. Obtained structures contained the same H1 and tip of H3 as the rough structure built using the PPfold program. These models were further improved using the Mfold program and then adjusted manually based on comparison of homologous positions in the alignment similarly to ITS1.
The predicted 2D RNA structures of ITS1 and ITS2 were obtained in a dot bracket notation and were visualized and drawn using VARNA: Visualisation Applet for RNA program (Darty et al. 2009).
Identification and mapping of base-pair changes in 2D structures of ITS
We identified three types of substitutions in the aligned ITS sequences. The first involves compensatory base changes (double-sided substitution, CBCs) that occur when both nucleotides of a paired site mutate (i.e. G=C ↔ C=G, A-U or U-A). Because these canonical base pairs are isosteric (Leontis et al. 2002) they can be substituted for each other within stems without causing structural perturbations. The second type of substitution, the hemi-compensatory base change (one-sided substitution, hCBC), entails the change of a canonical base pair to a ‘wobble’ base pair (i.e. G=C → G/U). This type of substitution, which is classified as near-isosteric (Stombaugh et al. 2009), retains base pairing in RNA molecules but produces minor structural perturbations in the helix structure because the canonical G=C pair and wobble G/U pair are not members of the same isosteric family (Leontis et al. 2002). Finally, we identified non-compensatory base changes (non-CBCs) that involve the replacement of a canonical or a wobble pair with any non-canonical pair. Once all existing substitutions were identified among analysed strains, the unique substitutions were mapped onto the 2D structures of predicted models of ITS1 and ITS2 of B. barbirostris.
RESULTS
Phylogenetic results
In the first analysis, the alignment consisted of 70 combined nuc18S, nuc28S and rpb2 sequences of members of the Sordariomycetes, each with 5 028 characters after introduction of gaps. The alignment had 2 390 distinct alignment patterns (ML analysis conducted by RAxML); the ML tree is shown in Fig. 1. The genus Barbatosphaeria was resolved as a robust monophyletic clade (99 % ML BS / 1.0 PP) comprising several lineages that correspond to seven species. Tectonidula hippocrepida, the type species of the genus, grouped within the Barbatosphaeria clade at a basal position. The closest relatives to Barbatosphaeria are Ceratostomella and Xylomelasma, two genera with non-stromatic ascomata, long neck, pale brown non-septate ascospores and unknown asexual morphs, and a clade comprising members of the Ophiostomatales and Natantiella.
Fig. 1.
Multilocus phylogenetic analysis of the nuc18S-nuc28S-rpb2 sequences of Barbatosphaeria and members of the Sordariomycetes. Phylogram inferred from the ML analysis with RAxML using a GTRCAT model of evolution. Only high branch support is shown at the nodes, maximum likelihood bootstrap support (ML BS) ≥ 85% and Bayesian posterior probability (PP) ≥ 95%). Branches with a black circle symbol indicate ML BS = 100%, PP values = 1.0. Taxa in bold refer to new combinations or new taxonomic treatments; ‘T’ refers to ex-type strains of species of Barbatosphaeria
In the second analysis, thirteen combined ITS and β-tubulin sequences were assessed for eight Barbatosphaeria species. The multiple sequence alignment consisted of 1 140 characters after introduction of gaps. The alignment had 229 distinct alignment patterns (ML analysis); the ML tree is shown in Fig. 2. The genus Barbatosphaeria is shown as a strongly supported monophyletic clade (100/1.0). Tectonidula hippocrepida groups in the genus in a basal position; it is closely related to two other Barbatosphaeria species with non-septate ascospores.
Fig. 2.
Phylogenetic analysis of the combined ITS rDNA and β-tubulin sequences of Barbatosphaeria. Phylogram inferred from the ITS rDNA sequences with ML analysis using a GTRCAT model of evolution. Legend as in Fig. 1.
Consensus 2D structure of ITS1
The predicted 2D structure of ITS1 shown in Fig. 3 is modelled for B. barbirostris. The consensus 2D structure of ITS1 consisted of three helices and a variable single-stranded segment at the 3’-end. The 3’-end did not fold into a homologous structure in all strains analysed and could not be compared among taxa.
Fig. 3.
A partial secondary structure of ITS1 rRNA molecule of B. barbirostris showing three helices (H1–H3). All substitutions recorded among members of Barbatosphaeria are mapped on the 2D model. Parts of hairpin loops and three-way junctions highlighted with grey colour represent regions with variable number of nucleotides. Taxa highlighted with green colour refer to members of the B. arboricola clade.
The consensus 2D model of ITS1 contains three helices (H1, H2 and H3) separated by an adjacent single-stranded (junction) region. H1 is the most conserved of all three helices with a single substitution in the hairpin loop. H2 is highly variable; it exhibits one symmetrical and one asymmetrical internal loop and a hairpin loop. The differences highlighted in yellow colour in Fig. 3 correlate with ascospore septation. The changes at the bottom of the H2 stem, i.e. hCBC substitution of the first C=G pair by a newly formed U/G pair and a substitution of AAA by UUCU in the second internal loop on the 3’side of the H2 are present only in taxa with non-septate ascospores. H3 is the longest and the only branched duplex in the ITS1 consisting of a three-way junction. The two hairpin loops exhibit sequence variation. In the three arms labelled A, B and C two bp with CBCs, one with both CBC and hCBC, four with hCBCs and one with both hCBC and non-CBC substitutions are shown.
Consensus 2D structure of ITS2
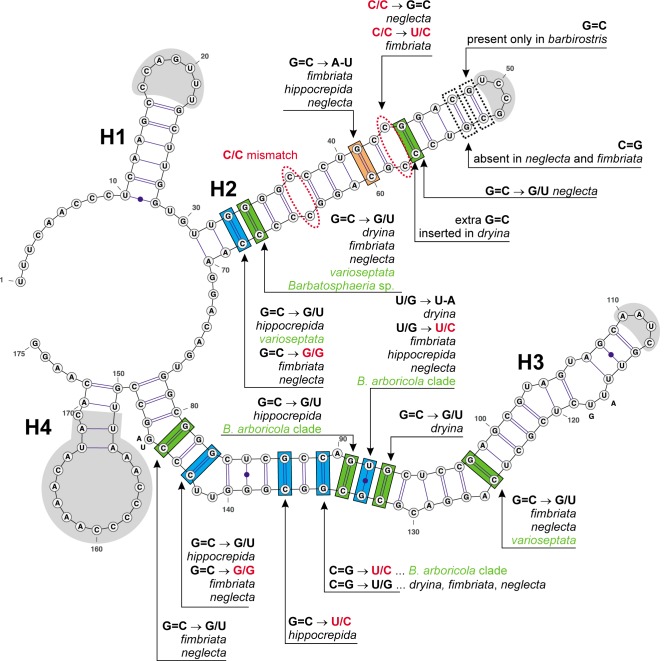
The consensus secondary structure of the ITS2 molecule mapped onto a predicted model of B. barbirostris (Fig. 4) is folded into a ring structure with four main helices (H1–H4) separated by adjacent single-stranded regions. H1 consists of a conserved stem and a hairpin loop, which is variable. H2 is composed of a long stem structure with two C/C (pyrimidine/pyrimidine) mismatches, of which only the first is conserved in all taxa. Barbatosphaeria barbirostris possesses the longest H2. This duplex contains the only CBC substitutions detected in ITS2, two bp with hCBC and one with both hCBC and non-CBC. H3 is the longest duplex in the ITS2. It consists of one internal loop, six bulges and a hairpin loop. This helix contains four bp with hCBCs, three with both hCBCs and non-CBCs and one with a single non-CBC substitution. The hairpin loop is of a variable length. The fourth duplex H4 is short, having the first two bp conserved in the majority of species; the remaining part of the stem and a hairpin loop show significant variability.
Fig. 4.
A partial secondary structure of ITS2 rRNA molecule of B. barbirostris showing four helices (H1–H4). All substitutions recorded among members of Barbatosphaeria are mapped on the 2D model. Parts of hairpin loops highlighted with grey colour represent regions with variable number of nucleotides. Legend to symbols and colours as in Fig. 3.
The variability of the B. arboricola clade at the RNA structural level
This clade comprises five morphologically similar strains belonging to B. arboricola, B. varioseptata and Barbatosphaeria sp. While three strains of B. arboricola always had 1-septate ascospores, in B. varioseptata and Barbatosphaeria sp. the ascospores were characterised by a delayed formation of one or two additional septa followed by increased length in the former.
