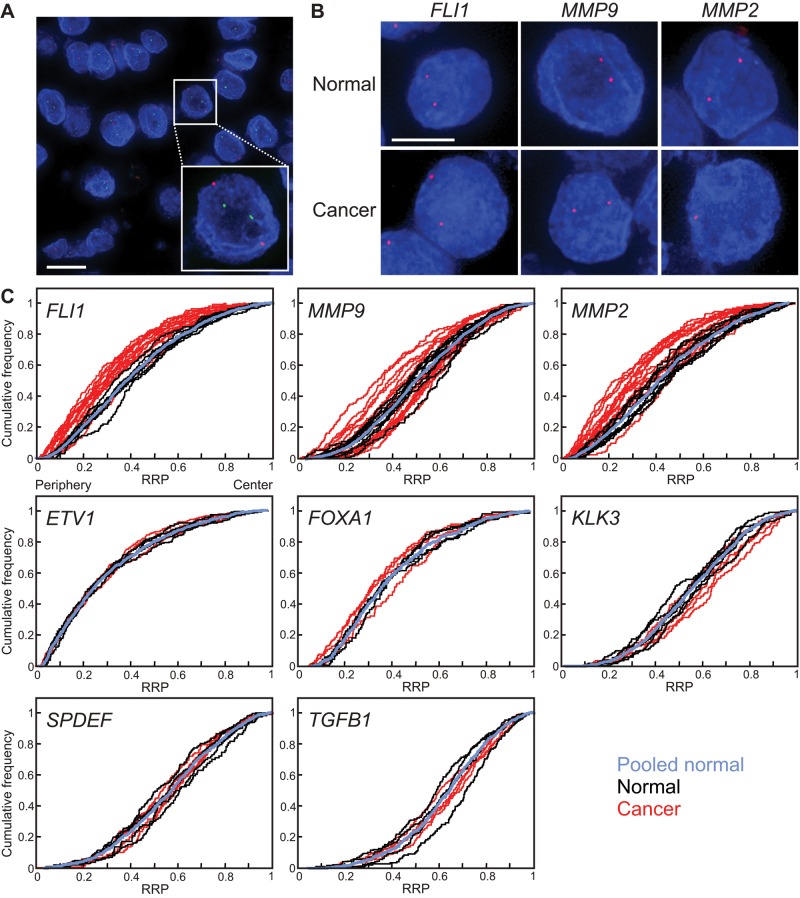
The spatial organization of the genome is altered in prostate cancer compared to normal tissue in a gene-specific manner. The repositioning of two genes, FLI1 and MMP9, is specific to cancer, and the positioning patterns of these genes may serve as diagnostic biomarkers.
Abstract
Genes occupy preferred spatial positions within interphase cell nuclei. However, positioning patterns are not an innate feature of a locus, and genes can alter their localization in response to physiological and pathological changes. Here we screen the radial positioning patterns of 40 genes in normal, hyperplasic, and malignant human prostate tissues. We find that the overall spatial organization of the genome in prostate tissue is largely conserved among individuals. We identify three genes whose nuclear positions are robustly altered in neoplastic prostate tissues. FLI1 and MMP9 position differently in prostate cancer than in normal tissue and prostate hyperplasia, whereas MMP2 is repositioned in both prostate cancer and hyperplasia. Our data point to locus-specific reorganization of the genome during prostate disease.
INTRODUCTION
The interphase nucleus is a highly organized organelle, with genes and chromosomes occupying preferred locations within the cell nucleus (Ferrai et al., 2010; Meaburn et al., 2016). Although the spatial organization of the genome is largely conserved among individuals (Borden and Manuelidis, 1988; Wiech et al., 2005; Murata et al., 2007; Meaburn et al., 2009; Timme et al., 2011), nuclear location is not an inherent feature of a locus, since gene loci can occupy distinct positions depending on context or conditions. For example, gene positions may differ among tissue types during differentiation or as cells become quiescent or senescent (Bridger et al., 2000; Boyle et al., 2001; Kosak et al., 2002; Parada et al., 2004; Meaburn and Misteli, 2008; Meaburn et al., 2016; Takizawa et al., 2008a; Ferrai et al., 2010; Peric-Hupkes et al., 2010; Foster et al., 2012). Although the molecular mechanisms that govern the nuclear position of a gene are unknown, changes in gene positioning are sometimes, but not always, correlated with functional changes, such as altered transcription levels or replication timing or with epigenetic changes to chromatin (Kosak et al., 2002; Williams et al., 2006; Hiratani et al., 2008; Meaburn and Misteli, 2008; Meaburn et al., 2016; Takizawa et al., 2008a; Morey et al., 2009; Ferrai et al., 2010; Peric-Hupkes et al., 2010; Towbin et al., 2012; Kind et al., 2013; Therizols et al., 2014; Rafique et al., 2015).
There is accumulating evidence that spatial reorganization of the genome is a pathological feature, with repositioning of individual genes or genomic regions detected in a wide range of diseases, including epilepsy (Borden and Manuelidis, 1988), laminopathies (Meaburn et al., 2007; Mewborn et al., 2010; Mehta et al., 2011), parasitic or viral infection (Li et al., 2010; Knight et al., 2011; Arican-Goktas et al., 2014), Down syndrome (Paz et al., 2015), endometriosis (Mikelsaar et al., 2014), and cancer (Cremer et al., 2003; Murata et al., 2007; Meaburn and Misteli, 2008; Meaburn et al., 2009; Zeitz et al., 2013). For example, in cultured fibroblasts from laminopathy patients, some chromosomes, such as chromosomes (HSAs) 13 and 18, occupy altered nuclear positions, although others, including HSAX and HSA4, are unaffected (Meaburn et al., 2007; Mewborn et al., 2010; Mehta et al., 2011). Similarly, in brain tissue from epilepsy patients, the centromere of HSAX is relocated but 1q12, 9q12, and Y12 are not (Borden and Manuelidis, 1988).
Spatial genome reorganization has been associated with cancer. HSA18 and HSA19, which in many cell types are peripherally and internally located, respectively, broadly maintained their positions in cell lines derived from various cancers, including colon and cervical cancer, melanoma, and Hodgkin’s disease, although they tend to be in closer proximity, and a small fraction of nuclei have inverted the locations of these two chromosomes (Boyle et al., 2001; Cremer et al., 2003; Meaburn et al., 2007). Similarly, HSA19 is also shifted to a more peripheral position in tumors of a subset of thyroid cancer patients (Murata et al., 2007). Moreover, HSA18 and the BCL2 gene are shifted to a more peripheral position in invasive cervical squamous carcinoma compared with the apical, but not basal, layer of nonneoplastic squamous epithelium (Wiech et al., 2009). In both an in vitro model of breast cancer and breast cancer tissues, ∼40% of the tested genes were in altered locations (Meaburn and Misteli, 2008; Meaburn et al., 2009). Of importance, most of these gene-repositioning events are specific to the oncogenic state and do not occur in benign breast disease (Meaburn et al., 2009). Not only might the position of a genomic region change in relation to the nuclear edge or center (radial positioning) in cancer, but its position relative to its neighbors might also be affected. For instance, the group of loci that are in close spatial proximity to the IGFBP3 locus is significantly changed in breast cancer cell lines compared with a normal control (Zeitz et al., 2013).
Spatial genome reorganization in cancer, however, is not a global event. The positions of many genomic regions remain conserved in cancer (Kozubek et al., 2002; Parada et al., 2002; Wiech et al., 2005; Meaburn and Misteli, 2008; Meaburn et al., 2009; Timme et al., 2011). For example, the radial position of HSA8 in pancreatic cancer (Timme et al., 2011) and HSA10 in most thyroid cancers (Murata et al., 2007) is similar to that in normal tissue, and the position of the majority of genes analyzed in breast cancer (Wiech et al., 2005; Meaburn and Misteli, 2008; Meaburn et al., 2009) and leukemia (Kozubek et al., 2002) are unaffected. Similarly, the preferred clustering of chromosomes 12, 14, and 15 is maintained in a mouse lymphoma cell line compared with cultured spleenocytes (Parada et al., 2002).
