Repair of double-strand breaks in mammalian mitochondria depends on microhomology-mediated end joining (MMEJ). Classical NHEJ is not detectable in mitochondria. DNA ligase III, but not ligase IV or ligase I, is involved in mitochondrial MMEJ. The protein machinery involved in miitochondrial MMEJ includes CtIP, FEN1, ligase III, MRE11, and PARP1.
Abstract
Mitochondrial DNA (mtDNA) deletions are associated with various mitochondrial disorders. The deletions identified in humans are flanked by short, directly repeated mitochondrial DNA sequences; however, the mechanism of such DNA rearrangements has yet to be elucidated. In contrast to nuclear DNA (nDNA), mtDNA is more exposed to oxidative damage, which may result in double-strand breaks (DSBs). Although DSB repair in nDNA is well studied, repair mechanisms in mitochondria are not characterized. In the present study, we investigate the mechanisms of DSB repair in mitochondria using in vitro and ex vivo assays. Whereas classical NHEJ (C-NHEJ) is undetectable, microhomology-mediated alternative NHEJ efficiently repairs DSBs in mitochondria. Of interest, robust microhomology-mediated end joining (MMEJ) was observed with DNA substrates bearing 5-, 8-, 10-, 13-, 16-, 19-, and 22-nt microhomology. Furthermore, MMEJ efficiency was enhanced with an increase in the length of homology. Western blotting, immunoprecipitation, and protein inhibition assays suggest the involvement of CtIP, FEN1, MRE11, and PARP1 in mitochondrial MMEJ. Knockdown studies, in conjunction with other experiments, demonstrated that DNA ligase III, but not ligase IV or ligase I, is primarily responsible for the final sealing of DSBs during mitochondrial MMEJ. These observations highlight the central role of MMEJ in maintenance of mammalian mitochondrial genome integrity and is likely relevant for deletions observed in many human mitochondrial disorders.
INTRODUCTION
Maintenance of genomic integrity is of prime importance for cellular function and survival in all organisms. Endogenous and exogenous insults generate DNA damage in the nucleus and other organelles of living cells (Boesch et al., 2011; Gostissa et al., 2011). Of the various types of DNA damage, DNA double-strand breaks (DSBs) pose the most serious threat to the genome (Deriano and Roth, 2013), and failure to repair such lesions could lead to chromosomal rearrangement, disease, or cell death (Khanna and Jackson, 2001; Lieber et al., 2006; Nambiar et al., 2008; Srivastava et al., 2012; Bunting and Nussenzweig, 2013). DSBs can be generated by exogenous factors such as ionizing radiation and chemotherapeutic agents. Endogenous mediators of DSBs include free radicals and enzymatic processes (Weterings and Chen, 2008; Lieber, 2010).
Repair of DSBs is mediated by either the classical nonhomologous DNA end-joining (NHEJ) pathway or homologous recombination (HR). HR restores the original DNA sequence using DNA from an intact sister chromatid as a template. Although error-prone, NHEJ also helps to maintain genome function (Moore and Haber, 1996; Jackson, 2002; Hefferin and Tomkinson, 2005; Wyman and Kanaar, 2006; Corneo et al., 2007; Yan et al., 2007; Lieber, 2010). Classical NHEJ (C-NHEJ) repairs DSBs in all phases of the cell cycle except the M phase, whereas HR activity is restricted to late S and G2 phases (Lieber et al., 2003; Ciccia and Elledge, 2010; Orthwein et al., 2014).
It is now evident that an alternative NHEJ (A-NHEJ) operates in human and yeast cells (Riballo et al., 2004; Wang et al., 2006), which may be genetically deficient for one or more factors that are critical for C-NHEJ (Boulton and Jackson, 1996; Kabotyanski et al., 1998; Li et al., 2008). The junctions created by A-NHEJ are often associated with larger deletions and use of microhomology, and hence it is alternatively described as microhomology-mediated end joining (MMEJ; Deriano and Roth, 2013). It is likely that MMEJ is indeed a subset of A-NHEJ.
Proteins that mediate MMEJ participate in end resection and stabilization of paired intermediates using microhomologies. In addition, specific nucleases remove 5′ and 3′ overhangs to enable ligation by DNA ligase I or ligase III. Although the precise mechanistic aspects of the pathway are not entirely clear, the enzymatic and microhomology requirements of MMEJ have been described (Nussenzweig and Nussenzweig, 2007; Sharma et al., 2015). Recent studies suggest that MMEJ is operational even when C-NHEJ is functionally active (Nussenzweig and Nussenzweig, 2007; Sharma et al., 2015).
The MMEJ pathway is considered less faithful than C-NHEJ and has been implicated in generation of chromosomal rearrangements in progenitor B-cell tumors of mice doubly deficient for XRCC4 or ligase IV and p53. Such observations have not yet been confirmed in human cells (Roth, 2002; Zhu et al., 2002; Ghezraoui et al., 2014). It is not clear whether MMEJ represents a defined and linear pathway or a myriad of pathways that use microhomologies (Deriano and Roth, 2013).
Mitochondria are essential organelles that generate energy through oxidative phosphorylation and are often described as the powerhouses of the cell (Wallace, 2005). Uniquely, mitochondria encapsulate their own DNA (mtDNA) and are semiautonomous. The mammalian mitochondrial genome comprises of 2–10 copies of circular, negatively supercoiled, double-stranded DNA. Mitochondrial DNA is just as prone to damage as nuclear DNA and relies on import of nuclear proteins involved for repair (Bohr, 2002).
Mitochondrial DNA is more prone to damage due to frequent exposure to reactive oxygen species (ROS) generated by mitochondrial oxidative phosphorylation and lack of protective histones (Chen et al., 2011). Consequently, mtDNA accumulates mutations at a 10- to 50-fold higher rate than nuclear DNA (Yakes and Van Houten, 1997; Hudson et al., 1998; Michikawa et al., 1999; Pakendorf and Stoneking, 2005). The importance of mtDNA integrity is evidenced by the increasing mutations, deletions, inversions, and complex rearrangements linked to various heritable human disease syndromes (Falk and Sondheimer, 2010). In addition, mtDNA rearrangements and large-scale deletions are associated with cancer and aging (www.mitomap.org/MITOMAP). More than 100 unique mtDNA deletions have been identified in patients with mitochondrial dysfunction (Holt et al., 1988; Servidei, 2003; www.mitomap.org/bin/view.pl/MITOMAP/DeletionsMultiple). Of interest, 85% of mtDNA deletions are flanked by short, directly repeated sequences and are implicated in aging (Eshaghian et al., 2006; Meissner et al., 2008), cancer (Abnet et al., 2004; Wu et al., 2005; Tseng et al., 2006; Chen et al., 2011), Kearns–Sayre syndrome, myopathies, progressive external ophthalmoplegia (Schon et al., 1989; Degoul et al., 1991; Ballinger et al., 1992), diabetes, and deafness (Ballinger et al., 1992).
Although base excision repair has been extensively studied in the context of mtDNA, very little is known about DSB repair in mitochondria. Because DSBs are a prerequisite for many DNA alterations in mitochondria, the mechanisms underlying lesional repair in mitochondria are of great importance. Preliminary studies have suggested a potential role for HR activity in mitochondria (Kajander et al., 2001; D’Aurelio et al., 2004). However, other DSB repair pathways, such as C-NHEJ and MMEJ, have yet to be explored in the context of mtDNA. The observed microhomology regions associated with deletions in patients indicate a potential role for NHEJ in maintenance of the mitochondrial genome. In addition, the protein machinery involved in mitochondrial DSB repair requires further characterization.