Two ITS sequences of B. arboricola, CBS 114120 and CBS 127690, were identical and showed 99.8 % similarity with the ITS sequence of the ex-type strain CBS 127689. A 5.6 % divergence was found between ITS sequences of B. arboricola (ex-type strain) and B. varioseptata and 4.2 % divergence between B. arboricola and Barbatosphaeria sp.
In the ITS1, one CBC is unique for all strains of this species-complex, and one hCBC is present among strains of B. arboricola (in CBS 127689, Fig. 3). The single CBC present in H2 and H3 of ITS2 is shared among Barbatosphaeria species with septate ascospores. On the other hand, in the B. arboricola clade several hCBCs vary significantly among strains linked to sexual morphs with 1-septate and 1–3-septate ascospores, respectively (Fig. 4), and support the existence of at least two species. Barbatosphaeria varioseptata is characterized by three hCBCs and Barbatosphaeria sp. by one hCBC compared to B. arboricola. The other differences among species of B. arboricola clade exist in the variable areas of a hairpin loop of H3 of ITS1 and of hairpin loops of H1, H2 and H4 of ITS2.
The variability of the B. neglecta clade at the RNA structural level
The B. neglecta clade comprises two species, i.e. B. fimbriata and B. neglecta. Their ITS sequences showed 5.0 % divergence. The whole clade is characterized by one CBC in ITS1 (shared with B. hippocrepida), four hCBCs and three non-CBCs in ITS2. In ITS1 we detected one CBC and three hCBCs in H2 and H3 that characterise these strains (Fig. 3, 4). In ITS2, in a position of the second C/C mismatch in H2 (Fig. 4), the C/C pair changes into a canonical pair C=G in B. neglecta, while in B. fimbriata (M.R. 3694), the substitution leads to another non-canonical pair U/C. In H2 we also see a single hCBC that characterizes B. neglecta. Other differences are in the stem of H4 of ITS2. Following the premise of the CBC species concept, these two strains cannot be regarded conspecific and they were introduced as two individual, though closely related species. This conclusion is supported by differences in the sexual morphology. Both species clearly differ in the size of asci and ascospores.
TAXONOMY
Barbatosphaeria Réblová, Mycologia 99: 727. 2007
= Tectonidula Réblová, Mycol. Res. 113: 998. 2009.
Asexual morphs. Ramichloridium- and sporothrix-like.
Ascomata non-stromatic dark brown to black, usually aggregated in circular groups, occasionally in rows or solitary, growing between periderm and wood, often associated with old stromata of other ascomycetes. When in circular groups, venter lying horizontally, necks are decumbent to perpendicular, converging, piercing the periderm in a group. Venter globose to subglobose, easily detached from the substratum, with cylindrical neck. Venter and neck are sparsely covered with dark olive to red-brown pubescence that disappears with age, mostly necks and bottom part of venter remain covered with hairs. Ascomatal wall leathery to fragile, 2-layered. Ostiolum periphysate. Paraphyses abundant, persistent, longer than the asci. Asci arising from croziers on elongating ascogenous hyphae, floating freely in the centrum at maturity; unitunicate, clavate, tapering toward the slender stipe; basal part of the stipe conspicuously swollen, often remaining attached to the ascogenous hyphae after the mature ascus is liberated. Ascospores ellipsoid, oblong, subcylindrical, straight, sometimes curved, allantoid, U- to horseshoe-shaped or ¾ circular, hyaline, 1- to several-septate.
Notes — Based on the results of both phylogenies (Fig. 1, 2), Tectonidula is a member of Barbatosphaeria and is placed in its synonymy. Two new combinations for the former Tectonidula species are proposed.
The main features of ascomata, asci and ascospores that are common to all species are listed under the general description of Barbatosphaeria and are not repeated in the descriptions of individual species. Ascospores tend to group in the middle of the sporiferous part of ascus, leaving the apex empty. They are often arranged 2–3-seriately or in a fascicle. Because the ascus stipe is degrading with age and sometimes not well visible, the asci should always be measured in the pars sporifera, which makes the ascus size fairly comparable among all species.
Barbatosphaeria arboricola Réblová, sp. nov. — MycoBank MB810126; Fig. 5
Fig. 5.
Barbatosphaeria arboricola. a–c. Circular groups of ascomata, arrow indicates upper part of necks covered with pubescence; d. vertical section of the ascomatal wall; e. asci; f. ascospores; g. paraphyses (a–c. M.R. 3085; d–g. holotype PRM 924373). — h–l: Culture of sporothrix-like asexual morph; h. colony; i. conidia; j–l. conidiophores (ex-type culture CBS 127689, all on PDA at 25°C); d–f, l. DIC; g, i–k. PC. — Scale bars: a–c = 1000 μm, d = 50 μm, e–g, j–l = 10 μm, i = 5 μm.
Asexual morph. Sporothrix-like.
Etymology. Arbor (L), meaning tree, and -cola (L), meaning dwelling in a tree, refers to this species occurring between cortex and wood on the branches of trees.
Diagnosis — Differing from B. barbirostris by longer asci, (29–) 30–36(–40) μm long in pars sporifera and 6.0–7.5(–8.0) μm wide, and longer ascospores, (7.7–)8.0–9.5(–10.0) × 1.8–2.2(–2.5) μm.
Ascomata dark brown to black, aggregated in circular groups of 5–15, occasionally solitary. Venter globose to subglobose, 400–650 μm high, 350–550 μm diam, often lying horizontally, collapsing on a dorsal side upon drying; necks cylindrical, straight to slightly flexuous decumbent 600–2000 μm long, 100–120 μm wide, converging radially, piercing the periderm together. Venter and neck sparsely covered with red-brown pubescence that disappears with age, only the necks remaining covered with hairs in the upper part; hairs septate, smooth-walled, obtuse at the ends, 3.0–3.5 μm wide. Ascomatal wall 60–80 μm thick, leathery to fragile, 2-layered. Outer layer consisting of thick-walled, brown to red-brown, polyhedral to elongate cells of textura prismatica, towards the interior grading into several layers of thin-walled subhyaline to hyaline flattened cells. Ostiolum periphysate. Paraphyses septate, wider near the base c. 4.0–5.0 μm, tapering to 2.0–3.5 μm toward the tip. Asci clavate in sporiferous part, sometimes slightly swollen in the middle when ascospores are clustered in a fascicle, tapering toward the stipe, (29–)30–36(–40) μm long in pars sporifera, 6.0–7.5(–8.0) μm wide (in broadest part) (mean ± SD = 33.2 ± 2.9 × 7.4 ± 0.6 μm), with a stipe 10–18 μm long, 8-spored; ascal apex broadly rounded to obtuse, with a non-amyloid apical annulus c.1.0 μm high, 2.0–2.3 μm wide. Ascospores ellipsoid to oblong, sometimes slightly curved, (7.7–)8.0–9.5(–10.0) × 1.8–2.2(–2.5) μm (mean ± SD = 8.6 ± 0.5 × 2.0 ± 0.2 μm), hyaline, smooth, 1-septate, non-constricted, arranged obliquely 2–3-seriately or in a fascicle in the sporiferous part of the ascus.
Culture characteristics — Colonies slow growing, 12–17 mm diam on PDA after 21 d at 25 °C. Aerial mycelium velvety, zonate, dark beige to grey (oac703) in the centre, later consequently pale beige (oac816) with dark grey (oac725) zones, darker toward the margin of the colony. Sporulation widespread with a uniform layer of conidiophores. Conidial masses whitish. Conidiophores of sporothrix-like developing early from germinating ascospores, 2.5–13 × 1.0–2.0 μm (mean ± SD = 8.4 ± 4.1 × 1.5 ± 0.3 μm), micronematous to semimacronematous, unbranched, cylindrical or of irregular shape, hyaline. Conidiogenous cells polyblastic, intercalary or terminal, bearing 1–3 denticles producing conidia holoblastically. Conidia (2.7–)3.0–3.5 × 0.8–1.0 μm (mean ± SD = 3.2 ± 0.3 × 0.9 ± 0.1 μm), aseptate, hyaline, oblong, slightly curved to suballantoid, slightly tapering at the base. Chlamydospores not observed.