Prostate cancer is highly prevalent. In the United States alone, >220,000 new cases are diagnosed each year, with one in seven men developing prostate cancer during their lifetime, and there are >27,500 deaths from it annually, accounting for 9% of all male cancer deaths (Siegel et al., 2015). A major challenge in the treatment of prostate cancer patients is the difficulty of distinguishing indolent from aggressive cancer. As a result, a large number of patients receive unnecessary treatment for cancers that will likely remain asymptomatic during their lifetime (Draisma et al., 2009; Cooperberg et al., 2010). There is also a subset of patients with high-risk prostate cancer who are undertreated and are not receiving the treatment their cancer requires to ensure disease-free survival (Cooperberg et al., 2010). Emerging evidence suggests that changes in nuclear architecture features, such as nuclear shape or global levels and patterns of histone modifications and nuclear lamin proteins, may have a clinical value for prostate cancer detection and prognosis (Veltri and Christudass, 2014). However, whereas spatial (re)organization of the genome has been proposed to have diagnostic potential for breast cancer (Meaburn and Misteli, 2008; Meaburn et al., 2009), little is known about spatial genome organization in prostate cancer.
We performed an unbiased screen of the position of 40 genes in Formalin-fixed, paraffin- embedded (FFPE) human prostate tissues. Although we find limited variability in gene positioning patterns among individuals and general conservation of positioning patterns in cancer, we identified three genes that undergo disease-specific repositioning in prostate cancer.
RESULTS
Comprehensive mapping of gene positioning in prostate tissue
Analogous to our approach with regard to breast cancer (Meaburn et al., 2009), we screened the positioning patterns of 40 genes (Supplemental Table S1) in prostate specimens to identify genes that reposition in prostate cancer. To this end, to map comprehensively the spatial position of genes in normal and malignant prostate tissue, we combined fluorescence in situ hybridization (FISH) with quantitative imaging to map the nuclear positions of our set of genes in 4- to 5-μm-thick FFPE human prostate tissues (Supplemental Table S2). The panel of tissues used includes 30 prostate carcinomas, five hyperplasic (nonmalignant abnormality) tissues, and 29 normal or normal adjacent to tumor (NAT) tissues (Supplemental Table S2). The eight genes previously identified to reposition in breast cancer tissues (Meaburn et al., 2009), do not reposition in prostate (unpublished data), and we were unable to identify any particular characteristics of repositioning genes in breast cancer that predicted repositioning potential (Meaburn et al., 2009). Therefore we screened a wide range of genes in prostate tissues, which included many of the most important genes related to prostate cancer, such as genes reported to be misregulated in prostate cancer or to participate in translocations, as well as a set of randomly selected genes, which were selected independently of known changes in activity or involvement in prostate cancer development or progression. The test set included genes from 21 different chromosomes (Supplemental Table S1). To map the nuclear locations of individual genes in a given tissue, we determined the radial position of each allele in typically 100–200 randomly selected epithelial nuclei on projections of three-dimensional image stacks using Euclidean distance transform (EDT) after automated FISH signal detection, as previously described (Meaburn et al., 2009; see Materials and Methods for details). For each candidate gene, the positions, normalized to the nuclear radius to eliminate nuclear size effects, of all measured alleles in a tissue were combined and represented as a cumulative relative radial distribution (RRD). Statistical difference between the cumulative RRDs of different samples was assessed using the nonparametric two-sample one-dimensional (1D) Kolmogorov–Smirnov (KS) test as previously described (Meaburn et al., 2009; see Materials and Methods).
To screen for genes that specifically reposition in prostate cancer, we used a previously described screening strategy (Meaburn et al., 2009). Briefly, gene positions were first compared in at least three normal and three cancerous prostate tissues (Table 1 and Supplemental Figure 1A). Genes that repositioned in the majority of cancers, in the absence of a large variability among normal tissues, were then screened in a larger number of tissues (Table 1 and Supplemental Figure 1A). Finally, the sensitivity and specificity of the repositioning events of the top hits were assessed by positioning these genes in hyperplasic tissues and determining false-positive and false-negative rates (Supplemental Figure 1A; see later discussion for details).
TABLE 1:
Cross-comparisons among individual tissues.
| Gene | Number of tissues | Percentage (number) of SD cross-comparisons among: | ||
|---|---|---|---|---|
| Normal | Cancer | Individual normal tissues | Individual normal and cancer tissues | |
| AR | 7 | 6 | 9.5 (2/21) | 16.7 (7/42) |
| BCL2 | 4 | 4 | 0.0 (0/6) | 0.0 (0/16) |
| BRCA2 | 4 | 3 | 0.0 (0/6) | 0.0 (0/12) |
| CCND1 | 4 | 4 | 50.0 (3/6) | 37.5 (6/16) |
| DCN | 5 | 3 | 20.0 (2/10) | 0.0 (0/15) |
| EGFR | 6 | 4 | 0.0 (0/15) | 0.0 (0/24) |
| ERG | 5 | 3 | 50.0 (5/10) | 40.0 (6/15) |
| ESR2 | 5 | 4 | 0.0 (0/10) | 0.0 (0/20) |
| ETV1 | 5 | 3 | 0.0 (0/10) | 0.0 (0/15) |
| FGFR1 | 7 | 5 | 28.6 (6/21) | 22.9 (8/35) |
| FGFR2 | 6 | 4 | 33.3 (5/15) | 41.7 (10/24) |
| FLI1 | 8 | 14 | 7.1 (2/28) | 60.7 (68/112) |
| FOXA1 | 4 | 4 | 0.0 (0/6) | 0.0 (0/16) |
| FUT4 | 4 | 3 | 0.0 (0/6) | 0.0 (0/12) |
| GREB1 | 4 | 3 | 16.7 (1/6) | 25.0 (3/12) |
| HOXA9 | 4 | 3 | 0.0 (0/6) | 8.3 (1/12) |
| KLK3 | 5 | 4 | 20.0 (2/10) | 30 (6/20) |
| LMNA | 4 | 3 | 50.0 (3/6) | 16.7 (2/12) |
| MATR3 | 4 | 3 | 33.3 (2/6) | 0.0 (0/12) |
| MMP1 | 3 | 3 | 33.3 (1/3) | 11.1 (1/9) |
| MMP14 | 3 | 3 | 33.3 (1/3) | 44.4 (4/9) |
| MMP2 | 8 | 16 | 7.1 (2/28) | 48.4 (62/128) |
| MMP9 | 10 | 19 | 17.8 (8/45) | 44.2 (84/190) |
| NPM1 | 3 | 3 | 33.3 (1/3) | 33.3 (3/9) |
| NUMA1 | 4 | 4 | 0.0 (0/6) | 12.5 (2/16) |
| PADI4/6 | 6 | 6 | 13.3 (2/15) | 5.6 (2/36) |
| PTEN | 3 | 3 | 0.0 (0/3) | 22.2 (2/9) |
| RAF1 | 8 | 5 | 32.1 (9/28) | 30.0 (12/40) |
| SATB1 | 5 | 4 | 20.0 (2/10) | 15.0 (3/20) |
| SERPINB2 | 4 | 4 | 16.7 (1/6) | 6.3 (1/16) |
| SLC45A3 | 5 | 0 | 0.0 (0/10) | ND |
| SP100 | 5 | 4 | 0.0 (0/10) | 20.0 (4/20) |
| SPDEF | 5 | 4 | 10.0 (1/10) | 5.0 (1/20) |
| TGFB1 | 4 | 4 | 50.0 (3/6) | 30.0 (5/16) |
| THBS1 | 5 | 4 | 20.0 (2/10) | 30.0 (6/20) |
| TIMP2 | 6 | 7 | 53.3 (8/15) | 33.3 (14/42) |
| TIMP3 | 6 | 4 | 60.0 (9/15) | 58.3 (14/24) |
| TMPRSS2 | 6 | 4 | 60.0 (9/15) | 50.0 (12/24) |
| VEGFA | 13 | 7 | 24.4 (19/78) | 47.3 (43/91) |
| VIM | 5 | 4 | 70.0 (7/10) | 40.0 (8/20) |
SD, significantly different, based on a KS test, p < 0.01. ND, not determined.