In the present study, we report that whereas classical NHEJ is undetectable in both rodents and human mitochondria, MMEJ is critical for DSB repair in mtDNA. Whereas the minimum homology required for MMEJ was 5 nucleotides (nt), an increase in microhomology length enhanced efficiency. Robust induction of several proteins involved in DSB repair was observed in mitochondria of rodent tissues and human cells. Immunoprecipitation and inhibitor studies in rodents showed that the observed MMEJ in mitochondria depends on CtIP, FEN1, MRE11, and poly(ADP-ribose) polymerase (PARP1). Antisense RNA-mediated knockdown of DNA ligases in HeLa cells showed that ligase III, but not ligase IV or ligase I, is important for MMEJ in mitochondria.
RESULTS
Mitochondrial proteins do not support joining of DSBs by C-NHEJ
To test whether mammalian mitochondria possess the protein machinery to repair double-stranded DNA breaks through C-NHEJ, we studied mitochondrial protein extracts in a DSB repair assay. Mitochondrial protein extracts from the brain, testes, spleen, and kidney of rats were analyzed for purity by immunoblotting using specific markers (Figure 1, A and B). Results confirmed the presence of the specific mitochondrial markers VDAC and cytochrome C proteins in mitochondrial extracts, but not cytoplasmic extracts (Figure 1A). Expression of β-actin served as loading control (Figure 1B). To exclude nuclear contamination of the mitochondrial extracts, we tested for the presence of PCNA, a nucleus-specific protein. Whereas robust expression was seen in cell-free extracts (CFEs), PCNA was undetectable in mitochondrial extracts (Figure 1B).
FIGURE 1:
Comparison of efficiency of classical NHEJ in cell-free extracts and in mitochondrial extracts prepared from various rat tissues. (A, B) Evaluation of purity of mitochondrial extracts. Mitochondrial and cell-free extracts were prepared from rat tissues (brain, testes, spleen, kidney) and analyzed for specific markers by Western blotting. CE, cytoplasmic extract; ME, mitochondrial extract. CEs and MEs were probed with VDAC and cytochrome C (mitochondrial markers), with β-actin as loading control (A). PCNA and β-actin (nuclear and CFE markers, respectively) were used as the markers to test the purity of fractions. (C) Schematic representation of the oligomeric substrates (5′ compatible, noncompatible [5′-5 and 5′-3′], and blunt ends) mimicking endogenous DSBs used for the study. (D–G) Bar diagrams showing quantification of the end-joined products from mitochondrial extracts from the brain (D), testes (E), spleen (F), and kidney (G) using 5′-compatible, noncompatible, and blunt ends. WCE extracts from respective rat tissues were used as positive control. For NHEJ assay, 5-μg extracts were incubated with [γ-32P]ATP-labeled oligomeric DNA substrate for 2 h at 30°C (testis) or 37°C. The reaction products were deproteinized and resolved on 8% denaturing PAGE. For quantification, the highest photostimulated light unit (PSLU) value of end joining was taken as 100%, and the relative efficiency between different extracts was calculated. Experiments were repeated a minimum of three times.
The ability of mitochondrial proteins to support classical NHEJ was tested by incubating extracts with double-stranded (ds) oligomeric DNA that mimic endogenous DSBs (Figure 1C). We incubated [γ-32P]ATP end-labeled ds oligonucleotides containing unique end structures (5′ overhangs, noncompatible [5′-5 and 5′-3′] and blunt ends; 2 or 6 h) with mitochondrial protein extracts from testes, brain, spleen, and kidney of rats and resolved purified products in denaturing PAGE gels (Supplemental Figure S1A). Whole-cell extracts (WCEs) prepared in parallel from the respective tissues served as controls. Results showed that whereas WCEs catalyzed efficient end joining, resulting in dimers, trimers, and other multimers, mitochondrial proteins failed to do so irrespective of the type of DSB (Figure 1, D–G) or quantity of extracts used (Supplemental Figure S2, A and B). When incubated with CFE, the overall joining efficiency depended on the nature of DSB used and was maximal with 5′-compatible ends (Figure 1, D–G). In contrast, when mitochondrial extracts (MEs) were used, end joining was seen exclusively with testes ME using a 5′-compatible substrate (Figure 1E) that does not require any processing before ligation. Of importance, there was no detectable joining when mitochondrial proteins were incubated with noncompatible or blunt-end DNA substrates, irrespective of the source of tissue extracts (Figure 1, D–G). This does not, however, rule out the possibility of weak end joining at very high concentrations of MEs. Nonetheless, MEs possessed no classical end-joining activity at a range of concentrations for which CFEs showed proficient C-NHEJ–mediated repair. Thus our results show that MEs do not support end-to-end joining of nonligatable broken DNA, suggesting the lack of a functionally operative C-NHEJ in mitochondria.
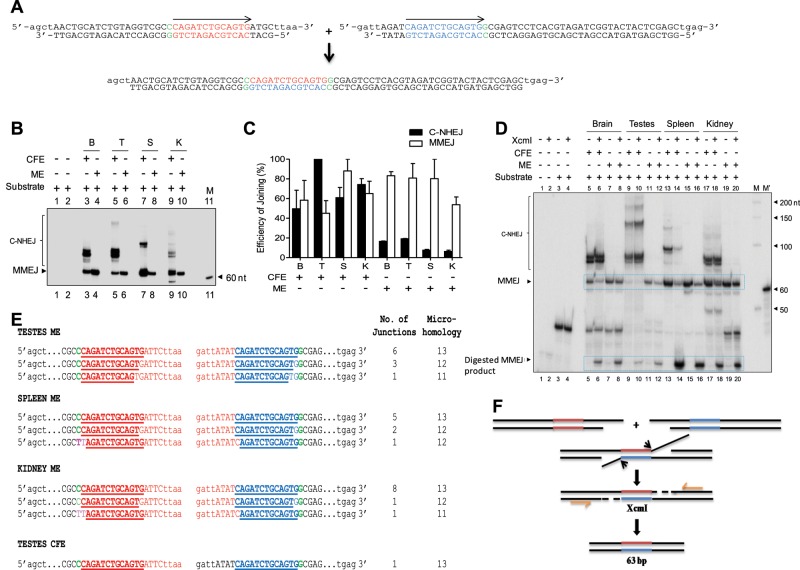
Mitochondrial extracts possess efficient MMEJ
Given that there was no detectable C-NHEJ in mitochondria, we wondered whether DSB repair was mediated through an alternative microhomology-dependent joining pathway. To test this hypothesis, we designed two oligomeric ds DNAs harboring 13-nt direct repeats (microhomology; Figure 2A). On joining by MMEJ, these microhomology regions can be cleaved using XcmI-mediated restriction endonuclease digestion. The sequential steps involved in the MMEJ assay are outlined in Supplemental Figure S1B. After “radioactive” PCR amplification, a 63–base pair fragment is expected if joining is mediated through MMEJ, whereas higher–molecular weight products indicate either C-NHEJ or MMEJ (Figure 2A). The MMEJ assay was performed by incubating 5 μg of mitochondrial protein extracts (derived from rat brain, testes, spleen, or kidney) with ds DNA containing DSBs flanked by direct repeats (2 h at 25°C), and the end-joined products were resolved in 8% denaturing PAGE gels. Of interest, a distinct 63-nt band (Figure 2B, arrow) was observed upon incubation with mitochondrial extracts (from the brain, testes, spleen, and kidney), suggesting functional microhomology-mediated end joining. Of interest, we did not find any major differences between individual tissue extracts in the efficiency of MMEJ catalyzed (Figure 2, B and C). Of importance, and consistent with the foregoing results, we did not observe any joined products that could be attributed to functional C-NHEJ in mitochondria. However, when cell-free extracts were used for the study, joined products consistent with both MMEJ and C-NHEJ were observed, in keeping with our previous observations (Sharma et al., 2015; Figure 2, B and C). Hence our results suggest that whereas cell-free extracts use both C-NHEJ and MMEJ for end joining of DSBs, mitochondrial extracts catalyze joining exclusively through MMEJ.