Specimens examined. CZECH REPUBLIC, Central Bohemia, distr. Mělník, Kokořínsko Protected Landscape Area, valley of the Pšovka brook between Olešno and Ráj, decaying wood of Populus tremula, 15 Nov. 2009, M. Réblová (PRM 924373, holotype, culture ex-type CBS 127689); ibid., Central Bohemia, distr. Rakovník, Tři stoly near Nové Strašecí, decaying bark and branch of Betula verrucosa, 31 Oct. 2009, M. Réblová M.R. 3085, culture CBS 127690. – SWEDEN, Uppland, Dalby parish, Jerusalem, bark of Betula pendula, 22 Jan. 1988, K. Holm & L. Holm, culture CBS 114120.
Notes — Barbatosphaeria arboricola is similar to B. barbirostris in 1-septate, ellipsoid to oblong ascospores, but differs from the latter by longer asci and ascospores; the ascospores of B. barbirostris are (5.0–)6.9–7.0 μm long, the asci measure 25–35 μm in pars sporifera (Réblová 2007). Barbatosphaeria arboricola is also similar to B. varioseptata in the shape of ascospores and asci, but the latter differs by longer ascospores with a delayed formation of two additional septa, while the asci are comparable in size to those of B. arboricola. The ascospores of morphologically similar Barbatosphaeria sp. represent an intermediate morphotype between B. arboricola and B. varioseptata. Its ascospores are mostly 1-septate but some may develop a second and occasionally a third septum while still in the asci. It differs from B. arboricola by shorter asci, while the sizes of the ascospores are comparable.
Barbatosphaeria barbirostris (Dufour) Réblová, Mycologia 99: 727. 2007 — Fig. 6
Fig. 6.
Barbatosphaeria barbirostris. a–c. Ascomata under periderm, arrow indicates upper part of the neck covered with pubescence; d. asci; e, f. ascospores (a, b from M.R. 3766; c from M.R. 649; d–f from M.R. 3767); d, f: DIC; e: PC. — Scale bars: a, b, c = 1000 μm, d = 10 μm, e, f = 5 μm.
Asexual morphs. Ramichloridium- and sporothrix-like.
Specimens examined. CZECH REPUBLIC, Southern Bohemia, Šumava Mts, Prášily, Mt Ždanidla, decaying wood of a Fagus sylvatica, 20 June 1995, M. Réblová M.R. 649; ibid., Novohradské hory Mts, Pohoří na Šumavě, Myslivna Mt, decaying wood of Fagus sylvatica, 6 Oct. 2012, M. Réblová M.R. 3766, M.R. 3767. For other examined material see Réblová (2007).
Notes — For full synonymy, description and illustrations of sexual and asexual morphs refer to Réblová (2007). The great variability in disposition of ascomata of B. barbirostris on the natural substratum, varying from small to large loose circular groups to rows or rarely a solitary growth, and simple morphology of 1-septate ellipsoid ascospores, resulted in the description of this species under four generic taxonomic synonyms and its combinations under eight ascomycete genera.
Barbatosphaeria dryina (Curr.) Réblová, comb. nov. — MycoBank MB810127; Fig. 7
Fig. 7.
Barbatosphaeria dryina. a. Group of ascomata growing between periderm and wood on old stromata; b. vertical section of the ascomatal wall; c. ascospores; d. paraphyses; e, f. asci (a, b. M.R. 3717; c. M.R. 3111; d–f. M.R. 3072). — g–j: Culture of sporothrix-like asexual morph. g. colony; h–j. conidiophores; k. conidia (CBS 127691, all on PDA at 25°C); b, c, f. DIC; d, e, h–k. PC. — Scale bars: a = 1000 μm, b = 50 μm, c–k = 10 μm.
Basionym. Sphaeria dryina Curr., Trans. Linn. Soc. London 22: 278. 1859.
≡ Calosphaeria dryina (Curr.) Nitschke, Pyrenomyc. Germ. 1: 94. 1867.
≡ Valsa (Incusae) dryina (Curr.) Berk. & Broome, Ann. Mag. Nat. Hist., ser. 3. 3: 366. 1859.
Asexual morph. Sporothrix-like.
Ascomata dark brown to black, growing between periderm and wood, often aggregated in circular or semi-circular groups of 5–9(–15), occasionally solitary. Venter globose to subglobose, 450–600 μm high, 400–600 μm diam, non-collapsing or collapsing dorsally upon drying; necks cylindrical, straight to slightly flexuous 900–1700 μm long, 150–250 μm wide, converging and piercing the periderm. Venter and neck covered with red-brown pubescence that disappears with age, only the necks remaining covered with hairs in the upper part; hairs septate, smooth-walled, obtuse at the ends, 3.5–4.5 μm wide. Ascomatal wall 55–80 μm thick, leathery to fragile, 2-layered. Outer layer consisting of thick-walled, brown or red-brown polyhedral to elongate cells of textura prismatica to epidermoidea, towards the interior grading into several layers of thin-walled subhyaline to hyaline flattened cells. Ostiolum periphysate. Paraphyses septate, wider near the base c. 4.5–6.0 μm, tapering to 2.0–3.0 μm toward the tip. Asci clavate in sporiferous part, tapering toward the stipe, 35–50(–52) μm long in pars sporifera, (8.0–)9.0–12.0 μm wide (in broadest part) (mean ± SD = 42.3 ± 3.4 × 10.5 ± 0.6 μm), with a stipe 16–25 μm long, 8-spored; ascal apex broadly rounded to obtuse, with a non-amyloid apical annulus c. 0.5–1 μm wide, 2 μm high. Ascospores oblong to subcylindrical and strongly curved, (7.5–)8.0– 10.5 × (2.2–)2.5–3.0 μm (mean ± SD = 8.9 ± 1.2 × 2.4 ± 0.2 μm), hyaline, smooth, 1-septate, non-constricted, arranged obliquely 2–3-seriately in the sporiferous part of the ascus.
Culture characteristics — Colonies slow growing, 11–16 cm diam on PDA after 21 d at 25 °C. Aerial mycelium, velvety, zonate, white (oac909) to beige (oac816) in the centre, later dark grey (oac902) with pale grey (oac725) zones, always darker toward the margin of the colony. Sporulation widespread with a uniform layer of conidiophores. Conidial masses whitish. Conidiophores of sporothrix-like developing early from germinating ascospores, 2.5–5.5 × 1.0–1.5 μm (mean ± SD = 3.3 ± 1.2 × 1.3 ± 0.3 μm), micronematous to semimacronematous, unbranched, flask-shaped to cylindrical or of irregular shape, hyaline. Conidiogenous cells polyblastic, intercalary or terminal, bearing 1–4 denticles producing conidia holoblastically. Conidia 3.5–5.2 × 0.8–1.0 μm (mean ± SD = 4.4 ± 0.7 × 0.9 ± 0.1 μm), aseptate, hyaline, suballantoid to oblong, curved, slightly tapering at the base. Chlamydospores not observed.
Specimens examined. CZECH REPUBLIC, Southern Moravia, distr. Břeclav, Milovice, Milovická stráň Nature Reserve, north slopes of Mt Špičák 293 masl, decaying wood and bark of a deciduous tree, 18 Nov. 2009, M. Réblová M.R. 3072, culture CBS 127691; ibid., Southern Moravia, distr. Břeclav, Valtice, Rendez-vous Nature Reserve, 21 Nov. 2009, wood of Quercus sp., M. Réblová M.R. 3111, culture CBS 137796. – ENGLAND, Surrey, Weybridge, wood of Quercus sp., Sept. 1856, F. Currey, K 158687, syntype. – ITALY, Liguria, Ossegna, Colli di Ossegna, 20 Sept. 2013, decaying wood and bark of a branch of Quercus sp., M. Réblová M.R. 3717, culture CBS 137798.
Notes — This species is distinct among other members of Barbatosphaeria by relatively large, 1-septate, oblong, strongly curved ascospores. It often occurs in association with other periderm-inhabiting ascomycetes, e.g. Enchnoa infernalis, Caudospora taleola or Diatrypella quercina. Based on the ascospore morphology, it can be compared with B. fagi, which differs by more U- to horseshoe-shaped ascospores half the size (Samuels & Candoussau 1996).
Barbatosphaeria fagi (Cand. & Samuels) Réblová, comb. nov. — MycoBank MB810128
Basionym. Calosphaeria fagi Cand. & Samuels, Nova Hedwigia 62: 57. 1996.
= Tectonidula fagi (Samuels & Cand.) Réblová, Mycol. Res. 113: 1000. 2009.