Conservation of positioning patterns in morphologically normal prostate tissues
It is unknown how similar positioning patterns are between prostates from different individuals, yet knowing the level of interindividual variability in the positioning pattern of a given gene is critical in determining whether any repositioning detected in cancer tissues is specific to cancer or simply a product of interindividual variability. Potential variability of positioning patterns is particularly relevant in the prostate, since the position of several genes has been shown to be sensitive to androgen (Lin et al., 2009; Mani et al., 2009). To determine the degree of variability in gene localization among individuals, we compared the positioning pattern of our set of 40 genes among morphologically normal prostate tissues. For most of the genes analyzed, the radial distribution of the gene among normal prostate tissues was similar (Table 1, Figures 1 and 2, and Supplemental Figures S1 and S2). For 12 of 40 (30.0%) genes, the RRDs were statistically identical among all individuals (p > 0.01; Table 1, Figures 1 and 2, and Supplemental Figures S1 and S2). An additional 20 genes (50.0%), although not identical among all comparisons, were very similarly positioned among most individuals, with <33.4% of cross-comparisons being statistically significantly different (p < 0.01; Table 1, Figures 1 and 2, and Supplemental Figures S1 and S2). The position of 20% of genes (8 of 40; CCND1 [3 of 6 cross-comparisons], ERG [5 of 10], LMNA [3 of 6], TGFB1 [3 of 6], TIMP2 [8 of 15], TIMP3 [9 of 15], TMPRSS2 [9 of 15] and VIM [7of 10]) were highly variable among individuals, with 50–70% of the cross-comparisons among normal tissues being significantly different (Table 1, Figures 1 and 2, and Supplemental Figures S1 and S2).
FIGURE 1:
Gene-specific spatial reorganization of the genome in prostate cancer. (A, B) Gene loci were detected by FISH in FFPE prostate tissue sections. Blue, 4′,6-diamidino-2-phenylindole nuclear counterstain. Projected image stacks. (A) EGFR (red) and SPDEF (green) gene loci, in a malignant prostate tissue. Bar, 10 μm. (B) FLI1, MMP9, and MMP2 gene loci (red) in normal and cancer tissues. Bar, 5 μm. (C) Cumulative RRDs for the indicated genes in prostate cancer (red), normal prostate tissues (black), and the pooled normal distribution (PND, blue). The positioning patterns of some but not all genes are different in prostate cancer compared with normal tissues. RRP, relative radial position.
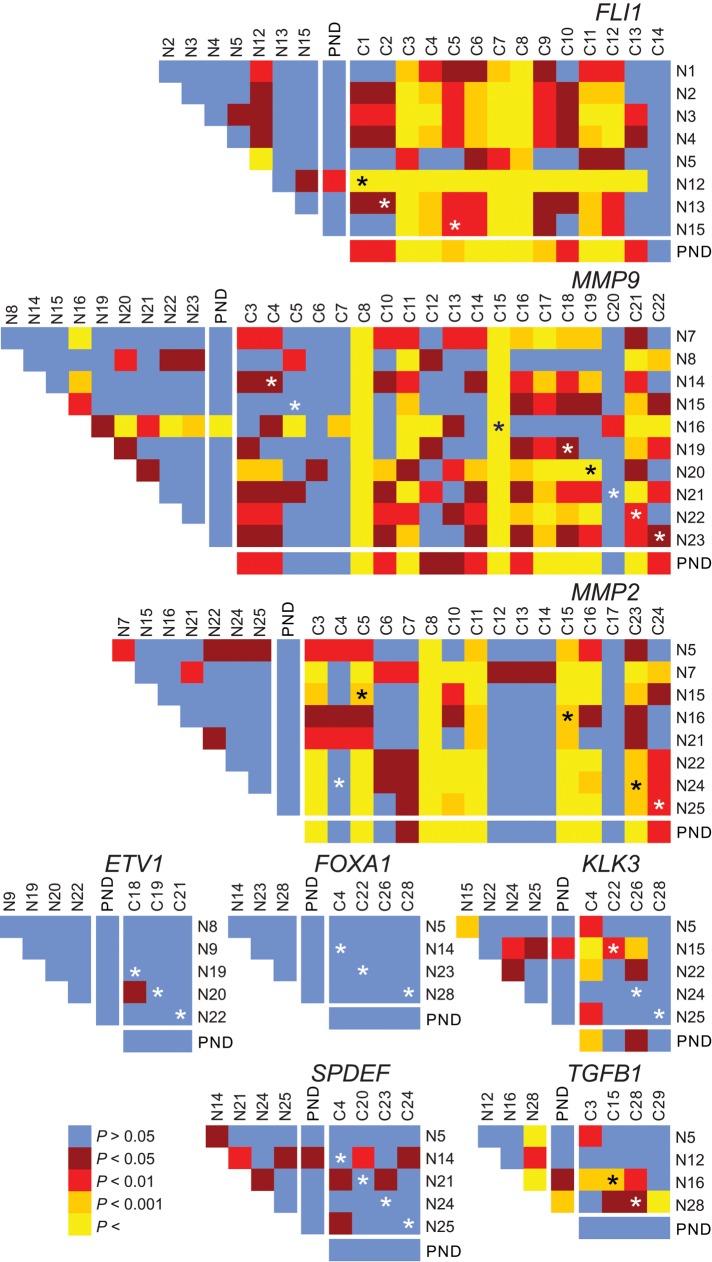
FIGURE 2:
Gene-specific repositioning in prostate cancer. Heat maps representing the pairwise statistical comparisons of radial positioning patterns of indicated genes among tissues using the two-sample 1D KS test. Although positioning patterns are statistically similar (blue, brown) among most normal tissues (N1–N28; see Supplemental Table S2), they can be divergent (red, orange, yellow) in cancer tissues (C1–C24; see Supplemental Table S2). Black and white asterisks indicate a cross-comparison between a normal and cancer specimens from the same individual. PND, pooled normal distribution.
Because the position of a gene in a given tissue can be significantly different from some but not all other tissues, as an additional measure of conservation in positioning patterns among individuals, we compared the RRD of each gene in each normal tissue to a pooled normal distribution (PND). The PND is used as a standardized “average” position of a gene within normal tissues and was generated by combining the RRDs from all normal tissues analyzed into a single distribution for each gene, as previously described (Meaburn et al., 2009; see Materials and Methods). Again we found a high level of similarity among individuals (Table 2, Figures 1 and 2, and Supplemental Figures S1 and S2). For 26 of the 40 (65.0%) genes, no individual’s normal tissue was significantly different from the PND, and an additional eight (20.0%) genes had only a single normal tissue significantly different to the PND (7.7–25% of normal tissues; Table 2, Figures 1 and 2, and Supplemental Figures S1 and S2). On the other hand, three genes (3 of 40; 7.5%)—FGFR1 (2 of 7 normal tissues), TMPRSS2 (2 of 6), and RAF1 (3 of 8)—had approximately one-third (28.6–37.5%) of normal tissues significantly different from the PND, and an additional three genes (CCND1 [2 of 4], TIMP2 [3 of 6], and TIMP3 [4 of 6]) were significantly different from the PND in 50.0–66.7% of normal tissues (Table 2 and Supplemental Figures S1 and S2). Taken together, these data demonstrate that the radial positions of most genes are conserved among prostate tissue from different individuals. However, the position of a subset of genes is variable among individuals.