FIGURE 2:
Evaluation of microhomology-mediated DNA end joining in MEs prepared from rat tissues. (A) Schematic presentation of the microhomology (13 nt) containing substrate used and expected MMEJ reaction product (63 base pairs) after PCR amplification. Region of microhomology is indicated in blue and red in the substrates. (B) Denaturing PAGE profile showing MMEJ observed after PCR amplification catalyzed by MEs from rat brain, testes, spleen, and kidney. Cell-free extracts from respective tissues served as the control. The arrow indicates expected 63-nt-long dimeric product resulting from MMEJ. Other bands above the MMEJ product are due to microhomology-independent NHEJ products. M, 60-nt marker. For MMEJ assay, 5-μg extracts were incubated with oligomeric ds DNA substrates for 2 h at 25°C, and the end-joined products were PCR amplified using [γ-32P]ATP- labeled primers and resolved on 8% denaturing PAGE. (C) Bar diagram showing quantification of the end-joined products based on multiple experiments. The efficiency of the joining was calculated. For quantification, the highest PSLU value of C-NHEJ or MMEJ was taken as 100%, and the relative efficiency within different extracts was calculated. (D) XcmI digestion of MMEJ products of mitochondrial extracts from the brain, testes, spleen, and kidney. XcmI digestion depends on recreation of the microhomology region upon joining, as indicated in A. MMEJ products were incubated with XcmI (5 U/10 μl reaction) at 37°C for 2 h, and the products were resolved on 8% denaturing PAGE. (E) Cloning and sequencing of MMEJ junctions derived from mitochondria of testes, spleen, and kidney. The nucleotides in blue represent the retained microhomology region, and red shows deletion. Green represents the junction between random sequence and the start of microhomology. Pink indicates mutations. All NHEJ junctions shown are from different clones and are derived from independent PCR and transformations. (F) Mechanism of MMEJ based on sequence analysis.
To confirm the requirement of microhomology for end joining catalyzed by mitochondrial proteins, we performed an XcmI restriction enzyme digestion (Figure 2D). On digestion with XcmI, a band corresponding to 63 nt formed due to MMEJ was cleaved both in the case of MEs and CFEs, showing microhomology-mediated end joining, albeit with different efficiency (Figure 2D). To confirm further the use of microhomology at the sequence level, we PCR amplified, cloned, and sequenced the joined junctions. Sequencing analyses of a number of independent clones showed that in all cases the deletion used the direct repeats in mitochondria (Figure 2, E and F). Perfect microhomology was seen in 68% of clones (19 of 28, derived from testes, spleen, and kidney), whereas 32% were joined with one or two mismatches (Figure 2E). These mismatches or mutations likely explain the partial cleavage of the joined products by XcmI digestion.
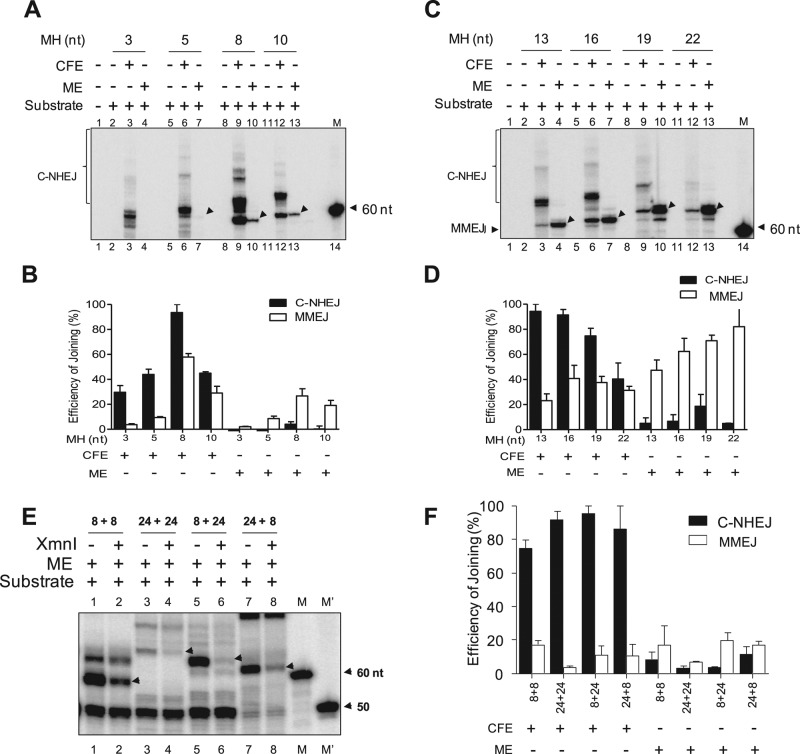
Efficiency of MMEJ in mitochondria depends on the length of microhomology
Because efficient MMEJ was observed in mitochondrial extracts, we next focused on the effect of microhomology length on joining. To address this, we performed a joining assay with oligomeric DNA substrates possessing DSBs flanked by microhomology lengths of 3, 5, 8, 10, 13, 16, 19, and 22 base pairs, using mitochondrial protein extracts prepared from rat testes. Results showed that the efficiency of MMEJ improved with increasing length of microhomology (Figure 3, A–D). Of interest, a microhomology length of 3 nt did not support MMEJ in mtDNA, and the efficiency was weak when 5-nt microhomology was used (Figure 3, A and B). A microhomology length–dependent increase in MMEJ efficiency was also observed with cell-free extracts, which is consistent with our previous study (Sharma et al., 2015). Taken together, our results demonstrate that, unlike whole cells, mitochondria are devoid of functional C-NHEJ but instead possess MMEJ activity, which is dependent on the length of microhomology.
FIGURE 3:
Evaluation of length and position of microhomology on efficiency of MMEJ in mitochondria. (A) Gel profile showing efficiency of MMEJ in mitochondrial extracts when length of microhomology is 3, 5, 8, or 10 nt. (B) Bar diagram showing quantification of MMEJ and C-NHEJ products shown in A. Graph is a cumulative representation of three independent repeats. For quantification, highest PSLU value of C-NHEJ or MMEJ was taken as 100%, and the relative efficiency between different extracts or substrates was calculated. (C) Denaturing PAGE profile showing efficiency of MMEJ in mitochondrial extracts when the length of the microhomology is 13, 16, 19, or 22 nt. (D) Bar graphs representing quantification of the C-NHEJ and MMEJ products shown in C. In all cases, cell-free extracts from testes served as the control. For quantification, the highest PSLU value of C-NHEJ or MMEJ was taken as 100%, and the relative efficiency between different extracts or substrates was calculated. (E) Comparison of MMEJ efficiency of mitochondrial extracts when an 8-nt microhomology region was positioned at different lengths from a DSB. Here, 8+8 indicates that the distance between the microhomology and DSB is 8 nt in both substrates; 24+24, that the distance between microhomology and DSB is 24 nt in each substrate; and while 8+24 and 24+8 that the distance is either 8 or 24 nt as indicated. DNA substrates were designed in such a way that microhomology- mediated joining creates a restriction site for XmnI. The band at 50 nt is contributed by random primer extension of unused DNA substrates where radiolabeled primer can bind. (F) Bar diagram showing quantification of the end-joined products of the gels shown in E. In all cases, MMEJ products are indicated by arrows, and C-NHEJ products are bracketed. For quantification, the highest PSLU value of C-NHEJ or MMEJ was taken as 100%, and the relative efficiency between different extracts or substrates was calculated. M′, γ-32P–labeled 50-nt ladder; M, labeled 60-nt oligomer.