Asexual morphs. Ramichloridium- and sporothrix-like.
Notes — For description and illustrations of sexual and asexual morphs refer to Samuels & Candoussau (1996).
Barbatosphaeria fimbriata Réblová, sp. nov. — MycoBank MB810129; Fig. 8
Fig. 8.
Barbatosphaeria fimbriata. a, b. Ascomata growing between periderm and wood, young (a) densely covered with pubescence and old (b) glabrous; c. vertical section of the ascomatal wall, arrow indicates a superficial layer of interwoven hyphae; d. young asci with swollen bulbous base attached to ascogenous hyphae; e. asci; f. ascospores; g. paraphyses (a, c, d–g. holotype PRM 924374; b. M.R. 3694); c, e. DIC; d, f, g. PC. — Scale bars: a, b = 1000 μm, c = 50 μm, d–g = 10 μm.
Asexual morph. Sporothrix-like.
Etymology. Fimbriatus (L), meaning fringed (fimbriated), referring to hairs covering ascomata.
Diagnosis — Differing from B. neglecta by shorter asci, 12.5–18(–21) μm long in pars sporifera and 4.0–4.5(–5.0) μm wide, and shorter ascospores (3.5–)4.0–4.5(–5.0) × 1.0–1.3 μm.
Ascomata dark brown, growing between periderm and wood, aggregated in circular groups of 6–18, often associated with stromata of other fungi. Venter globose to subglobose, 350–400 μm high, 300–400 μm diam, lying horizontally, collapsing dorsally upon drying; necks cylindrical, straight to slightly flexuous 1000–1500 μm long, 120–150 μm wide, converging radially. Venter and neck covered with brown pubescence that tends to disappear with age, bottom part of venter mostly remaining covered with hairs. When young, hairs densely interwoven forming a layer of textura intricata around ascomata up to 40 μm thick. Hairs septate, smooth-walled, obtuse at the ends, 2.0–3.5 μm wide. Ascomatal wall 35–55 μm thick, leathery to fragile, 2-layered. Outer layer consisting of thick-walled, brown, polyhedral to elongate cells of textura prismatica to textura angularis, towards the interior grading into several layers of thin-walled subhyaline to hyaline flattened cells. Ostiolum periphysate. Paraphyses septate, wider near the base c. 4.0–5.0 μm, tapering to 2.5–3.0 μm toward the tip. Asci clavate in sporiferous part, tapering toward the stipe, 12.5–18(–21) μm long in pars sporifera, 4.0–4.5(–5.0) μm wide (in broadest part) (mean ± SD = 14.6 ± 2.3 × 4.3 ± 0.3 μm), with a stipe 8.5–13 μm long, 8-spored; ascal apex obtuse, with a non-amyloid apical annulus c. 1.0 μm high, 1.5–1.8 μm wide. Ascospores oblong to subcylindrical, slightly curved, (3.5–)4.0–4.5(–5.0) × 1.0–1.3 μm (mean ± SD = 4.0 ± 0.4 × 1.0 ± 0.2 μm), hyaline, smooth, non-septate, arranged obliquely 2–3-seriately in the sporiferous part of the ascus.
Specimens examined. CZECH REPUBLIC, Southern Moravia, distr. Břeclav, Valtice, Rendez-vous Nature Reserve, decaying wood and bark of a branch of Quercus sp., M. Réblová M.R. 3694. – FRANCE, Midi-Pyrénées, Ariège, Rimont, Grand Bois, 850 m asl, decaying bark of Fagus sylvatica, associated with Eutypella quaternata, 7 Mar. 2003, J. Fournier J.F. 03036 (PRM 924374, holotype).
Notes — Barbatosphaeria fimbriata is closely related to B. neglecta, from which it differs by shorter asci and ascospores. For differences at the RNA structural level, see Results. The living culture of B. fimbriata isolated from M.R. 3694 and included in phylogenetic analyses is no longer viable.
Barbatosphaeria hippocrepida (Réblová) Réblová, comb. nov. — MycoBank MB810130
Basionym. Tectonidula hippocrepida Réblová, Mycol. Res. 113: 998. 2009.
Asexual morph. Sporothrix-like.
Specimen examined. NEW ZEALAND, Westlands, Mount Aspiring National Park, Haast Pass, Historical Bridal Track, decayed bark and wood of a branch of Nothofagus sp., 31 Mar. 2005, M. Réblová (PDD 81440, holotype, ex-type culture ICMP 17630, CBS 123471).
Notes — For description and illustrations of sexual and asexual morphs refer to Réblová & Štěpánek (2009). Barbatosphaeria hippocrepida was collected in New Zealand and it is known only from a single collection. It is remarkably similar to Ceratostomella cyclospora (Kirschstein 1907), which was later placed into the genus Calosphaeria by Petrak (1924). Its type material is apparently lost (pers. comm. E. Gerhardt, Berlin Museum). It fits the description of B. hippocrepida apart from the ascoma and partly the ascus morphology. The long-necked ascomata of C. cyclospora are rarely solitary, mostly congregated by two or in larger groups in valsoid formation, partially or entirely immersed in bark or decorticated wood, and larger c. 1 mm diam (Petrak 1924). The ascomata of B. hippocrepida are 400–650 μm diam, arranged in circular groups between cortex and wood, lying horizontally with radially converging necks and with typical dorsal flattening of the venter with a central glabrous area surrounded by warts (Réblová & Štěpánek 2009). The asci of C. cyclospora are shorter in pars sporifera (20–25 μm) than those of B. hippocrepida (27–33 μm), and do not possess the conspicuous invagination at the ascal apex. It is likely, that C. cyclospora represents another name for B. hippocrepida or a species closely related to it. Without studying representative material we refrain from making any formal changes.
Barbatosphaeria neglecta Réblová, sp. nov. — MycoBank MB810131; Fig. 9
Fig. 9.
Barbatosphaeria neglecta. a. Ascomata; b. vertical section of the ascomatal wall; c, d. asci; e. young asci with swollen bulbous base attached to ascogenous hyphae; f. paraphyses; g. ascospores (holotype PRM 924375). — h–l: Culture of sporothrix-like asexual morph. h. colony; i, j. conidia; k, l. conidiophores (ex-type CBS 127693, all on PDA at 25°C); b, c, i, k. DIC; d–f, g, j, l. PC. — Scale bars: a = 1000 μm, b = 50 μm, c–f, k, l = 10 μm, g, i, j = 5 μm.
Asexual morph. Sporothrix-like.
Etymology. Neglectus (L), meaning neglected or not chosen, referring to the inconspicuous, easily overlooked morphology of this species.
Diagnosis — Differing from B. fimbriata by longer asci, 22–27 μm long in pars sporifera and 4.5–5.5 μm wide, and longer ascospores (4.8–)5.5–6.5(–6.9) × 1.0–1.5 μm.
Ascomata dark brown to black, growing between periderm and wood, aggregated in circular groups of 3–6, sometimes solitary. Venter globose to subglobose, 320–400 μm high, 300–430 μm diam, lying horizontally or upright; necks cylindrical, straight to slightly flexuous 500–1500 μm long, 100–120 μm wide. Venter and neck sparsely covered with brown or red-brown pubescence that disappears with age, only bottom part of venter remaining covered with hairs; hairs septate, smooth-walled, obtuse at the ends, 3.0–4.5 μm wide. Ascomatal wall 35–47 μm thick, leathery to fragile, 2-layered. Outer layer consisting of thick-walled, brown to red-brown polyhedral to elongate cells of textura prismatica, towards the interior grading into several layers of thin-walled subhyaline to hyaline flattened cells. Ostiolum periphysate. Paraphyses septate, wider near the base c. 3.5–4.5 μm, tapering to 2.0 μm toward the tip. Asci clavate in sporiferous part, tapering toward the stipe, 22–27 μm long in pars sporifera, 4.5–5.5 μm wide (in broadest part) (mean ± SD = 24.7 ± 2.5 × 5.4 ± 0.4 μm), with a stipe 8.0–14 μm long, 8-spored; ascal apex broadly rounded to obtuse, with a non-amyloid apical annulus c. 1.0 μm high, 2.0 μm wide. Ascospores oblong to subcylindrical, sometimes slightly curved, (4.8–)5.5–6.5(–6.9) × 1.0–1.5 μm (mean ± SD = 6.1 ± 0.5 × 1.4 ± 0.2 μm), hyaline, smooth, aseptate, arranged obliquely 2–3-seriately in the sporiferous part of the ascus.