TABLE 2:
Comparison of individual tissues to a pooled normal distribution.
| Gene | Percentage (number) of SD cross-comparisons between: | |
|---|---|---|
| Individual normal tissues and pooled normal | Individual cancer tissues and pooled normal | |
| AR | 0.0 (0/7) | 33.3 (2/6) |
| BCL2 | 0.0 (0/4) | 0.0 (0/4) |
| BRCA2 | 0.0 (0/4) | 33.3 (1/3) |
| CCND1 | 50.0 (2/4) | 25.0 (1/4) |
| DCN | 0.0 (0/5) | 0.0 (0/3) |
| EGFR | 0.0 (0/6) | 0.0 (0/4) |
| ERG | 20.0 (1/5) | 33.3 (1/3) |
| ESR2 | 0.0 (0/5) | 0.0 (0/4) |
| ETV1 | 0.0 (0/5) | 0.0 (0/3) |
| FGFR1 | 28.6 (2/7) | 40.0 (2/5) |
| FGFR2 | 16.7 (1/6) | 50.0 (2/4) |
| FLI1 | 12.5 (1/8) | 92.9 (13/14) |
| FOXA1 | 0.0 (0/4) | 0.0 (0/4) |
| FUT4 | 0.0 (0/4) | 0.0 (0/3) |
| GREB1 | 25.0 (1/4) | 0.0 (0/3) |
| HOXA9 | 0.0 (0/4) | 33.3 (1/3) |
| KLK3 | 20.0 (1/5) | 25.0 (1/4) |
| LMNA | 0.0 (0/4) | 33.3 (1/3) |
| MATR3 | 0.0 (0/4) | 0.0 (0/3) |
| MMP1 | 0.0 (0/3) | 0.0 (0/3) |
| MMP14 | 0.0 (0/3) | 66.7 (2/3) |
| MMP2 | 0.0 (0/8) | 56.3 (9/16) |
| MMP9 | 10.0 (1/10) | 68.4 (13/19) |
| NPM1 | 0.0 (0/3) | 66.7 (2/3) |
| NUMA1 | 0.0 (0/4) | 50.0 (2/4) |
| PADI4/6 | 0.0 (0/6) | 16.7 (1/6) |
| PTEN | 0.0 (0/3) | 0.0 (0/3) |
| RAF1 | 37.5 (3/8) | 40.0 (2/5) |
| SATB1 | 0.0 (0/5) | 25.0 (1/4) |
| SERPINB2 | 0.0 (0/4) | 0.0 (0/4) |
| SLC45A3 | 0.0 (0/5) | ND |
| SP100 | 0.0 (0/5) | 25.0 (1/4) |
| SPDEF | 0.0 (0/5) | 0.0 (0/4) |
| TGFB1 | 25.0 (1/4) | 0.0 (0/4) |
| THBS1 | 0.0 (0/5) | 50.0 (2/4) |
| TIMP2 | 50.0 (3/6) | 28.6 (2/7) |
| TIMP3 | 66.7 (4/6) | 50.0 (2/4) |
| TMPRSS2 | 33.3 (2/6) | 75.0 (3/4) |
| VEGFA | 7.7 (1/13) | 57.1 (4/7) |
| VIM | 0.0 (0/5) | 25.0 (1/4) |
SD, significantly different, based on a KS test, p < 0.01. ND, not determined.
Identification of repositioned genes in prostate cancer
Having established limited variability in gene positioning among individuals, we screened for genes that reposition in prostate cancer. We compared the RRDs of 39 genes in multiple cancer tissues to individual normal tissues and to the PND (Tables 1 and 2, Figures 1 and 2, and Supplemental Figures S1 and S2). The position of nine genes (23.1%) was indistinguishable in all cross-comparisons among individual normal and cancer tissues, and an additional 19 genes (48.7%) had only limited variability between normal and cancer tissues, with 5.0–33.3% of cross-comparisons reaching significance. Nine genes (23.1%; CCND1 [6 of 16 comparisons], ERG [6 of 15], VIM [43 of 91], FGFR2 [10 of 24], MMP9 [84 of 190], MMP14 [4 of 9], VEGFA [43 of 91], MMP2 [62 of 128], and TMPRSS2 [12 of 24]) were differently positioned in 37.5–50.0% of cross-comparisons. Two genes (5.1%; TIMP3 [14 of 24] and FLI1 [68 of 112]) were repositioned in ∼60% of cross-comparisons between normal and malignant prostate tissues (Table 1, Figures 1 and 2, and Supplemental Figures S1 and S2). For most genes, the number of significantly different cross-comparisons between normal and cancer specimens was not greater than that of the cross-comparisons among normal tissues, suggesting that the repositioning was not cancer specific (Table 1). For example, whereas TIMP3 was differently positioned in 58.3% (14 of 24) of the cross-comparisons between individual normal and cancer tissues, 60.0% (9 of 15) of cross-comparisons were statistically different when normal tissues were cross-compared with each other (Table 1 and Supplemental Figure S2). Taking into consideration the degree of positioning changes in all cross-comparisons and the interindividual variability of candidates, we identified three genes—MMP2, MMP9, and FLI1—that exhibited robust differential radial positioning between normal and cancer tissues. For these genes, the percentage of cross-comparisons between normal and cancer tissues was higher by >25% than when normal tissues were compared with each other (48.4 vs. 7.1%, 44.2 vs. 17.8%, and 60.7 vs. 7.1%, respectively; Table 1 and Figure 2).
Repositioning of FLI1, MMP9, and MMP2 in prostate cancers was confirmed by analysis of their distributions in individual cancer samples to their PNDs. Most genes in the test set displayed limited repositioning in prostate cancer tissues compared with their PNDs. Two-thirds (26) of genes were repositioned in 0–33.3% of cancer tissues compared with the PND, and six genes repositioned in 40–50% of cancers (Table 2, Figures 1 and 2, and Supplemental Figures S1 and S2). As observed in the cross-comparison analysis, FLI1, MMP9, and MMP2 showed robust repositioning in prostate cancer, and their distributions were distinct from their PNDs in 92.9% (13 of 14 of cancer tissues), 68.4% (13 of 19), and 56.3% (9 of 16) of cancers, respectively (Table 2 and Figures 1 and 2). Although an additional four genes were also repositioned in >50% of cancers compared with the PND (VEGF, 57.1% [4 of 7]; MMP14, 66.7% [2 of 3]; NPM1, 66.7% [2 of 3]; and TMPRSS2, 75% [3 of 4]; Table 2 and Supplemental Figures S1 and S2), we classified these genes as low-priority hits and did not pursue them further because the percentage of cross-comparisons between normal and cancer tissues was not significantly higher than the variability detected among normal tissues and visual inspection of the cumulative frequency distribution plots revealed that only a small number of the distributions in cancer tissues fell outside the range of the normal tissues (Supplemental Figure S1).
In the vast majority (96 of 125; 76.8%) of cases, there was a similar repositioning behavior when a cancer was compared with its matched NAT or with the PND (Figure 2 and Supplemental Figure S2). This observation suggests that repositioning, if present in a patient, is limited to the cancer tissue itself and that comparison to the PND is suitable to assess whether a gene is repositioned in cancer, even when no normal tissue from the same individual is available.
We were unable to define any feature of a prostate cancer that predicted the likelihood of a gene repositioning within it. The propensity of the three marker genes to reposition in a given cancer was not linked to changes in copy number (Supplemental Table S3), nor was the variation in their positioning patterns correlated with the clinicopathologically defined aggressiveness of the cancer (Supplemental Table S2 and Figure 3).