Relative positioning of the microhomology region with the DSB affects MMEJ efficiency
Use of microhomology by alternative NHEJ leads to extensive deletions at the junctions (Sharma et al., 2015). Thus we sought to understand the effect of spacing between the microhomology region and DNA termini. For this, we designed distinct oligomeric DNA substrates containing 8-nt microhomology regions but positioned internally either 8 or 24 nt from the terminus. These microhomology regions can be cleaved by XmnI restriction digest of the microhomology- mediated joining products (as described earlier for XcmI in the case of 13-nt microhomology substrates). A joining assay using mitochondrial protein extracts from rat testes showed efficient MMEJ joining when the microhomology was positioned 8 nt from the DSB (Figure 3, E and F). MMEJ efficiency was significantly compromised when the microhomology was positioned 24 nt from the DSB (Figure 3, E and F). However, when the microhomology was 24 nt from the terminus in one of the DNA substrates, we observed efficient MMEJ (Figure 3, E and F). A similar pattern of MMEJ efficiency was observed when the foregoing permutations of substrates were incubated with cell-free testes extracts. Of importance, despite the remarkable overall efficiency of C-NHEJ with cell-free extracts, it was undetectable in the case of mitochondrial extracts (Figure 3C). Furthermore, XmnI digestion confirmed that the band corresponding to MMEJ indeed used microhomology for joining (Figure 3E). In addition, sequencing analyses of the joined junctions confirmed the use of microhomology regions (Supplemental Figure S3, A and B). In summary, our results suggest that mitochondrial extracts can catalyze MMEJ and the efficiency of the joining depends on the distance between the microhomology and DSB termini.
MMEJ catalyzed by mitochondrial extracts depends on CtIP, FEN1, ligase III, MRE11, and PARP1
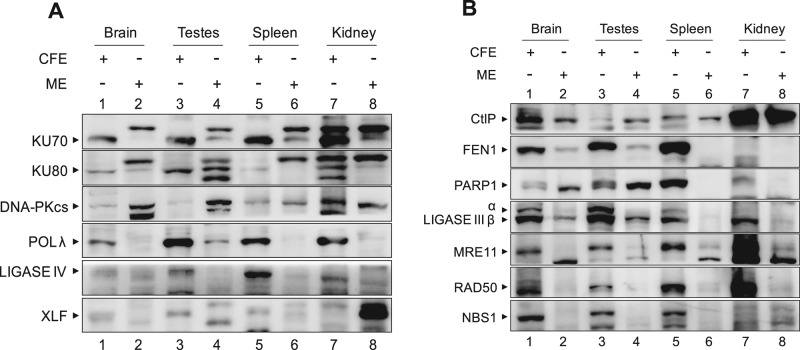
To understand the mechanisms of DSB joining in mitochondria, we examined the presence of critical C-NHEJ and MMEJ proteins in mitochondrial extracts by Western blotting (Figure 4, A and B). Mitochondrial extracts from the brain, testes, spleen, and kidney were analyzed for expression of classical NHEJ proteins using β-actin as loading control (Figure 1B). Immunoblots showed no bands corresponding to the expected positions of KU70, KU80, POL λ, ligase IV, and XLF in most of the mitochondrial extracts, whereas robust expression was observed in cell-free extracts (Figure 4A). The abundance of DNA-dependent protein kinase catalytic subunit (DNA-PKcs), however, was comparable in mitochondrial and respective cell-free extracts. Of interest, we observed variable expression of PARP1 in mitochondrial tissue extracts (Figure 4B). In addition, we detected expression of ligase III and CtIP in all the mitochondrial extracts tested, whereas FEN1 expression was restricted to brain and testes. Although weak expression of MRE11 and RAD50 was observed in mitochondrial extracts from the tissues tested, NBS1 was undetectable (Figure 4B). For many proteins, we found multiple or variable band patterns in MEs compared with their CFE counterparts. To test the identity of these bands, we performed Western blots of proteins such as KU70, KU80, DNA-PK, and ligase IV after immunoprecipitation (Supplemental Figure S4). Although most of the bands identified with respective antibodies suggest specificity, further characterization of the significance of the multiple observed bands is beyond the scope of this study.
FIGURE 4:
Immunoblot analyses showing the presence and level of expression of DSB repair proteins in mitochondria of various rat tissues. (A) Comparison of level of expression of C-NHEJ proteins in mitochondrial extracts and CFE prepared from rat tissues. The ∼30-μg extracts from the brain, testes, spleen, and kidney were resolved on 8–10% SDS–PAGE, transferred to polyvinylidene fluoride membrane, and probed using the respective antibodies. (B) Presence and expression of MMEJ and other DSB repair proteins in mitochondrial extracts of various tissues and corresponding whole-cell extracts.
To characterize the protein machinery involved in observed MMEJ, we performed immunoprecipitation (IP) of distinct DSB proteins from mitochondrial extracts prepared from rat testes. Immunodepletion was confirmed by Western blot analysis, and β-actin served as the loading control (Figure 5A and Supplemental Figure S5, A and B). A significant reduction in microhomology-mediated joining (13 nt) was observed upon incubation with extracts after CtIP, FEN1, ligase III, MRE11, or PARP1 was immunodepleted (Figure 5, B and C). There was no significant reduction in the MMEJ efficiency when classical NHEJ proteins such as DNA-PKcs, KU70, and KU80 were immunodepleted (Figure 5, B and C). However, studies using complementation of proteins to the depleted extracts will be required to delineate the order and specificity of proteins involved in catalyzing MMEJ in the context of mtDNA.
FIGURE 5:
Efficiency of mitochondrial MMEJ after immunodepletion and inhibitor-based inactivation of C-NHEJ and alternate NHEJ proteins. (A) Bar diagrams showing the quantification of efficiency of immunodepletion in mitochondrial extracts. β-Actin was used as loading control. (B) Evaluation of efficiency of MMEJ in mitochondrial extracts after immunodepletion assayed using 13-nt microhomology substrates. (C) Bar diagram showing quantitation of MMEJ efficiency in immunodepleted rat tissue mitochondrial extracts based on multiple gels. For quantification, the relative intensity of the MMEJ band between different samples was calculated and expressed as PSLU. (D–I) Comparison of the effect of repair inhibitors against ATM kinase (KU55933), DNA-PK (NU7026), ligase I (L82), PI3K (wortmannin), MRE11 (mirin), and PARP1 (3-ABA) on MMEJ of 13-nt microhomology substrates. For quantification, the relative intensity of the MMEJ band between different samples was calculated and is expressed as PSLU (bar graphs). We preincubated 1- to 2-μg rat tissue mitochondrial extracts with inhibitors with increasing concentration (10 μM to 10 mM) for 30 min on ice and subjected them to MMEJ assay for 2 h at 25°C. The end-joined products were PCR amplified using γ-32P–labeled primers and resolved on 8% denaturing PAGE. In the case of control reactions, for which inhibitor was not added, and equal concentration of DMSO was used (see Supplemental Figure S7 for details). *p < 0.05, **p < 0.01, ***p < 0.001.