Culture characteristics — Colonies slow growing, 13–16 mm diam on PDA after 21 d at 25 °C. Aerial mycelium white (oac909) to beige (oac816), velvety, zonate, brown-grey (oac770) near the centre, consequently pale and dark zones changing, always becoming darker toward the margin of the colony. Centre of the colony around the inoculum block without aerial mycelium. Sporulation widespread with a uniform layer of conidiophores. Conidial masses whitish. Conidiophores of the sporothrix-like developing early from germinating ascospores, 1.5–4.2 × 0.9–1.2 μm (mean ± SD = 3.0 ± 1.4 × 1.1 ± 0.3 μm), micronematous to semimacronematous, unbranched, cylindrical or irregular shape, hyaline. Conidiogenous cells polyblastic, intercalary or terminal, bearing 1–3 denticles producing conidia holoblastically. Conidia 3.2–4.2(–5.0) × 0.9–1.2(–1.4) μm (mean ± SD = 3.8 ± 0.6 × 1.1 ± 0.2 μm), aseptate, hyaline, oblong, slightly curved, slightly tapering at the base. Chlamydospores not observed.
Specimen examined. CZECH REPUBLIC, Central Bohemia, distr. Mělník, Kokořínsko Protected Landscape Area, valley of the Pšovka brook between Olešno and Ráj, decaying wood of Betula verrucosa, 15 Nov. 2009, M. Réblová (PRM 924375, holotype, culture ex-type CBS 127693).
Notes — Barbatosphaeria neglecta forms a monophyletic clade with the morphologically similar B. fimbriata. Both species possess non-septate ascospores and differ by size of ascospores and asci. For differences at the RNA structural level, see Results.
Barbatosphaeria varioseptata Réblová, sp. nov. — MycoBank MB810132; Fig. 10a–i
Fig. 10.
Barbatosphaeria spp. a–i: Barbatosphaeria varioseptata. a. Ascomata growing between periderm and wood; b. vertical section of the ascomatal wall; c, d. ascospores; e. paraphyses with young asci attached to the ascogenous hyphae; f. asci (holotype PRM 924376). — g–i: Culture of sporothrix-like asexual morph. g. colony; h. conidiophores; i. conidia (ex-type CBS 137797, all on PDA at 25°C). — j–l: Barbatosphaeria sp. j, k. asci; l. ascospores (M.R. 3730). b–d, f, k, l. DIC; e, h–j. PC. — Scale bars: a = 1000 μm, b = 50 μm, c–f, h, j–l = 10 μm, i = 5 μm.
Asexual morph. Sporothrix-like.
Etymology. Varius (L), meaning changing, variable, septum (L), meaning partition, refers to the variably septate ascospores.
Diagnosis — Differing from B. arboricola and B. barbirostris by longer ascospores, (8.5–)9.5–11.0(–11.5) × 2.0–2.2(–2.5) μm with a delayed formation of two additional septa.
Ascomata dark brown to black, aggregated in circular groups of 8–15. Venter globose to subglobose, 350–500 μm high, 400–500 μm diam, lying horizontally, collapsing dorsally upon drying; necks cylindrical, straight to slightly flexuous decumbent 800–2000 μm long, 100–150 μm wide, converging radially. Venter and neck covered with brown or red-brown pubescence that disappears with age, only the necks and bottom part of venter remaining sparsely covered; hairs septate, smooth-walled, obtuse at the ends, 3.5–4.5 μm wide. Ascomatal wall 45–70 μm thick, leathery to fragile, 2-layered. Outer layer consisting of thick-walled, brown to red-brown polyhedral to elongate cells of textura prismatica, towards the interior grading into several layers of thin-walled subhyaline to hyaline flattened cells. Ostiolum periphysate.Paraphyses septate, wider near the base c. 3.5–4.5 μm, tapering to 2.5–3.0 μm toward the tip. Asci clavate in sporiferous part, tapering toward the stipe, (29–)34–44(–55) μm long in pars sporifera, 7.0–8.0(–10.0) μm wide (in broadest part) (mean ± SD = 39.5 ± 5.9 × 7.9 ± 1.1 μm), with a stipe 12–30 μm long, 8-spored; ascal apex broadly rounded to obtuse, with a non-amyloid apical annulus c. 1.0–1.2 μm high, 2.0–2.9 μm wide. Ascospores oblong to subcylindrical, (8.5–)9.5–11.0(–11.5) × 2.0–2.2(–2.5) μm (mean ± SD = 10.2 ± 0.9 × 2.2 ± 0.1 μm), hyaline, smooth, 1-septate, at maturity with up to three septa, non-constricted, arranged obliquely 2–3-seriately in the sporiferous part of the ascus.
Culture characteristics — Colonies slow growing, 12–18 cm diam on PDA after 21 d at 25 °C. Aerial mycelium white (oac909) to beige (oac816), velvety, zonate, light pink (oac697) at the centre, later with pale and dark grey zones alternating, darker toward the margin of the colony. Sporulation widespread with a uniform layer of conidiophores. Conidial masses whitish. Conidiophores of the sporothrix-like developing early from germinating ascospores, 2.5–5.6 × 0.9–1.5 μm (mean ± SD = 3.7 ± 1.4 × 1.1 ± 0.2 μm), micronematous to semimacronematous, unbranched, cylindrical or of irregular shape, hyaline. Conidiogenous cells polyblastic, intercalary or terminal, bearing 1–3 denticles producing conidia holoblastically. Conidia 2.5–3.5 × 0.9–1.1 μm (mean ± SD = 3.0 ± 0.5 × 1.0 ± 0.1 μm), aseptate, hyaline, oblong, slightly curved, slightly tapering at the base. Chlamydospores not observed.
Specimen examined. CZECH REPUBLIC, Southern Moravia, distr. Břeclav, Milovice, Milovický les Nature Reserve, north slopes of Mt Špičák 293 m asl, decaying wood and bark of a branch of Quercus sp., 18 Nov. 2010, M. Réblová (PRM 924376, holotype, culture ex-type CBS 137797).
Notes — Barbatosphaeria varioseptata is positioned at the basal branch of the B. arboricola clade (Fig. 1, 2). It can be distinguished from the morphologically similar B. arboricola by longer, 3-septate ascospores and slightly longer asci. This species is also well-defined by presence of several hCBC substitutions in the H2 and H3 helices of ITS2 (Fig. 4).
Barbatosphaeria sp. — Fig. 10j–l
Asexual morph. Sporothrix-like.
Ascomata dark brown to black, aggregated in circular groups of 10–15 around old stromata of Eutypella quaternata. Venter globose to subglobose, 400–450 μm high, 400–500 μm diam, lying horizontally, often collapsing dorsally upon drying; necks cylindrical, slightly flexuous decumbent 1000–1500 μm long, 100–150 μm wide, converging radially. No pubescence observed on venter and neck. Ascomatal wall 43–50 μm thick, leathery to fragile, 2-layered. Outer layer consisting of thick-walled, brown to red-brown polyhedral to elongate cells of textura prismatica, towards the interior grading into several layers of thin-walled subhyaline to hyaline flattened cells. Ostiolum periphysate. Paraphyses septate, wider near the base c. 4.5–6.5 μm, tapering to 2.5–3.0 μm toward the tip. Asci clavate in sporiferous part, tapering toward the stipe, 24–28 μm long in pars sporifera, 6.0–7.0 μm wide (in broadest part) (mean ± SD = 26.2 ± 1.7 × 6.3 ± 0.3 μm), with a stipe 10–16 μm long, 8-spored; ascal apex obtuse, with a non-amyloid apical annulus c. 0.8–1.0 μm high, 2.5–2.9 μm wide. Ascospores ellipsoid to oblong, 7.5–8.6(–9.0) × 1.2–2.0 μm (mean ± SD = 8.1 ± 0.5 × 1.8 ± 0.2 μm), hyaline, smooth, 1-septate, with delayed formation of one, rarely two septa, non-constricted, arranged obliquely 2–3-seriately in the sporiferous part of the ascus.
Specimen examined. SLOVAK REPUBLIC. Central Slovakia, distr. Žilina, Malá Fatra Mts, Kunerad near Rajecké Teplice, decaying bark and wood of Fagus sylvatica, 5 July 2012, M. Réblová M.R. 3730.