FIGURE 3:
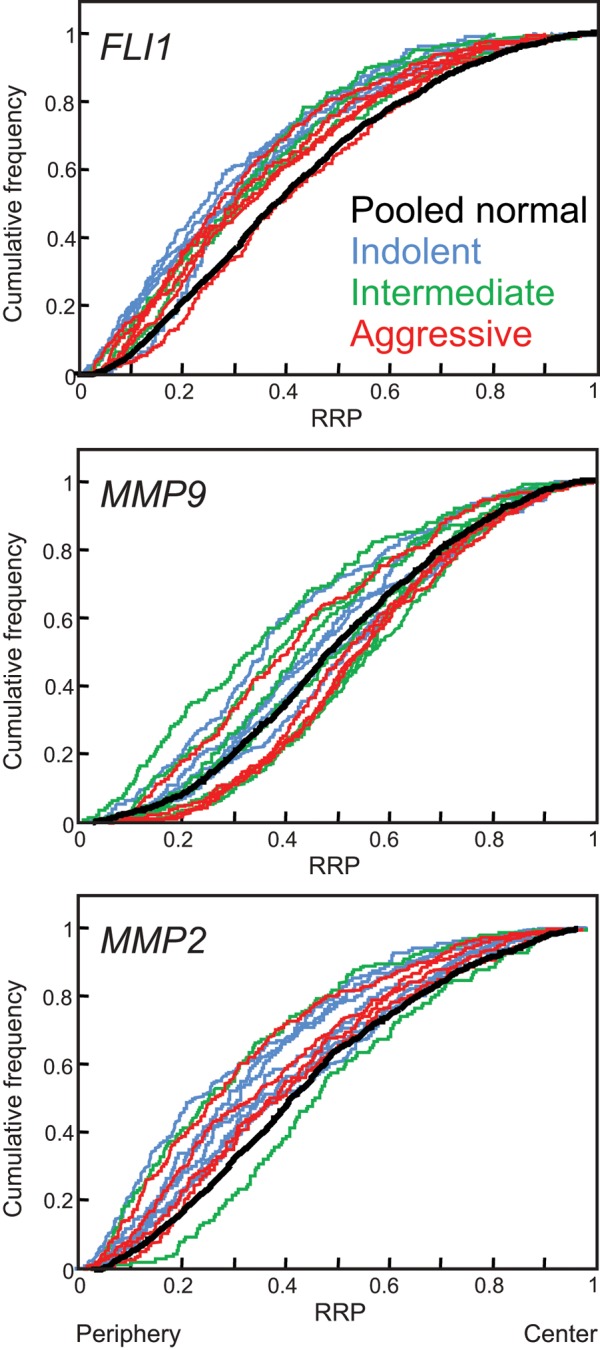
Gene repositioning in prostate cancer is not linked to prognostic status. Cumulative RRDs are color coded according to the prognostic status of the cancer: blue, indolent prostate cancers (Gleason score ≤6, without metastasis); green, intermediate cancers (Gleason score 7, without metastasis); red, aggressive cancers (Gleason score ≥8, and/or metastasis). The pooled normal distribution (black) is included for comparison. RRP, relative radial position.
Cancer specificity of FLI1 and MMP9 repositioning
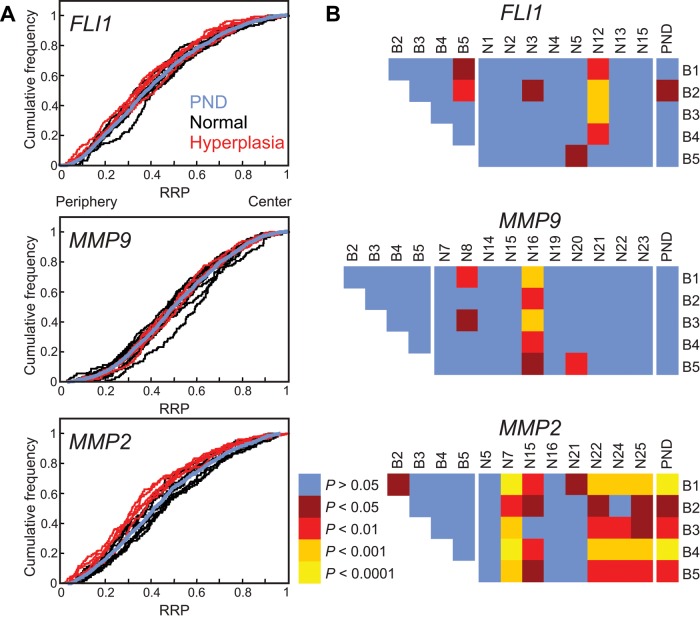
Given that loci can alter their nuclear location in diseases other than cancer (Borden and Manuelidis, 1988; Meaburn et al., 2007, 2009), we sought to determine whether the repositioning of FLI1, MMP9, and MMP2 was specific to cancer or was a more general feature of prostate disease. We compared the radial positioning patterns of these genes in five benign hyperplastic tissues with the positioning patterns of normal prostate tissues and among multiple hyperplasia tissues (Table 3 and Figure 4). MMP9 and MMP2 were positioned identically in all hyperplastic tissues (Table 3 and Figure 4). The position of FLI1 is similarly conserved among hyperplastic tissues, with only a single cross-comparison reaching significance (1 of 10; Table 3 and Figure 4). FLI1 and MMP9 did not reposition in hyperplasia compared with normal tissues, since the positioning of both genes was indistinguishable from the PND in all five hyperplasia tissues (Table 3 and Figure 4), and only 10% (4 of 40) and 12% (6 of 50) of the cross-comparisons among individual normal tissues reached significance, respectively. These findings, combined with the low repositioning rate in normal tissue, suggests that FLI1 and MMP9 repositioning is specific to cancer. In contrast, compared with normal tissues, MMP2 was repositioned in both prostate cancer and hyperplasia. MMP2 was in a significantly different position in 80% (4 of 5) of the hyperplastic tissues compared with the PND and in 45% (18 of 40) of cross-comparisons between hyperplasic and normal tissues (Table 3 and Figure 4), suggesting that the repositioning of MMP2 is common in prostate disease and not limited to malignancy. We had access to both hyperplastic and cancer tissue from the same prostate (B1 and C3, respectively; Supplemental Table S2). In keeping with the comparisons between hyperplasia and normal tissue, FLI1 and MMP9 occupied significantly different positions in the cancer and hyperplasic tissue (p = 0.004 and 0.002, respectively), whereas MMP2 was in a similar position in the two disease states (p = 0.516) for this individual.
TABLE 3:
Comparison of gene positioning in benign tissues.
| Gene | Percentage (number) of SD cross-comparison among: | ||
|---|---|---|---|
| Individual hyperplasia tissues | Individual normal and hyperplasia tissues | Individual hyperplasia tissues and pooled normal | |
| FLI1 | 10.0 (1/10) | 10.0 (4/40) | 0.0 (0/5) |
| MMP9 | 0.0 (0/10) | 12.0 (6/50) | 0.0 (0/5) |
| MMP2 | 0.0 (0/10) | 45.0 (18/40) | 80.0 (4/5) |
SD, significantly different, based on a two-sample 1D KS test, p < 0.01.
FIGURE 4:
Gene positioning in benign disease. Positions of indicated genes were compared between hyperplasia (B1–B5) and normal prostate tissue (N1–N25). (A) Cumulative RRDs for the indicated genes in hyperplasia (red), normal tissues (black), and the pooled normal distribution (PND; blue). (B) Pairwise statistical comparisons of RRDs between hyperplasic tissues and normal tissues and among hyperplasic tissues, using the two-sample 1D KS test. All three genes are similarly positioned between hyperplasic tissues and only MMP2 repositions in hyperplasic compared with normal tissues. RRP, relative radial position.