We next studied the effects of established chemical inhibitors of DSB repair on MMEJ of mtDNA. Mitochondrial extracts were preincubated with inhibitors of phosphoinositide 3-kinase (PI3K; wortmannin), DNA-PK (NU7026), pATM (KU55933), ligase I (L182), MRE11 (mirin), and PARP1 (3-aminobenzamide [3-ABA]) and subjected to MMEJ assay. Results showed that there was no significant difference in MMEJ efficiency in the presence of wortmannin, NU7026, KU55933, and L182, ruling out the involvement of DNA-PKcs, ATM, and ligase I in MMEJ (Figure 5, D–F, I, and Supplemental Figures S6 and S7). Of interest, a concentration-dependent decrease in MMEJ efficiency was observed in the presence of mirin and 3-ABA, further confirming the roles of MRE11 and PARP1 in mitochondrial MMEJ (Figure 5, G and H, and Supplemental Figure S7, B and C). In addition, when wortmannin and NU7026 were used, both inhibitors inhibited C-NHEJ, whereas MMEJ was unaffected, indicating the specificity of the inhibitors (Supplemental Figure S6).
Hence our studies using IP experiments in conjunction with inhibitor studies suggest that the MMEJ in mitochondria is dependent on CtIP, FEN1, ligase III, MRE11, and PARP1.
DNA ligase III, but not ligase IV, is involved in mitochondrial MMEJ
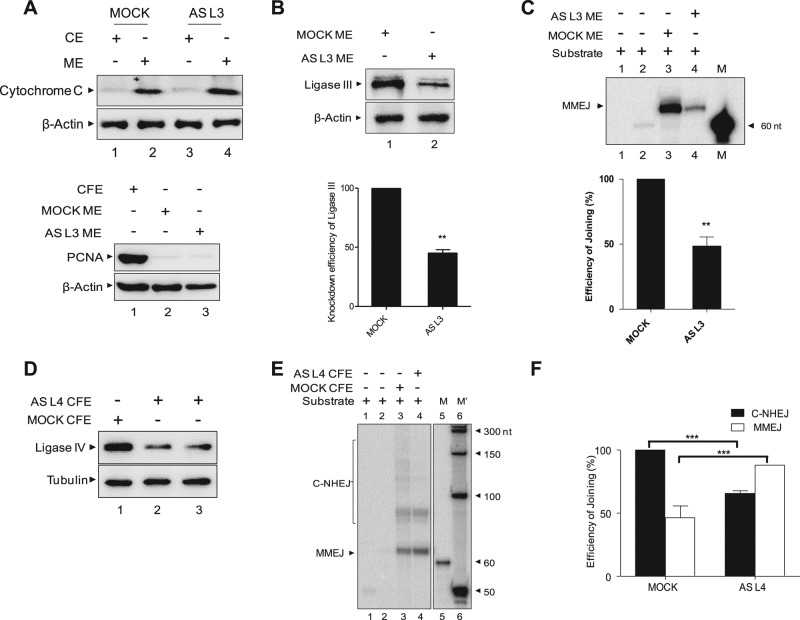
Because ligase I inhibitor did not show any effect on mitochondrial MMEJ, we focused our attention on the likely involvement of ligase III in catalyzing mitochondrial MMEJ. Owing to the lack of ligase III–knockout mice, we resorted to knockdown experiments in the HeLa cell line using human ligase III antisense plasmids. Purity and knockdown were confirmed in both mitochondrial and nuclear extracts by Western blot analysis (Figure 6, A and B). A significant reduction in microhomology-associated joining was observed when mitochondrial extracts prepared after ligase III knockdown was used (Figure 6C), in contrast to extracts prepared from a mock-transfected cell line. To additionally confirm that the mitochondrial MMEJ activity is indeed dependent on ligase III but not ligase IV in vivo, we knocked down the latter in HeLa cells using antisense RNA. The knockdown of ligase IV was confirmed by Western blotting in two independent transfections in both cell-free and mitochondrial extracts (Figure 6D). Of interest, ligase IV knockdown resulted in reduction of C-NHEJ–mediated product formation catalyzed by CFE but not MMEJ activity as compared with mock-transfected controls (Figure 6, E and F). Thus these results confirm that ligase III and not ligase IV is responsible for MMEJ in mammalian mtDNA.
FIGURE 6:
Mitochondrial DNA ligase III, but not ligase IV, is required for mitochondrial EJ inside the cells. (A) Immunoblot analyses showing the purity of cytosolic extracts (CEs) and mitochondrial protein extracts (MEs) prepared from HeLa cells transfected with antisense DNA ligase III plasmid. We used ∼15-μg extracts from HeLa cells (CE, ME, and CFE) for immunoblotting. Cytochrome C, PCNA, and β-actin were used as mitochondrial, nuclear, and CFE markers, respectively. (B) Immunoblot analysis showing knockdown of DNA ligase III from HeLa cells and its quantification. For quantification, expression in MOCK ME was taken as 100, and relative expression in AS L3 ME was calculated. β-Actin was used as a loading control. (C) MMEJ assay using HeLa mitochondrial extracts prepared from mock and antisense DNA ligase III–transfected cells on 13-nt microhomology–containing DNA substrates. Bar diagram showing relative reduction in MMEJ efficiency in antisense DNA ligase III–transfected cells compared with the mock transfection. (D) Immunoblot analysis showing knockdown of DNA ligase IV in HeLa cells transfected with mock and antisense DNA ligase IV plasmid. Tubulin was used as internal loading control. (E) Comparison of MMEJ in mitochondrial extracts after knockdown of ligase IV in HeLa. MMEJ activity in CFE served as the control. (F) Bar diagram showing relative reduction in C-NHEJ but not in MMEJ efficiency in antisense DNA ligase IV–transfected cells compared with the control transfection shown in D. *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
The integrity of mtDNA plays a critical role in maintaining cellular homeostasis, and efficient repair of mtDNA damage is important for cellular survival. When DSBs are induced in mitochondria by restriction endonucleases, both intramolecular and intermolecular recombination products with large deletions are observed (Fukui and Moraes, 2009). Although homologous recombination could be one of the potential pathways of mtDNA DSB repair to ensure stability of the genome, evidence to this end in mitochondria is lacking. The mechanisms underlying mitochondrial instability and the ensuing repair processes remain unclear. Various studies have suggested involvement of proteins such as retinoblastoma protein, BRCA1 (Coene et al., 2005), p53 (Ferecatu et al., 2009), CtBP (Kim and Youn, 2009), FEN (Liu et al., 2008; Szczesny et al., 2008; Kalifa et al., 2009), DNA ligase III (De and Campbell, 2007; Gao et al., 2011; Simsek et al., 2011), MRE11 (Dmitrieva et al., 2011; Kalifa et al., 2012), PARP1 (Rossi et al., 2009; Lapucci et al., 2011), and KU80 (Coffey and Campbell, 2000) in the maintenance of mtDNA integrity over and above their roles in ensuring nuclear genomic homeostasis.
Mitochondria possess efficient MMEJ but lack classical NHEJ
Our results provide compelling evidence that mitochondrial protein extracts prepared from distinct rat tissues and HeLa cells possess efficient microhomology-mediated joining activity. Of interest, the mitochondrial extracts were unable to catalyze joining through the classical NHEJ pathway, irrespective of the type of DSB studied. The efficiency of the MMEJ improved when the length of the microhomology region was increased, which is consistent with our recent study (Sharma et al., 2015). In addition, it appears that the spacing between the microhomology region and DSB is likely critical in determining joining efficiency. When microhomology was placed farther from the DNA terminus, joining efficacy was low.