Notes — Barbatosphaeria sp. is characterised by 1–2-septate, rarely 3-septate ascospores and, besides B. varioseptata, represents another morphotype with ascospores containing more than one septum. It groups with members of the B. arboricola clade (Fig. 1, 2). It differs from B. varioseptata by shorter asci and ascospores. The material is rather old and insufficient as a type collection. The culture is no longer viable. We prefer to not describe this taxon until fresh and mature material is collected.
KEY TO ACCEPTED SPECIES IN BARBATOSPHAERIA
1. Ascospores non-septate ......................... 2
1. Ascospores with 1–3 septa ....................... 4
2. Ascospores oblong to subcylindrical, slightly curved .... 3
2. Ascospores allantoid, U- to horseshoe shaped ......................................... B. hippocrepida
3. Ascospores longer than 5.0 μm; asci longer than 21 μm in pars sporifera .......................... B. neglecta
3. Ascospores up to 5.0 μm long; asci up to 21 μm long in pars sporifera ..............................B. fimbriata
4. Ascospores 1–3-septate ......................... 5
4. Ascospores 1-septate ........................... 6
5. Ascospores longer than 9 μm; asci longer than 30 μm in pars sporifera ........................... B. varioseptata
5. Ascospores up to 9 μm; asci up to 30 μm in pars sporifera .............................. Barbatosphaeria sp.
6. Ascospores straight, ellipsoid, oblong to subcylindrical .. 7
6. Ascospores strongly curved or U- to horseshoe-shaped . 8
7. Ascospores longer than 7 μm ............ B. arboricola
7. Ascospores up to 7 μm long ............ B. barbirostris
8. Ascospores oblong, strongly curved, longer than 5 μm .......................................... B. dryina
8. Ascospores U- to horseshoe shaped, up to 5 μm long ............................................ B. fagi
DISCUSSION
Taxonomy of Barbatosphaeria
Barbatosphaeria is positioned in the Sordariomycetidae in a well-supported clade (97/1.0) comprising members of the Ophiostomatales, Annulatascaceae, Papulosaceae and several small or monotypic genera from freshwater or terrestrial habitats. These taxa are distantly related and share simple and indistinct morphology of dark ascomata with carbonised wall, long neck or beak and hyaline ascospores (e.g. Hyde 1992, Ho et al. 1999, Wong & Hyde 1999, Wong et al. 1999, Raja et al. 2003, Shearer et al. 2003, Tsui et al. 2003, Campbell & Shearer 2004, Arzanlou et al. 2007, Ferrer et al. 2012, Liu et al. 2012, Jaklitsch et al. 2013). Many of these fungi have never been cultivated. If asexual morphs are known, the dominant conidiogenesis is holoblastic, with often denticulate conidiogenous cells. Some of these genera, especially those closely related and morphologically similar to Barbatosphaeria, are shown in Fig. 1. Mostly nuc28S sequences of members of this clade are available in GenBank, but this gene alone does not offer sufficient phylogenetic resolution and internal support for classification at higher systematic levels. It is probable that fungi so far described and placed in this clade represent only a fraction of the actual diversity of this group in nature and they are a mere mosaic of morphological traits.
Barbatosphaeria can be compared with members of Lentomitella and Natantiella. The ascomata of these genera never form circinate groups or loose circular formations; they grow solitarily or in large irregular groups with the venter and most of the neck deeply immersed in decorticated often soft decaying wood. Based on the morphology of asci and ascospores, Natantiella ligneola differs from members of Barbatosphaeria by larger, ellipsoid ascospores and robust ascomata. Its asexual morph is unknown, only sterile mycelium was formed in the axenic culture (Réblová & Štěpánek 2009). Lentomitella differs by non-stipitate asci with a larger apical annulus, ellipsoid to fusiform ascospores and phaeoisaria-like asexual morph (Réblová 2006).
Barbatosphaeria including Tectonidula is shown as a strongly supported clade (100/1.0 in the ITS-β-tubulin analysis and 99/1.0 in the multigene analysis) comprising nine species; eight of them are studied in our phylogenies using sequences of five nuclear loci. Tectonidula is a monotypic genus characterised by allantoid, strongly curved, U- to horseshoe shaped ascospores and is represented by T. hippocrepida in our analyses. In previous studies, when B. barbirostris and T. hippocrepida were analysed as the only representatives of both genera, they formed a monophyletic clade but with low (based on nuc28S gene; Réblová & Štěpánek 2009) to medium support (based on 3-gene analysis; Jaklitsch et al. 2013, Réblová et al. 2014). In the multigene analysis that contains other members of Barbatosphaeria, T. hippocrepida is shown at the basal position (Fig. 1) in the Barbatosphaeria clade, while in the combined ITS-β-tubulin phylogeny it is together with B. fimbriata and B. neglecta basal to all other species with septate ascospores (Fig. 2).
Species of Barbatosphaeria are well distinguishable based on the morphology of ascospores, particularly the size, shape and number of septa in combination with the size of the ascus. The ascus shape, i.e. truncate to broadly-rounded at the apex, clavate or cylindrical-clavate in the sporiferous part with a long, slender tapering stipe, is similar in all species. Ascomata of Barbatosphaeria grow between cortex and wood, hidden from our sight, often associated with dead stromata and ascomata of other periderm-inhabiting fungi, especially those of members of the Diaporthales or Diatrypaceae. Asexual morphs with holoblastic denticulate conidiogenesis were experimentally proven for all species of Barbatosphaeria (Samuels & Candoussau 1996, Réblová 2007, this study). Both ramichloridium- and sporothrix-like asexual morphs were obtained in B. barbirostris and B. fagi; other species develop only micronematous or semimacronematous sporothrix-like conidiophores in axenic culture.
Two Barbatosphaeria species with aseptate, oblong, slightly curved ascospores, B. fimbriata and B. neglecta, are remarkably similar to Calosphaeria parasitica. Based on a revision of the holotype (Herbarium Genève, G00127176) and other herbarium material of C. parasitica, ascomata of this species are associated with ascomata of Eutypella quaternata, they grow outside its ascomata or inside the ascomatal cavity. Calosphaeria parasitica differs from Barbatosphaeria by glabrous ascomata, broadly clavate asci with broadly rounded, thickened apex without an apical annulus and ascospores arranged 3–4-seriately or in a fascicle directly below the ascal apex. The herbarium material of C. parasitica was too old to confirm the presence of subtending cells arising from ascogenous hyphae and bearing the asci, a character typical of members of the Calosphaeriales.
Secondary structure of ITS
Comparison of 2D structures of ITS1 and ITS2 among 13 Barbatosphaeria strains confirmed the existence of eight species. Although an experimental 3D structure of ITS1 and ITS2 has not been yet determined, the study of the 2D structure of both ITS molecules using comparative methods has been used in eukaryotes to define species complex groups (e.g. Bridge et al. 2008, Rodriguez-Martínez et al. 2012). The helices H2 of ITS1 and H2 and H3 of ITS2 were also detected in other fungi and used to delimit groups of asexual morphs linked to Chaetosphaeria in the Chaetosphaeriaceae (Réblová & Winka 2000) and to study genera and families of the Chaetothyriales (Réblová et al. 2013). Molecular morphometrics, a phylogenetic approach based on 2D elements, was successfully applied using ITS sequences to the Erysiphaceae (Takamatsu et al. 1998) and Lycoperdaceae (Krüger & Gargas 2004, 2008).
In ITS1 of Barbatosphaeria, only helix H3 possesses a homologous structure with significant variability and could be compared among all strains. The ITS2 consisting of four helices maintains a similar core structure in all strains and can be used for comparisons.
The 2D structure of the ITS2 has been widely investigated (Coleman & Mai 1997, Goertzen et al. 2003, Schultz et al. 2005, Coleman & van Oppen 2008, Ullrich et al. 2010, Caisová et al. 2011). The number of four helices (H1–H4), a pyrimidine-pyrimidine mismatch in H2 and the occurrence of the YGGUY motif in helix H3 represent hallmarks, which are universal among eukaryotes. Additional helices may be positioned on the ring structure anywhere between helices 1–4 in different groups of organisms (Coleman 2007, Réblová et al. 2013). The pyrimidine-pyrimidine mismatch in H2, i.e. C/C after the sixth base pair in our study, was present in all Barbatosphaeria species (Fig. 4). We detected also a second C/C mismatch in the upper half of H2, but it was lost in B. neglecta and B. fimbriata. The evolutionary motif was not detected among strains belonging to a single genus. A larger comparative analysis including strains of several genera from different families would be necessary to detect the third hallmark of ITS2.