High-confidence detection of prostate cancers using multiplexed positioning biomarkers
Because FLI1 and MMP9 repositioning occurs predominantly in cancer tissues, we determined their false-positive and false-negative rates as a preliminary step toward assessing their potential as biomarkers for prostate cancer. A false positive was defined as repositioning (p > 0.01; KS test) of the marker gene in a nonmalignant (normal or hyperplastic) tissue compared with the PND, thus incorrectly classifying the tissue as cancer. Conversely, a false negative was defined as lack of repositioning (p < 0.01) of the marker gene in a cancer, leading to incorrect classification of the tissue as nonmalignant. As expected due to its high rate of repositioning in hyperplasia, MMP2 had a high false-positive rate (30.8% [4 of 13 nonmalignant tissues]; Figures 2 and 4 and Table 4). This finding, combined with its high false-negative rate (43.8% [7 of 16 cancer tissues]; Figure 2 and Table 5), suggests that MMP2 would not be a useful biomarker for prostate cancer. In contrast, FLI1 had low false-negative and false-positive rates (7.1% [1 of 14 cancer tissues] and 7.7% [1 of 13 nonmalignant tissues], respectively; Figures 2 and 4 and Tables 4 and 5). MMP9 had a fairly high false-negative rate (31.6% [6 of 19 cancer tissues]; Figure 2 and Table 5) but a low false-positive rate (6.7% [1 of 15 nonmalignant tissues]; Figures 2 and 4 and Table 4).
TABLE 4:
False-positive rates.
| Gene | Normal-tissue false positives | Benign-tissue false positives | Total false positives |
|---|---|---|---|
| FLI1 | 12.5% (1/8) | 0.0% (0/5) | 7.7% (1/13) |
| MMP9 | 10.0% (1/10) | 0.0% (0/5) | 6.7% (1/15) |
| MMP2 | 0.0% (0/8) | 80.0% (4/5) | 30.8% (4/13) |
The percentage (number) of tissues that give a false-positive result. A false positive is scored when a gene has a statistically significant different RRD in a normal or hyperplastic tissue compared with the pooled normal (p < 0.01; KS test).
TABLE 5:
False-negative rates for single and multiplexed genes.
| Gene(s) | False-negative rate |
|---|---|
| Single gene | |
| FLI1 | 7.1% (1/14) |
| MMP9 | 31.6% (6/19) |
| MMP2 | 43.8% (7/16) |
| Multiplexed genes | |
| FLI1 and MMP9 | 0.0% (0/11) |
| FLI1 and MMP2 | 9.1% (1/11) |
| MMP9 and MMP2 | 28.6% (4/14) |
The percentage (number) of tissues that give a false-negative result. A false negative is scored when the RRD of a gene in a cancer tissue is similar to that of the pooled normal distribution (KS test, p > 0.01). For multiplexed genes, a false positive is scored when neither gene in the pair is repositioned compared with the pooled normal distribution (p > 0.01, KS test). Only tissues in which both genes of the pair were positioned are included.
None of the genes we tested repositioned in all prostate cancer tissues. However, at least one repositioned gene was present in every prostate cancer tissue analyzed (30 of 30 cancers; Figure 2 and Supplemental Figure S2). Given the low false-positive rate of FLI1 and MMP9, we explored the possibility of using these two genes in a multiplex format for the detection of cancer samples. When analyzed together, FLI1 or MMP9, or both, repositioned in all (11 of 11) cancers (Figure 2 and Supplemental Table S4). Consequently, multiplexing FLI1 and MMP9 reduced the false-negative rate to 0% (0 of 11; Table 5). We conclude that in combination, FLI1 and MMP9 are strong potential biomarkers for prostate cancer.
DISCUSSION
This study was designed to identify gene loci that undergo repositioning during prostate carcinogenesis. Spatial mapping of the radial position of 40 genes by FISH identified two genes that robustly reposition in prostate cancer and a third gene that repositions in both prostate cancer and benign prostate disease. The vast majority of analyzed genes, however, did not undergo significant cancer-specific repositioning, pointing to considerable conservation of spatial genome organization in prostate cancer.
In keeping with previous studies on positioning of select genes and of whole chromosomes in brain (Borden and Manuelidis, 1988), pancreas (Timme et al., 2011), thyroid (Murata et al., 2007), and breast (Wiech et al., 2005; Meaburn et al., 2009), we find that the spatial positioning of most genes tested in normal prostate tissue is generally well conserved among individuals. Of 40 genes analyzed, only three (CCND1, TIMP2, and TIMP3) were highly variable among individuals. A prerequisite of a useful cancer biomarker is conservation among individuals in nonmalignant tissues because such variability leads to low sensitivity and specificity of the marker. Therefore some genes will not be suitable for diagnostic purposes due to variable positioning patterns among individuals. The variability in the position of some genes in normal prostate tissue is higher than previously detected for 15 genes in morphologically normal breast tissue (Meaburn et al., 2009). The reasons for this difference between the two tissue types are not known. One possibility is that some glands within normal prostate tissues have preneoplastic features. For example, proliferative inflammatory atrophy, which is regarded as a histologically distinctive pattern of benign glands, has some of the molecular features of prostate neoplasia (De Marzo et al., 1999). In contrast, there is no reported counterpart lesion of mammary glands. Another possibility is that these prostate glands could have included low-grade prostatic intraepithelial neoplasia, which is challenging to recognize in dark-field FISH preparations. In addition, some genes could be more susceptible to interindividual variations in their positioning patterns, which could be either a general feature of the locus or tissue specific. For example, the position of CCND1 was similar among normal breast tissues (Meaburn et al., 2009) but varied among individuals in the prostate, pointing to a potential tissue-specific driver to interindividual positioning patterns. Because CCND1 is a cell cycle regulator and epithelial cells are largely quiescent in normal breast and prostate tissues, a direct link between CCND1 function and tissue specificity seems unlikely.
We identified three genes that consistently reposition in prostate cancer. The repositioning of FLI1 and MMP9 was specific to cancer, as it did not occur in benign disease, in line with earlier observations in breast, where gene repositioning was generally limited to cancerous tissue (Meaburn et al., 2009). In contrast, MMP2 repositioned in cancer as well as in hyperplasic prostate tissue. Our findings are in line with previous studies in other types of cancer, for which repositioning of some loci and concurrent conservation in the positioning patterns of others have been reported (Croft et al., 1999; Parada et al., 2002; Cremer et al., 2003; Wiech et al., 2005, 2009; Murata et al., 2007; Meaburn and Misteli, 2008; Meaburn et al., 2009; Harewood et al., 2010). Of interest, FLI1 and MMP9 also reposition in breast cancer (unpublished data), suggesting that they are multitissue markers of cancer. Not all genes that reposition in cancer are general markers of a malignant state, however, since an additional eight genes that reposition in breast cancer (Meaburn et al., 2009) do not reposition in prostate cancer (unpublished data).