To ensure that the unique observation in our study was not due to potential nuclear contamination in the mitochondrial extracts, we paid particular attention to exclude this possibility. First, mitochondrial proteins were isolated using two independent methods, and in each case, purity was confirmed by immunoblotting using specific mitochondrial and nuclear markers. Second, irrespective of the method used, mitochondrial protein extracts exhibited similar DSB repair activity (robust MMEJ but not C-NHEJ). Third, we believe that even trace nuclear protein contamination would have resulted in C-NHEJ, which was only evident with CFE. As further confirmation, the mitochondrial extracts prepared from human HeLa cells showed efficient MMEJ but were devoid of C-NHEJ activity (limited to DSBs with 5′-compatible overhangs; unpublished data). We hence propose that the observed exclusive MMEJ activity is indeed of mitochondrial origin and not due to any nuclear contamination.
Mitochondrial MMEJ depends on ligase III
In our studies, we observed mitochondria-specific expression of ligase III, PARP1, CtIP, MRE11, and RAD50 in rat tissues. Immunoprecipitation and antisense RNA-mediated knockdown in conjunction with specific inhibitors demonstrated that the DNA ligase responsible for MMEJ in mitochondria is ligase III. This observation is in line with previous reports suggesting that ligase III is indispensible for mtDNA integrity (Gao et al., 2011; Simsek et al., 2011; Oh et al., 2014). Our study now suggests the additional involvement of PARP1, CtIP, and MRE11 in mitochondrial MMEJ.
Although we detected the presence of proteins responsible for MMEJ by immunoblotting of mitochondria, the mechanism by which these proteins are transported to mitochondria remains ill defined. Most of them lack a conventional mitochondrial localization signal (MLS), despite their presence in mitochondria. Of interest, ligase III, which we show is critical for MMEJ in mitochondria, is known to have an alternative translation initiation mechanism, which produces proteins with or without MLS from the same mRNA (Lakshmipathy and Campbell, 1999). Although we could not identify any obvious MLS sequences in other proteins involved in mitochondrial MMEJ, the existence of additional mitochondrial mechanism(s) cannot be ruled out. It is hence intriguing that we did observe multiple bands/differences for individual proteins by Western blot analyses in MEs compared with their CFE counterparts. Although IP confirmed the specificity of these bands, the significance of this observation is worth pursuing.
Base excision repair (BER) has been well studied in the repair that follows oxidative damage in mitochondria (Svilar et al., 2011). Isoforms of the nuclear BER proteins have been reported in mitochondria in diverse organisms (Boesch et al., 2011). Therefore it is possible that, as with BER proteins, some of the nuclear DSB repair proteins are also targeted to resolve breaks in mitochondrial DNA.
Mitochondrial DNA deletions in human patients can be explained by MMEJ-mediated repair
By using a cloning and sequencing strategy, we showed the use of microhomology (direct repeats) by mitochondrial proteins before joining. Even when an exact microhomology was not used, a portion of the direct repeat was used for the joining. The observed deletions at the MMEJ junctions and efficient use of microhomology regions by mitochondrial proteins bore a striking similarity to deletions observed within the mitochondrial DNA of patients with mitochondrial dysfunction. Of interest, among >100 types of mitochondrial DNA deletions reported, at least 85% are flanked by short, directly repeated sequences and are implicated in aging, cancer, mitochondrial syndromes, and other diseases (Schon et al., 1989; Degoul et al., 1991; Ballinger et al., 1992; Abnet et al., 2004; Wu et al., 2005; Eshaghian et al., 2006; Tseng et al., 2006; Meissner et al., 2008; Chen et al., 2011). One of the hallmarks of the observed mitochondrial genome rearrangement was interstitial deletion between two direct repeat sequences in such a way that one of the repeats was retained after breakage and rejoining. Hence the characteristic features of the observed MMEJ in mitochondria are similar to those observed during mitochondrial deletions in patient samples. During MMEJ, an exonuclease-mediated deletion of one of the two strands of DNA from DSB exposes the microhomology region, followed by annealing of the repeat, removal of the flap by endonuclease, and final sealing by ligase III, resulting in a rearranged product (Sharma et al., 2015).
A majority of mtDNA deletions observed in vivo span direct DNA repeat sequences 5–13 base pairs in length (Tanhauser and Laipis, 1995; Van Tuyle et al., 1996). In our studies, we found that a minimum length of 5-nt microhomology was necessary for efficient MMEJ in mitochondria. In addition, efficiency of the joining improved when the length of the microhomology was increased to 18 base pairs, which correlates with the in vivo data. The length of the deletions typically observed in mammalian mtDNA in vivo ranges from 2 to 7 kb (Anderson et al., 1981; Queen and Korn, 1984). These large deletions in patients could be the result of independent DNA strand breaks occurring in close vicinity to inverted repeats joined by MMEJ. Hence our observation of small microhomology use is particularly relevant. Given the collective results, the MMEJ observed in our studies provides a potential mechanism for the genesis of mitochondrial DNA deletions seen in specific mitochondrial disorders. We suggest that understanding the pathological basis for these breaks warrants further investigation.
MATERIALS AND METHODS
Plasmid constructs
The antisense expression plasmids pREP-hLIGIII and pREP-hLIGIV were kind gifts from Colin R. Campbell (University of Minnesota).
Oligomers
The oligomers were purified using 8–15% denaturing PAGE as described (Nambiar et al., 2013). The 5′ end labeling of the oligomeric DNA was performed as previously described (Nambiar et al., 2013), using T4 polynucleotide kinase in appropriate buffer containing [γ-32P]ATP at 37°C for 1 h. The labeled substrates were purified using G-25 Sephadex columns and stored at −20°C until use. See Supplemental Table S1 for the oligomers used.
Preparation of DNA substrates for C-NHEJ and MMEJ
For C-NHEJ studies, oligomeric DNA substrates (75 base pairs) containing 5′-compatible overhangs were prepared by annealing a γ-32P-labeled TSK1 with cold complementary oligomer TSK2 (1:3 ratio) by slow annealing in the presence of 100 mM NaCl and 1 mM EDTA as described (Naik et al., 2010). Double-stranded DNA containing 5′-5′ and 5′-3′ noncompatible overhangs were prepared by mixing radiolabeled TSK1 with cold complementary oligomers VK11 and VK13, respectively. Blunt-ended DNA substrate was prepared by annealing radiolabeled VK7 with cold VK8. For MMEJ studies, substrates containing 13-nt microhomology were prepared by annealing SS69 with SS70 and SS71 with SS72, respectively, as mentioned (Chiruvella et al., 2012; Sharma et al., 2015). The 8-nt microhomology region substrates where the microhomology region was positioned 8 nt from DNA ends were generated by annealing SS56 with SS57 and SS67 with SS68, respectively. The 8-nt microhomology substrates positioned 24 and 8 nt, respectively, from DNA ends were prepared by annealing SS67 with SS68 and SS103 with SS104. The 8-nt microhomology substrates positioned 8 and 24 nt, respectively, from DNA ends were prepared by annealing SS56 with SS57 and SS105 with SS106. The 8-nt microhomology substrates where the microhomology region was positioned 24 nt away from DNA ends were prepared by annealing SS103 with SS104 and SS105 with SS106, respectively. After end joining, junctions were PCR amplified using radiolabeled primer SS60 and unlabeled SS61.
Preparation of cell-free extracts
Cell-free extracts were prepared in ice-cold conditions as described (Baumann and West, 1998). Refer to the Supplemental Methods for more details.