Recording all existing substitutions and mapping them onto predicted 2D models of ITS1 and ITS2 of B. barbirostris allowed us to significantly improve the alignment of ITS sequences of members of Barbatosphaeria and to study the evolutionary history of ITS1 and ITS2.
The CBC species concept
Examination of all positions in ITS1 and ITS2 revealed that some base pairs with double-sided (CBC), one-sided (hCBC) and non-compensatory (non-CBC) substitutions uniquely characterize certain clades and species lineages and represent non-homoplasious synapomorphies within Barbatosphaeria (Fig. 3, 4).
The CBC hypothesis, which has been used to delimit biological species, has been tested in Chlorophyta and representatives of the Fagales (Coleman 2000, 2003, Coleman & Vacquier 2002) and also by employing up to 100 000 2D models from the ITS2 database (Schultz et al. 2006, Müller et al. 2007, Selig et al. 2007, Koetschan et al. 2010). The CBC species concept is based on co-evolution of nucleotides that maintain an RNA transcript in the two most conserved helices H2 and H3 of the ITS2 (Coleman & Mai 1997, Fabry et al. 1999, Coleman 2000, 2005, Amato et al. 2007). Even a single CBC in these duplexes indicates sexual incompatibility. The original hypothesis defined a CBC clade as including organisms lacking CBCs in these conserved helices that differ from other CBC clades by as little as a single CBC. In contrast, CBCs in H1 and H4 of ITS2, and hCBCs in H2 and H3 may still allow mating between two organisms and therefore are less significant in species delimitation (Coleman 2000). CBC clades usually fall into one or more Z clades encompassing groups of organisms producing compatible gametes that can form zygotes, but which are separated by various pre- and postzygotic isolation mechanisms (Coleman 2000). Therefore, the Z clades rather than the CBC clades contain ‘biological species’ or represent a diverging population of one to several morphotypes capable of interbreeding.
In ITS2 the differences within the species that were represented by more than one strain are seen only in hairpin loops (Fig. 4), while the 2D structure of H2 and H3 helices remains unchanged. The variability among individual species is displayed in the 2D arrangement of the two main duplexes and existence of three types of substitutions.
The CBC substitutions in ITS1 and ITS2 of Barbatosphaeria mark different events. In the third and sixth bp of C-arm of H3 of ITS1 (Fig. 3) we detected two CBCs, which define three species-complexes, i.e. C=G and G=C base pairs in species with 1-septate ascospores (B. barbirostris, B. dryina) with the following compensatory base changes C=G → U/A and G=C in species with aseptate ascospores (B. fimbriata, B. neglecta, B. hippocrepida), C=G and G=C → U/A in species with 1–3-septate ascospores (B. arboricola, B. varioseptata, Barbatosphaeria sp.). Additionally, B. dryina possesses a species-unique insertion of three base pairs at the end of this duplex, i.e. two canonical C=G and one non-canonical U/C. The single CBC in the B-arm of H3 of ITS1 is between two species of the B. neglecta clade. In ITS2 we detected only one CBC in H2. The CBC is on the eleventh base pair and defines two groups, i.e. species with septate and aseptate ascospores, respectively. The individual clades within Barbatosphaeria represent the so-called Z clades (see above), defined by hCBCs and non-CBCs.
The rapidly evolving hCBC and the less frequent non-CBC substitutions may occur in organisms undergoing ecological adaptations or specialisation to certain habitats or substrates and are followed for example by changes in morphology (Caisová et al. 2011). The hCBCs and non-CBCs in conserved areas of ITS1 and ITS2 vary significantly within Barbatosphaeria. Of the seven bp with hCBCs detected in H3 of ITS1, four were species-unique, i.e. B. dryina (two bp) and B. fimbriata (two bp). The remaining hCBCs varied among species or strains of a single species (Fig. 3). The one non-CBC is limited to B. hippocrepida. In ITS2 we observed ten bp with hCBCs in H2 and H3 helices, four of them displayed both hCBC and non-CBC substitutions (Fig. 4). Five hCBCs characterise B. neglecta, four B. dryina and B. fimbriata, three are in B. hippocrepida and B. varioseptata, and one characterises the whole B. arboricola clade. The non-CBCs occurred in species with non-septate ascospores and in all members of the B. arboricola clade. The variability at the RNA structure level among strains of B. arboricola and B. neglecta clades is fully described in the Results section of this paper.
Acknowledgments
This study was supported by the Project of the National Foundation of the Czech Republic (GAP 506/12/0038), the project CEITEC (CZ.1.05/1.1.00/02.0068) and as a long-term research development project of the Institute of Botany, Academy of Sciences No. RVO 67985939, and of the Institute of Microbiology, Academy of Sciences No. RVO 61388971. Access to the MetaCentrum computing facilities provided under the program ‘Projects of Large Infrastructure for Research, Development, and Innovations’ LM2010005 funded by the Ministry of Education, Youth, and Sports of the Czech Republic and the access to the CERIT-SC computing and storage facilities provided under the programme Center CERIT Scientific Cloud, part of the Operational Program Research and Development for Innovations, reg. no. CZ. 1.05/3.2.00/08.0144 are acknowledged. We thank Walter Gams for his helpful editorial suggestions and Jacques Fournier for providing valuable material of B. fimbriata.
REFERENCES
- Amato A, Kooistra WHCF, Ghiron JHL, et al. 2007. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 158: 193–207. [DOI] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, et al. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge PD, Schlitt T, Cannon PF, et al. 2008. Domain II hairpin structure in ITS1 sequences as an aid in differentiating recently evolved animal and plant pathogenic fungi. Mycopathologia 166: 1–16. [DOI] [PubMed] [Google Scholar]
- Caisová L, Marin B, Melkonian M. 2011. A close-up view on ITS2 evolution and speciation – a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evolutionary Biology 11:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J, Shearer CA. 2004. Annulusmagnus and Ascitendus, two new genera in the Annulatascaceae. Mycologia 96: 822–833. [DOI] [PubMed] [Google Scholar]
- Coleman AW.2000The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151: 1–9. [DOI] [PubMed] [Google Scholar]
- Coleman AW.2003ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends in Genetics 19: 370–375. [DOI] [PubMed] [Google Scholar]
- Coleman AW.2005Paramecium aurelia revisited. Journal of Eukaryotic Microbiology 52: 68–77. [DOI] [PubMed] [Google Scholar]
- Coleman AW.2007Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Research 35: 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AW, Mai JC. 1997. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. Journal of Molecular Evolution 45: 168–177. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Oppen MJH van. 2008. Secondary structure of the rRNA ITS2 region reveals key evolutionary patterns in acroporid corals. Journal of Molecular Evolution 67: 389–396. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Vacquier VD. 2002. Exploring the phylogenetic utility of ITS sequences for animals: a test case for abalone (Haliotis). Journal of Molecular Evolution 54: 246–257. [DOI] [PubMed] [Google Scholar]
- Côté CA, Greer CL, Peculis BA. 2002. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8: 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2008. A fissitunicate ascus mechanism in the Calosphaeriaceae, and novel species of Jattaea and Calosphaeria on Prunus wood. Persoonia 20: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry S, Köhler A, Coleman AW. 1999. Intraspecies analysis: comparison of ITS sequence data and gene intron sequence data with breeding data for a worldwide collection of Gonium pectorale. Journal of Molecular Evolution 48: 94–101. [DOI] [PubMed] [Google Scholar]
- Ferrer A, Miller AN, Sarmiento C, et al. 2012. Three new genera representing novel lineages of Sordariomycetidae (Sordariomycetes, Ascomycota) from tropical freshwater habitats in Costa Rica. Mycologia 104: 865–879. [DOI] [PubMed] [Google Scholar]