We did not identify any common features shared among repositioning genes in prostate cancer. For example, the genes that reposition map to different chromosomes (FLI1, HSA11; MMP9, HSA20; and MMP2, HSA16). The repositioning of FLI1, at least, is unlikely to reflect whole-chromosome movements, since of five genes located on HSA11 (FLI1, CCND1, FUT4, MMP1, and NUMA1), only FLI1 robustly repositioned in prostate cancer. In addition, biological function or known relevance to tumorigenesis of genes does not predict repositioning behavior. Many of the genes we positioned that are commonly implicated in prostate cancer, such as KLK3 (prostate-specific antigen), AR, PTEN, TGFB1, VEGFA, and BCL2 (Bok and Small, 2002; Ferte et al., 2010; Lawrence et al., 2010; Shen and Abate-Shen, 2010), and the commonly translocated genes in prostate cancer, the androgen-regulated TMPRSS2 and the ETS transcription factors ERG and ETV1 (Tomlins et al., 2005), do not spatially reposition in prostate cancers. Yet the ETS transcription factor FLI1, which forms rare translocations in prostate cancer (Paulo et al., 2012), does. MMP9 and MMP2, which belong to the same class of matrix metallopeptidases (MMPs) and are involved in the breakdown of extracellular matrix, which is important to cancer- associated processes such as angiogenesis and metastasis (Egeblad and Werb, 2002), reposition in prostate cancer, and yet the potential functional relevance of the cancer-association gene repositioning is unclear. Although an increase in MMP9 and MMP2 expression has been linked to poor prognosis in prostate cancer, both genes are predominantly expressed by the stromal cells and not by the tumor cells (Egeblad and Werb, 2002). Other MMP family members, such as MMP14, and the genomic loci containing a cluster of MMP genes, including MMP1, did not reposition. Moreover, genes coding the MMP-regulating proteins TIMP2, TIMP3 (Egeblad and Werb, 2002), and SPDEF (Johnson et al., 2010), another ETS transcription factor, do not reposition in prostate cancer, further reducing a link between function and cancer-associated gene repositioning. A lack of a link between gene function and an altered radial position in cancer is in line with multiple studies that find no correlation between changes in gene expression and changes in radial positioning patterns, including a cell culture model of early breast cancer (Williams et al., 2006; Meaburn and Misteli, 2008; Takizawa et al., 2008b; Morey et al., 2009; Harewood et al., 2010; Shachar et al., 2015). Although we focused on radial positioning patterns to identify cancer-specific spatial genome-repositioning events, relative positioning of loci to other genomic loci or nuclear landmarks, such as nuclear bodies, may be a promising alternative parameter to characterize the spatial reorganization of gene loci in cancer.
Repositioning did not correlate with gene copy number or the presence of translocations. For example, FLI1 repositioned in both cancers with and without amplified FLI1, and we found FLI1 repositioning in 13 of 14 prostate cancers, yet the SLC45A3-FLI1 translocation occurs in only 1 in 200 prostate cancers (Paulo et al., 2012). Although it is possible that translocations could affect the position of genomic regions beyond the translocating region, even on chromosomes not involved in the translocation, our data demonstrate that in the background of genetic instability, it is still possible to use gene positioning for cancer diagnostics.
As previously observed for breast cancer (Meaburn et al., 2009), we find that combinatorial analysis of multiple genes results in detection of cancer with high accuracy. Whereas neither MMP9 nor FLI1 alone was repositioned in all prostate tumor samples, at least one of the two was repositioned in all cancer samples analyzed. Our observation of cancer-specific repositioning of these two genes, combined with their relatively low false-positive detection rate, makes MMP9 and FLI1 strong candidates for combinatorial use in cancer diagnosis.
MATERIALS AND METHODS
Tissues
To detect individual genes, FISH was performed on a panel of 4- to 5-μm-thick FFPE human prostate tissue sections, which consisted of 30 prostate cancers, 29 normal and NAT prostate tissues, and five hyperplasic tissues (Supplemental Table S2). Prostate tissues were obtained from the University of Washington (Seattle, WA) under the guidelines and approval of the Institutional Review Board of the University of Washington (#00-3449). These specimens were reviewed by a single genitourinary pathologist (L.D.T.). Additional tissues were purchased from US Biomax (Rockville, MD) and Imgenex (San Diego, CA; Supplemental Table S2). All specimens were deidentified.
FISH
FISH probes were produced by using nick translation to label DNA purified from bacterial artificial chromosome clones (BACPAC Resources Center [Oakland, CA]; Supplemental Table S1) with either biotin- or digoxigenin-conjugated dUTPs (Roche, Indianapolis, IN), as detailed in Meaburn (2010). An identical dual-probe FISH procedure was performed on all specimens as previously described (Meaburn et al., 2009), with the exceptions that 1) the 60°C slide baking step, before xylene (Avantor Performance Materials, Center Valley, PA) treatment, was omitted, 2) the time the tissue sections were incubated in 0.25 mg/ml Proteinase K (Sigma-Aldrich, St. Louis, MO) was increased to 15–20 min, and 3) 0.5 mg/ml Proteinase K was often required for single-tissues slides obtained from Biomax (but not tissue microarray slides).
Image acquisition and FISH analysis
Image accusation was performed essentially as described in Meaburn et al. (2009). Briefly, tissue sections were imaged with an IX70 (Olympus, Waltham, MA) DeltaVision (Applied Precision-GE Healthcare Bio-Sciences, Pittsburgh, PA) system, using a 60×/1.42 numerical aperture oil objective lens (Olympus) and an auxiliary magnification of 1.5. Image stacks were acquired with a 0.5-μm step interval along the z-axis. If mixed morphologies were present in a tissue section, before imaging, the tissue was visually inspected at low resolution (10× lens; Olympus) to identify malignant, normal, or hyperplastic regions. In these cases, hematoxylin and eosin–stained tissue slides that had been marked by a pathologist (L.D.T.) were used as a guide to ensure correct classification of different regions of a tissue. Image stacks were deconvolved and converted into maximum intensity projections using SoftWoRx (Applied Precision).
Image analysis was performed as previously described (Meaburn et al., 2009). Images were contrast enhanced based on visual inspection, and individual nuclei were manually delineated in Photoshop 7.0 (Adobe, San Jose, CA), with each nucleus saved in a separate image file. To determine the relative radial positioning of a gene in a given tissue, 91–263 randomly selected interphase epithelial nuclei were run though a custom-made image analysis software package (P. Gudla and S. Lockett, National Cancer Institute, Bethesda, MD; Meaburn et al., 2009) in MATLAB (MathWorks, Natick, MA) using the PRTools and DIPImage toolboxes (Delft University of Technology, Delft, Netherlands). The software automatically identified FISH signals (>99% of FISH signals detected, with a false-positive rate of <1%; Takizawa et al., 2008a) and determined both the binary EDT of the geometric gravity center of each FISH signal and the number of FISH signals in a nucleus for both genes visualized by FISH. In EDT, each pixel in a nucleus is assigned a value that equals the shortest distance to the nuclear edge. EDT values are then normalized to the maximum EDT value of a given nucleus to account for variations in nuclear size. Using this method, no assumption regarding nuclear shape or size is made when determining the radial position of a gene, allowing accurate comparisons between tissues even if there are differences or irregularities in nuclear shape or size. The relative radial positions of individual nuclei from the same specimen for each histologic entity were combined to generate a RRD for a gene. All alleles in a nucleus were included for analysis, and nuclei were analyzed regardless of the number of alleles present, unless there were no signals for either gene. Because some nuclei contained FISH signals for only one gene, when the number of nuclei analyzed is indicated, only nuclei containing FISH signals for the indicated gene are included. To generate PNDs, normalized EDTs from all nuclei from all normal tissues analyzed for a given gene were combined into a single data set (see Table 1 for the number of normal tissues analyzed for each gene and Supplemental Table S1 for the number of nuclei used to generate each gene’s PND). Finally, cumulative RRDs from different samples were statistically compared using the nonparametric two-sample 1D KS test. The null hypothesis that the samples are from the same distribution was rejected if p < 0.01.
Supplementary Material
Acknowledgments
We thank Prabhakar Gudla and Stephen Lockett for help with image analysis; Olufunmilayo Agunloye for technical assistance; Delft University of Technology for providing the DIPImage and PRTools toolboxes; and Tatiana Karpova for microscopy support. We also thank Kathy Doan for help in identifying and obtaining tissue used for this study. Fluorescence imaging was performed at the National Cancer Institute Fluorescence Imaging Facility (Bethesda, MD). This work was supported by Department of Defense Idea Award W81XWH-12-1-0224, the National Cancer Institute, National Institutes of Health, under Pacific Northwest Prostate Cancer SPORE Grant P50CA097186, and, in part, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations used:
- EDT
Euclidean distance transform
- FFPE
Formalin-fixed, paraffin-embedded
- FISH
fluorescence in situ hybridization
- HSA
human chromosome
- KS
Kolmogorov–Smirnov
- NAT
normal adjacent to tumor
- PND
pooled normal distribution
- RRD
relative radial distribution.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0280) on November 12, 2015.