Preparation of mitochondrial protein extracts from brain, testes, spleen, and kidney of rats and HeLa cells
Mitochondrial protein extracts were prepared by two independent methods. In the first, mitochondria were isolated based on differential centrifugation as described (Maianski et al., 2004; Wieckowski et al., 2009), with modifications. In brief, the brain, testes, spleen, and kidney were dissected from 3- to 5-wk-old male Wister rats using standard dissection procedures, and extraction was done at 4°C or on ice. Each tissue was minced independently with a sterile scalpel and washed three times in ice-cold phosphate-buffered saline. Homogenization buffer (70 mM sucrose, 200 mM mannitol, 1 mM EDTA, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, and 0.5% bovine serum albumin per gram of tissue) was added and homogenized in a Dounce-type homogenizer applying 20–30 strokes. The homogenate was then centrifuged (3000 rpm, 10 min at 4°C) to remove nuclei, cellular debris, and intact cells. The supernatant contained the cytosol and mitochondria. This step was repeated at least three times to avoid nuclear contamination in the cytosolic fraction. The cytosolic fraction was then centrifuged at 12,000 rpm (30 min at 4°C) to pellet the mitochondria. The mitochondrial pellet was washed in suspension buffer (10 mM Tris-HCl, pH 6.7, 0.15 mM MgCl2, 0.25 mM sucrose, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM dithiothreitol [DTT]) twice and lysed in mitochondrial lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.2% Triton X-100, 2 mM ethylene glycol tetraacetic acid, 2 mM EDTA, 1 mM DTT, and 10% glycerol) by mixing end over end for 30 min at 4°C along with protease inhibitors (PMSF [1 mM] and aprotinin, pepstatin, and leupeptin [1 μg/ml]). Mitochondrial extract was centrifuged at 12,000 rpm (5 min), and the supernatant (mitochondrial fraction) was aliquoted, snap frozen, and stored at −80°C till use.
Alternatively, mitochondrial protein extracts were also prepared from rat tissues and HeLa cells as per manufacturer’s instructions using the Mitochondrial Extraction Kit (Imgenex) and differential centrifugation.
In vitro C-NHEJ assay
C-NHEJ reactions were performed as described earlier (Kumar et al., 2010; Sharma et al., 2011; Srivastava et al., 2012). Refer to the Supplemental Methods for more details.
MMEJ assay
MMEJ reactions were performed as described previously (Chiruvella et al., 2012; Sharma et al., 2015). Reactions were carried out in a volume of 20 μl by incubating DNA substrates containing 13-nt microhomology (SS69/70 and SS71/72) in a buffer containing 50 mM Tris-HCl (pH 7.6), 20 mM MgCl2, 1 mM DTT, 1 mM ATP, and 10% polyethylene glycol at 25°C for 2 h. We used 5-μg mitochondrial extracts per reaction. The MMEJ reactions were terminated by heat denaturation at 65°C for 5 min. The end-joined products were PCR amplified using radiolabeled primer SS60 and unlabeled SS61. The PCR products were resolved on 8% PAGE and quantified as described.
PCR amplification, cloning, and sequencing of end-joined junctions
After MMEJ reactions, end-joined junctions were PCR amplified, and products were resolved on 8% denaturing PAGE, dried, exposed, and scanned. The band of interest was cut out from PAGE and DNA eluted in a solution containing Tris/EDTA and NaCl (0.5 M). Gel-purified PCR products were ligated into a TA vector at 16°C for 16 h. Deproteinized ligated products were used for transformation of Escherichia coli. Plasmid DNA was isolated from the clones of interest. The presence of insert was verified by restriction enzyme digestion and confirmed by DNA sequencing (SciGenom, India).
Immunoprecipitation
IP experiments were performed as described, with modifications (Chiruvella et al., 2012). Protein G agarose beads (Sigma-Aldrich, St. Louis, MO) were activated by immersion in water and incubated in IP buffer (300 mM NaCl, 20 mM Tris-HCl, pH 8.0, 0.1% NP-40, 2 mM EDTA, 2 mM EGTA, and 10% glycerol) for 30 min on ice, after which the beads were conjugated with appropriate antibody at 4°C overnight to generate antibody–bead conjugate. The antibody–bead conjugates were then separated by centrifugation and incubated with rat tissue mitochondrial extracts at 4ºC overnight. The conjugate bound to target proteins was then separated and washed. Protein depletion in the resulting supernatant was confirmed by immunoblot analysis and quantified using Multi Gauge (version 3.0). These immunodepleted extracts were used for MMEJ assays (Sharma et al., 2015).
Evaluation of effect of DSB repair inhibitors on MMEJ
Various DSB repair inhibitors were prepared as follows. ATM kinase inhibitor KU55933, DNA-PK inhibitor NU7026, ligase I inhibitor L82, PI3K inhibitor wortmannin, and MRE11 inhibitor mirin (all from Sigma-Aldrich) were dissolved in dimethyl sulfoxide (DMSO) to make 100 mM stock solutions. For the PARP1 inhibitor 3-ABA (Sigma-Aldrich), 300 mM stock solutions were prepared in DMSO. For evaluation of the effect of inhibitors on MMEJ, 2-μg mitochondrial extracts (rat testes) were preincubated with increasing concentration of inhibitors (10 μM to 10 mM) for 30 min on ice and subjected to end joining (2 h at 25°C), and the products were analyzed as described. In the case of control reactions for which inhibitor was not added, the highest concentration of DMSO was used.
Antisense RNA-mediated knockdown of expression of ligase III and ligase IV
DNA ligase III and DNA ligase IV antisense expression plasmids (∼20 μg) were transfected into human HeLa cells using the calcium phosphate method (Raghavan et al., 2005; Naik and Raghavan, 2012; Kumari and Raghavan, 2015). Cells were grown for 48 h in MEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Sigma-Aldrich) and were harvested for preparation of small-scale whole-cell or mitochondrial extracts.
Preparation of small-scale mitochondrial extracts from HeLa cells
Small-scale extracts were prepared as described with slight modifications (Smeaton et al., 2007). Refer to the Supplemental Text for more details.
Supplementary Material
Acknowledgments
We thank N. Kalakonda, M. Nambiar, S. Sharma, M. Nishana, M. Pandey, and S. Vartak for suggestions and critical reading of the manuscript; M. Srivastava and V. Gopalakrishnan for cell culture and additional assistance; and S. Sharma for help in designing oligomers and their purification. We thank the Central Animal, Confocal, and Fluorescence-Activated Cell Sorting Facilities of the Indian Institute of Science for help. Financial assistance for the project to S.C.R. from the Council of Scientific and Industrial Research (37(1579)/13/EMR-II) and the Indian Institute of Science–Department of Biotechnology partnership program (DBT/BF/PR/INS/2011-12/IISc), Bangalore, is acknowledged. Additional financial support from R. Varadarajan is also acknowledged. T.S.K. and R.S. are supported by Senior Research Fellowships from the Indian Institute of Science.
Abbreviations used:
- Alt-NHEJ
alternative nonhomologous end-joining
- CEs
cytoplasmic extracts
- CFE
cell-free extract
- C-NHEJ
classical nonhomologous end-joining
- DSBs
double-strand breaks
- IP
immunoprecipitation
- MEs
mitochondrial extracts
- MH
microhomology
- MMEJ
microhomology-mediated end-joining
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
- NHEJ
nonhomologous end-joining
- nt
nucleotide
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0260) on November 25, 2015.