- Gams W, Hoekstra ES, Aptroot A. 1998. CBS course of mycology, 4th ed Baarn, The Netherlands; Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- Geiser DM, Frisvad JC, Taylor JW. 1998. Evolutionary relationships in Aspergillus section Fumigati inferred from partial β-tubulin and hydrophobin DNA sequences. Mycologia 90: 831–845. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertzen LR, Cannone JJ, Gutell RR, et al. 2003. ITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae. Molecular Phylogenetics and Evolution 29: 216–234. [DOI] [PubMed] [Google Scholar]
- Gutell RR.1993. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Research 21: 3051–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Gray MW, Schnare MN. 1993. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures. Nucleic Acids Research 21: 3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit 5.0.9: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Ho WWH, Tsui CKM, Hodgkiss IJ, et al. 1999. Aquaticola, a new genus of Annulatascaceae from freshwater habitats. Fungal Diversity 3: 87–97. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hyde KD. 1992. Tropical Australian freshwater fungi I. Annulatascus velatispora gen. et sp. nov., A. bipolaris sp. nov. and Nais aquaticola sp. nov. (Ascomycetes). Australian Systematic Botany 5: 117–124. [Google Scholar]
- Jaklitsch WM, Réblová M, Voglmayr H. 2013. Molecular systematics of Woswasia atropurpurea gen. et sp. nov. (Sordariomycetidae), a fungicolous ascomycete with globose ascospores and holoblastic conidiogenesis. Mycologia 105: 476–485. [DOI] [PubMed] [Google Scholar]
- Joseph N, Krauskopf E, Vera MI, et al. 1999. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Research 27: 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschstein W. 1907. Neue Märkische Ascomyceten. Verhandlungen des Botanischen Vereins der Provinz Brandenburg. 48: 39–61. [Google Scholar]
- Koetschan C, Förster F, Keller A, et al. 2010. The ITS2 database III – sequences and structures for phylogeny. Nucleic Acids Research 38: D275–D279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D, Gargas A.2004The basidiomycete genus Polyporus – an emendation based on phylogeny and putative secondary structure of ribosomal RNA molecules. Feddes Repertorium 115: 530–546. [Google Scholar]
- Krüger D, Gargas A.2008Secondary structure of ITS2 rRNA provides taxonomic characters for systematic studies – a case in Lycoperdaceae (Basidiomycota). Mycological Research 112: 316–330. [DOI] [PubMed] [Google Scholar]
- Larget B, Simon DL. 1999. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution 16: 750–759. [Google Scholar]
- Lee SB, Taylor JW. 1990. Isolation of DNA from fungal mycelium and single spores. In: Innis MA, Gelfand DH, Snisky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 282–287. Academic Press, San Diego. [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Research 30: 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hu DM, Cai L. 2012. Conlarium duplumascospora gen. et. sp. nov. and Jobellisia gregariusca sp. nov. from freshwater habitats in China. Mycologia 104: 1178–1186. [DOI] [PubMed] [Google Scholar]
- Mai JC, Coleman AW. 1997. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. Journal of Molecular Evolution 44: 258–271. [DOI] [PubMed] [Google Scholar]
- Müller T, Philippi N, Dandekar T, et al. 2007. Distinguishing species. RNA 13: 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J. 2008. MrModeltest2 v. 2.3 (Program for selecting DNA substitution models using PAUP*). Evolutionary Biology Centre, Uppsala, Sweden. [Google Scholar]
- Online Auction Color Chart Co. 2004. The online auction color chart. The new language of color for buyers and sellers. Online Auction Color Chart Company. [Google Scholar]
- Petrak F. 1924. Mykologische Notizen. VII. Annales Mycologici 22, 1–2: 1–182. [Google Scholar]
- Raja HA, Campbell J, Shearer CA. 2003. Freshwater ascomycetes: Cyanoannulus petersenii, a new genus and species from submerged wood. Mycotaxon 88: 1–17. [Google Scholar]
- Rambaut A, Suchard MA, Xie D, et al. 2013. MCMC trace analysis tool version v1.6.0. Available from http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Réblová M.2006Molecular systematics of Ceratostomella sensu lato and morphologically similar fungi. Mycologia 98: 63–93. [DOI] [PubMed] [Google Scholar]
- Réblová M.2007Barbatosphaeria gen. et comb. nov., a new genus for Calosphaeria barbirostris. Mycologia 99: 723–732. [PubMed] [Google Scholar]
- Réblová M, Gams W, Seifert KA. 2011. Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Studies in Mycology 68: 163–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Mostert L, Gams W, et al. 2004. New genera in the Calosphaeriales: Togniniella and its anamorph Phaeocrella, and Calosphaeriophora as anamorph of Calosphaeria. Studies in Mycology 50: 533–550. [Google Scholar]
- Réblová M, Réblová K. 2013. RNA secondary structure, an important bioinformatics tool to enhance multiple sequence alignment: a case study (Sordariomycetes, Fungi). Mycological Progress 12: 305–319. [Google Scholar]
- Réblová M, Štěpánek V.2009New fungal genera, Tectonidula gen. nov. for Calosphaeria-like fungi with holoblastic-denticulate conidiogenesis and Natantiella gen. nov. for three species segregated from Ceratostomella. Mycological Research 113: 991–1002. [DOI] [PubMed] [Google Scholar]
- Réblová M, Štěpánek V, Schumacher RK. 2014. Xylochrysis lucida gen. et sp. nov., a new lignicolous ascomycete (Sordariomycetidae) with holoblastic conidiogenesis. Mycologia 106: 564–572. [DOI] [PubMed] [Google Scholar]
- Réblová M, Untereiner W, Réblová K. 2013. Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLOS One 8, 5: e63547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Winka K. 2000. Phylogeny of Chaetosphaeria and its anamorphs based on morphological and molecular data. Mycologia 92: 939–954. [Google Scholar]
- Rodriguez-Martínez R, Rocap G, Logares R, et al. 2012. Low evolutionary diversification in a widespread and abundant uncultured protist (MAST-4). Molecular Biology and Evolution 29: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Samuels GJ, Candoussau F. 1996. Heterogeneity in the Calosphaeriales: a new Calosphaeria with ramichloridium- and sporothrix-like synanamorphs. Nova Hedwigia 62: 47–60. [Google Scholar]
- Schultz J, Maisel S, Gerlach D, et al. 2005. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11: 361–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Muller T, Achtziger M, et al. 2006. The internal transcribed spacer 2 database – a web server for (not only) low level phylogenetic analyses. Nucleic Acids Research 34: W704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig C, Wolf M, Muller T, et al. 2007. The ITS2 database II: homology modelling RNA structure for molecular systematics. Nucleic Acids Research 36: D377–D380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer CL, Crane JL, Fallah PM. 2003. A reassessment of two freshwater ascomycetes, Ceriospora caudae-suis and Submersisphaeria aquatica. Mycologia 95: 41–53. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. 2005. RaxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463. [DOI] [PubMed] [Google Scholar]
- Stombaugh J, Zirbel CL, Westhof E, et al. 2009. Frequency and isostericity of RNA base pairs. Nucleic Acids Research 37: 2294–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukosd Z, Knudsen B, Kjems J, et al. 2012. PPfold 3.0: Fast RNA secondary structure prediction using phylogeny and auxiliary data. Bioinformatics 28: 2691–2692. [DOI] [PubMed] [Google Scholar]
- Takamatsu S, Hirata T, Sato Y. 1998. Phylogenetic analysis and predicted secondary structures of the rDNA internal transcribed spacers of the powdery mildew fungi (Erysiphaceae). Mycoscience 39: 441–453. [Google Scholar]
- Tsui CKM, Hodgkiss IJ, Hyde KD. 2003. Three new species of Aquaticola (Ascomycetes) from tropical freshwater habitats. Nova Hedwigia 77: 161–168. [Google Scholar]
- Ullrich B, Reinhold K, Niehuis O, et al. 2010. Secondary structure and phylogenetic analysis of the internal transcribed spacers 1 and 2 of bush crickets (Orthoptera: Tettigoniidae: Barbitistini). Journal of Zoological Systematics and Evolutionary Research 48: 219–228. [Google Scholar]
- Vijaykrishna D, Mostert L, Jeewon R, et al. 2004. Pleurostomophora, an anamorph of Pleurostoma (Calosphaeriales), a new anamorph genus morphologically similar to Phialophora. Studies in Mycology 50: 387–395. [Google Scholar]
- Wong SW, Hyde KD. 1999. Proboscispora aquatica gen. et sp. nov., from wood submerged in freshwater. Mycological Research 103: 81–87. [Google Scholar]
- Wong SW, Hyde KD, Jones EBG. 1999. Ultrastructural studies of freshwater ascomycetes, Fluminicola bipolaris gen. et sp. nov. Fungal Diversity 2: 189–197. [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]