REFERENCES
- Arican-Goktas HD, Ittiprasert W, Bridger JM, Knight M. Differential spatial repositioning of activated genes in Biomphalaria glabrata snails infected with Schistosoma mansoni. PLoS Negl Trop Dis. 2014;8:e3013. doi: 10.1371/journal.pntd.0003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok RA, Small EJ. Bloodborne biomolecular markers in prostate cancer development and progression. Nat Rev Cancer. 2002;2:918–926. doi: 10.1038/nrc951. [DOI] [PubMed] [Google Scholar]
- Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10:149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28:1117–1123. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marzo AM, Marchi VL, Epstein JI, Nelson WG. Proliferative inflammatory atrophy of the prostate: implications for prostatic carcinogenesis. Am J Pathol. 1999;155:1985–1992. doi: 10.1016/S0002-9440(10)65517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. Gene positioning. Cold Spring Harb Perspect Biol. 2010;2:a000588. doi: 10.1101/cshperspect.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferte C, Andre F, Soria JC. Molecular circuits of solid tumors: prognostic and predictive tools for bedside use. Nat Rev Clin Oncol. 2010;7:367–380. doi: 10.1038/nrclinonc.2010.84. [DOI] [PubMed] [Google Scholar]
- Foster HA, Griffin DK, Bridger JM. Interphase chromosome positioning in in vitro porcine cells and ex vivo porcine tissues. BMC Cell Biol. 2012;13:30. doi: 10.1186/1471-2121-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L, Schutz F, Boyle S, Perry P, Delorenzi M, Bickmore WA, Reymond A. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 2010;20:554–564. doi: 10.1101/gr.103622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TR, Koul S, Kumar B, Khandrika L, Venezia S, Maroni PD, Meacham RB, Koul HK. Loss of PDEF, a prostate-derived Ets factor is associated with aggressive phenotype of prostate cancer: regulation of MMP 9 by PDEF. Mol Cancer. 2010;9:148. doi: 10.1186/1476-4598-9-148. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Knight M, Ittiprasert W, Odoemelam EC, Adema CM, Miller A, Raghavan N, Bridger JM. Non-random organization of the Biomphalaria glabrata genome in interphase Bge cells and the spatial repositioning of activated genes in cells co-cultured with Schistosoma mansoni. Int J Parasitol. 2011;41:61–70. doi: 10.1016/j.ijpara.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kozubek S, Lukasova E, Jirsova P, Koutna I, Kozubek M, Ganova A, Bartova E, Falk M, Pasekova R. 3D Structure of the human genome: order in randomness. Chromosoma. 2002;111:321–331. doi: 10.1007/s00412-002-0210-8. [DOI] [PubMed] [Google Scholar]
- Lawrence MG, Lai J, Clements JA. Kallikreins on steroids: structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr Rev. 2010;31:407–446. doi: 10.1210/er.2009-0034. [DOI] [PubMed] [Google Scholar]
- Li C, Shi Z, Zhang L, Huang Y, Liu A, Jin Y, Yu Y, Bai J, Chen D, Gendron C, et al. Dynamic changes of territories 17 and 18 during EBV-infection of human lymphocytes. Mol Biol Rep. 2010;37:2347–2354. doi: 10.1007/s11033-009-9740-y. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ. Fluorescence in situ hybridization on 3D cultures of tumor cells. Methods Mol Biol. 2010;659:323–336. doi: 10.1007/978-1-60761-789-1_25. [DOI] [PubMed] [Google Scholar]
- Meaburn K, Burman B, Misteli T. Spatial genome organization and disease. In: Dellaire G, Basett-Jones D, editors. The Functional Nucleus. New York: Springer; 2016. (in press) [Google Scholar]
- Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, Kill IR, Bridger JM. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187:801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta IS, Eskiw CH, Arican HD, Kill IR, Bridger JM. Farnesyltransferase inhibitor treatment restores chromosome territory positions and active chromosome dynamics in Hutchinson-Gilford progeria syndrome cells. Genome Biol. 2011;12:R74. doi: 10.1186/gb-2011-12-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, Dellefave L, Pytel P, Selig S, Labno CM, et al. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One. 2010;5:e14342. doi: 10.1371/journal.pone.0014342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikelsaar R, Paves H, Org K, Mikelsaar AV. Chromosome variant 1qh- and its influence on the 3D organization of chromosome 1 heterochromatin in interphase nucleus of patients with endometriosis. J Genet. 2014;93:219–223. doi: 10.1007/s12041-014-0340-9. [DOI] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Nakazawa T, Ohno N, Terada N, Iwashina M, Mochizuki K, Kondo T, Nakamura N, Yamane T, Iwasa S, et al. Conservation and alteration of chromosome territory arrangements in thyroid carcinoma cell nuclei. Thyroid. 2007;17:489–496. doi: 10.1089/thy.2006.0328. [DOI] [PubMed] [Google Scholar]
- Parada L, McQueen P, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;7:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12:1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- Paulo P, Barros-Silva JD, Ribeiro FR, Ramalho-Carvalho J, Jeronimo C, Henrique R, Lind GE, Skotheim RI, Lothe RA, Teixeira MR. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240–249. doi: 10.1002/gcc.20948. [DOI] [PubMed] [Google Scholar]
- Paz N, Felipe-Blanco I, Royo F, Zabala A, Guerra-Merino I, Garcia-Orad A, Zugaza JL, Parada LA. Expression of the DYRK1A gene correlates with its 3D positioning in the interphase nucleus of Down syndrome cells. Chromosome Res. 2015;23:285–298. doi: 10.1007/s10577-015-9467-7. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique S, Thomas JS, Sproul D, Bickmore WA. Estrogen-induced chromatin decondensation and nuclear re-organization linked to regional epigenetic regulation in breast cancer. Genome Biol. 2015;16:145. doi: 10.1186/s13059-015-0719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. Identification of gene positioning factors using high-throughput imaging mapping. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008a;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008b;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme S, Schmitt E, Stein S, Schwarz-Finsterle J, Wagner J, Walch A, Werner M, Hausmann M, Wiech T. Nuclear position and shape deformation of chromosome 8 territories in pancreatic ductal adenocarcinoma. Anal Cell Pathol (Amst) 2011;34:21–33. doi: 10.3233/ACP-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Veltri RW, Christudass CS. Nuclear morphometry, epigenetic changes, and clinical relevance in prostate cancer. Adv Exp Med Biol. 2014;773:77–99. doi: 10.1007/978-1-4899-8032-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiech T, Stein S, Lachenmaier V, Schmitt E, Schwarz-Finsterle J, Wiech E, Hildenbrand G, Werner M, Hausmann M. Spatial allelic imbalance of BCL2 genes and chromosome 18 territories in nonneoplastic and neoplastic cervical squamous epithelium. Eur Biophys J. 2009;38:793–806. doi: 10.1007/s00249-009-0474-5. [DOI] [PubMed] [Google Scholar]
- Wiech T, Timme S, Riede F, Stein S, Schuricke M, Cremer C, Werner M, Hausmann M, Walch A. Human archival tissues provide a valuable source for the analysis of spatial genome organization. Histochem Cell Biol. 2005;123:229–238. doi: 10.1007/s00418-005-0768-3. [DOI] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, Fisher AG. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Zeitz MJ, Ay F, Heidmann JD, Lerner PL, Noble WS, Steelman BN, Hoffman AR. Genomic interaction profiles in breast cancer reveal altered chromatin architecture. PLoS One. 2013;8:e73974. doi: 10.1371/journal.pone.0073974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.