REFERENCES
- Abnet CC, Huppi K, Carrera A, Armistead D, McKenney K, Hu N, Tang ZZ, Taylor PR, Dawsey SM. Control region mutations and the “common deletion” are frequent in the mitochondrial DNA of patients with esophageal squamous cell carcinoma. BMC Cancer. 2004;4:30. doi: 10.1186/1471-2407-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc Natl Acad Sci USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, He J, Huang Y, Zhao W. The generation of mitochondrial DNA large-scale deletions in human cells. J Hum Genet. 2011;56:689–694. doi: 10.1038/jhg.2011.97. [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Sebastian R, Sharma S, Karande AA, Choudhary B, Raghavan SC. Time-dependent predominance of nonhomologous DNA end-joining pathways during embryonic development in mice. J Mol Biol. 2012;417:197–211. doi: 10.1016/j.jmb.2012.01.029. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene ED, Hollinshead MS, Waeytens AA, Schelfhout VR, Eechaute WP, Shaw MK, Van Oostveldt PM, Vaux DJ. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell. 2005;16:997–1010. doi: 10.1091/mbc.E04-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey G, Campbell C. An alternate form of Ku80 is required for DNA end-binding activity in mammalian mitochondria. Nucleic Acids Res. 2000;28:3793–3800. doi: 10.1093/nar/28.19.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong S-Y, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–487. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- D’Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G. Heterologous mitochondrial DNA recombination in human cells. Hum Mol Genet. 2004;13:3171–3179. doi: 10.1093/hmg/ddh326. [DOI] [PubMed] [Google Scholar]
- De A, Campbell C. A novel interaction between DNA ligase III and DNA polymerase gamma plays an essential role in mitochondrial DNA stability. Biochem J. 2007;402:175–186. doi: 10.1042/BJ20061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degoul F, Nelson I, Amselem S, Romero N, Obermaier-Kusser B, Ponsot G, Marsac C, Lestienne P. Different mechanisms inferred from sequences of human mitochondrial DNA deletions in ocular myopathies. Nucleic Acids Res. 1991;19:493–496. doi: 10.1093/nar/19.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Dmitrieva NI, Malide D, Burg MB. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am J Physiol Regul Integr Comp Physiol. 2011;301:R632–R640. doi: 10.1152/ajpregu.00853.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghian A, Vleugels RA, Canter JA, McDonald MA, Stasko T, Sligh JE. Mitochondrial DNA deletions serve as biomarkers of aging in the skin, but are typically absent in nonmelanoma skin cancers. J Invest Dermatol. 2006;126:336–344. doi: 10.1038/sj.jid.5700088. [DOI] [PubMed] [Google Scholar]
- Falk MJ, Sondheimer N. Mitochondrial genetic diseases. Curr Opin Pediatr. 2010;22:711–716. doi: 10.1097/MOP.0b013e3283402e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferecatu I, Le Floch N, Bergeaud M, Rodriguez-Enfedaque A, Rincheval V, Oliver L, Vallette FM, Mignotte B, Vayssiere JL. Evidence for a mitochondrial localization of the retinoblastoma protein. BMC Cell Biol. 2009;10:50. doi: 10.1186/1471-2121-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2:1007–1012. doi: 10.1093/embo-reports/kve233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L, Quintana DF, Schiraldi LK, Phadnis N, Coles GL, Sia RA, Sia EA. Mitochondrial genome maintenance: roles for nuclear nonhomologous end-joining proteins in Saccharomyces cerevisiae. Genetics. 2012;190:951–964. doi: 10.1534/genetics.111.138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Kim JH, Youn HD. C-terminal binding protein maintains mitochondrial activities. Cell Death Differ. 2009;16:584–592. doi: 10.1038/cdd.2008.186. [DOI] [PubMed] [Google Scholar]
- Kumar TS, Kari V, Choudhary B, Nambiar M, Akila TS, Raghavan SC. Anti-apoptotic protein BCL2 down-regulates DNA end joining in cancer cells. J Biol Chem. 2010;285:32657–32670. doi: 10.1074/jbc.M110.140350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R, Raghavan SC. Structure-specific nuclease activity of RAGs is modulated by sequence, length and phase position of flanking double-stranded DNA. FEBS J. 2015;282:4–18. doi: 10.1111/febs.13121. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapucci A, Pittelli M, Rapizzi E, Felici R, Moroni F, Chiarugi A. Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Mol Pharmacol. 2011;79:932–940. doi: 10.1124/mol.110.070110. [DOI] [PubMed] [Google Scholar]
- Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maianski NA, Geissler J, Srinivasula SM, Alnemri ES, Roos D, Kuijpers TW. Functional characterization of mitochondria in neutrophils: a role restricted to apoptosis. Cell Death Differ. 2004;11:143–153. doi: 10.1038/sj.cdd.4401320. [DOI] [PubMed] [Google Scholar]
- Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, Oehmichen M. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more. Exp Gerontol. 2008;43:645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik AK, Lieber MR, Raghavan SC. Cytosines, but not purines, determine recombination activating gene (RAG)-induced breaks on heteroduplex DNA structures: implications for genomic instability. J Biol Chem. 2010;285:7587–7597. doi: 10.1074/jbc.M109.089631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik AK, Raghavan SC. Differential reaction kinetics, cleavage complex formation, and nonamer binding domain dependence dictate the structure-specific and sequence-specific nuclease activity of RAGs. J Mol Biol. 2012;415:475–488. doi: 10.1016/j.jmb.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Nambiar M, Kari V, Raghavan SC. Chromosomal translocations in cancer. Biochim Biophys Acta. 2008;1786:139–152. doi: 10.1016/j.bbcan.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Nambiar M, Srivastava M, Gopalakrishnan V, Sankaran SK, Raghavan SC. G-quadruplex structures formed at the HOX11 breakpoint region contribute to its fragility during t(10;14) translocation in T-cell leukemia. Mol Cell Biol. 2013;33:4266–4281. doi: 10.1128/MCB.00540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Oh S, Harvey A, Zimbric J, Wang Y, Nguyen T, Jackson PJ, Hendrickson EA. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair (Amst) 2014;21:97–110. doi: 10.1016/j.dnarep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- Pakendorf B, Stoneking M. Mitochondrial DNA and human evolution. Annu Rev Genomics Hum Genet. 2005;6:165–183. doi: 10.1146/annurev.genom.6.080604.162249. [DOI] [PubMed] [Google Scholar]
- Queen C, Korn LJ. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984;12:581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol Cell Biol. 2005;25:5904–5919. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB. Amplifying mechanisms of lymphomagenesis. Mol Cell. 2002;10:1–2. doi: 10.1016/s1097-2765(02)00573-7. [DOI] [PubMed] [Google Scholar]
- Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- Servidei S. Mitochondrial encephalomyopathies:gene mutation. Neuromuscul Disord. 2003;13:848–853. doi: 10.1016/s0960-8966(03)00209-8. [DOI] [PubMed] [Google Scholar]
- Sharma S, Choudhary B, Raghavan SC. Efficiency of nonhomologous DNA end joining varies among somatic tissues, despite similarity in mechanism. Cell Mol Life Sci. 2011;68:661–676. doi: 10.1007/s00018-010-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6:e1697. doi: 10.1038/cddis.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeaton MB, Miller PS, Ketner G, Hanakahi LA. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007;35:e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanhauser SM, Laipis PJ. Multiple deletions are detectable in mitochondrial DNA of aging mice. J Biol Chem. 1995;270:24769–24775. doi: 10.1074/jbc.270.42.24769. [DOI] [PubMed] [Google Scholar]
- Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- Van Tuyle GC, Gudikote JP, Hurt VR, Miller BB, Moore CA. Multiple, large deletions in rat mitochondrial DNA: evidence for a major hot spot. Mutat Res. 1996;349:95–107. doi: 10.1016/0027-5107(95)00165-4. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–483. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.