The microtubule plus-end tracking protein EB1 moves into flagella and accumulates at the tip independently of intraflagellar transport. EB1 dwell for seconds at the tip, indicating stable EB1-binding sites. Simulations show that diffusion to capture is an alternative mechanism to accumulate proteins in cilia.
Abstract
The microtubule (MT) plus-end tracking protein EB1 is present at the tips of cilia and flagella; end-binding protein 1 (EB1) remains at the tip during flagellar shortening and in the absence of intraflagellar transport (IFT), the predominant protein transport system in flagella. To investigate how EB1 accumulates at the flagellar tip, we used in vivo imaging of fluorescent protein–tagged EB1 (EB1-FP) in Chlamydomonas reinhardtii. After photobleaching, the EB1 signal at the flagellar tip recovered within minutes, indicating an exchange with unbleached EB1 entering the flagella from the cell body. EB1 moved independent of IFT trains, and EB1-FP recovery did not require the IFT pathway. Single-particle imaging showed that EB1-FP is highly mobile along the flagellar shaft and displays a markedly reduced mobility near the flagellar tip. Individual EB1-FP particles dwelled for several seconds near the flagellar tip, suggesting the presence of stable EB1 binding sites. In simulations, the two distinct phases of EB1 mobility are sufficient to explain its accumulation at the tip. We propose that proteins uniformly distributed throughout the cytoplasm like EB1 accumulate locally by diffusion and capture; IFT, in contrast, might be required to transport proteins against cellular concentration gradients into or out of cilia.
INTRODUCTION
Microtubules (MTs) are polar assemblies of α- and β-tubulin. The MT plus-end is more dynamic, and various proteins bind to the plus-end, promoting MT elongation or shortening (Akhmanova and Steinmetz, 2010). End-binding protein 1 (EB1) is a widely distributed plus-end tracking protein that binds directly to MTs in vitro. EB1 has been widely used to track the tips of growing MTs and as an indicator for the presence of GTP/GDP+Pi tubulin near the plus-end. While polymerized GTP and GDP+Pi tubulin are its preferred targets, EB1also binds to more subdistal regions of growing MTs, suggesting that its binding is not strictly coupled to the nucleotide state of tubulin but is also sensitive to the conformational state of tubulin in the MT lattice (Maurer et al., 2011).
The plus-ends of the axonemal microtubules are at the distal tips of cilia and flagella. At the tip reside several tip-binding proteins, including kinesin-13 (Piao et al., 2009; Vasudevan et al., 2015), kinesin-4/Kif-7 (He et al., 2014), Che-12/crescerin (Das et al., 2015), Cep104/FAP256 (Satish Tammana et al., 2013), and Spef1/CLAMP (Gray et al., 2009). Similarly, EB proteins have been shown to be present at the tips of motile 9+2 flagella, mammalian primary cilia, and sensory cilia in Caenorhabditis elegans (Pedersen et al., 2003; Hao et al., 2011; Schroder et al., 2011). These results reveal that, within both motile and primary cilia, EB1 accumulates on the tips of apparently static axonemal MTs in contrast with its behavior on singlet MTs in the cytoplasm, where EB1 binding is largely limited to the plus-ends of growing MTs. In Chlamydomonas reinhardtii, EB1 remains at the tips of steady-state and even shortening flagella (Pedersen et al., 2003). Axonemal MTs are distinct from singlet MTs in the cell body due to their high content of posttranslationally modified tubulin and their dense decoration with associated structures, including integral protein ribbons (Linck et al., 2014). Further, the A-tubules of the outer doublets and the central-pair singlet MTs are capped by material of largely unknown composition (Dentler and Rosenbaum, 1977; Satish Tammana et al., 2013). These biochemical and ultrastructural specializations might be the reason for the resistance to MT-depolymerizing drugs, the high biochemical stability, and the apparent lack of treadmilling and dynamic instability of the axonemal MTs (Marshall and Rosenbaum, 2001; Watanabe et al., 2004). However, a continual albeit low-level incorporation of tubulin was demonstrated for steady-state flagella of zygotes (Marshall and Rosenbaum, 2001). These observations raise the question of whether EB1 binding to the flagellar tip is similarly transient and dependent on the addition of fresh tubulin as described for its binding to cytoplasmic singlet MTs.
Many flagellar proteins require intraflagellar transport (IFT), a motor-driven bidirectional motility of proteins along the axonemal MTs, to efficiently enter flagella and move to the tip (Rosenbaum and Witman, 2002; Wren et al., 2013; Craft et al., 2015). Pedersen et al. (2003) showed that EB1 remains at the tips of flagella after IFT has been switched off in Chlamydomonas fla10-1, a temperature-sensitive mutant in the anterograde IFT motor kinesin-2 (Kozminski et al., 1995). Taking into account that EB1 remains present at the flagellar tips of nongrowing flagella, these observations raise the possibility that EB1 is firmly attached to the flagellar tip and, once deposited at the tip via IFT, remains in that location in the absence of IFT. Alternatively, EB1 at the tip could be continuously exchanged by an IFT-independent mechanism.
In this study, we used in vivo imaging in C. reinhardtii to elucidate the dynamics of fluorescent protein (FP)-tagged EB1 in flagella. The FP tag (green fluorescent protein [GFP] or mNeonGreen [NG]) was fused to the C-terminus of EB1; such fusions are thought to be neutral with respect to EB1 dimerization and microtubule plus-end tracking but do interfere with the binding of some EB1-interacting proteins (Skube et al., 2010; Sen et al., 2013). C. reinhardtii tends to adhere with its two flagella to a cover glass, allowing for the tracking of single fluorescent particles in flagella by total internal reflection fluorescence (TIRF) microscopy (Lechtreck, 2013). IFT transport of EB1-FP was essentially absent and EB1-FP entered flagella by diffusion and dwelled transiently at the tip. In simulations, these two distinct phases of EB1 mobility in flagella were sufficient to explain its accumulation at the tip. Our data show that proteins can rapidly accumulate at the flagellar tip in an IFT-independent manner.
RESULTS
EB1 and EB1-GFP show a similar subcellular distribution
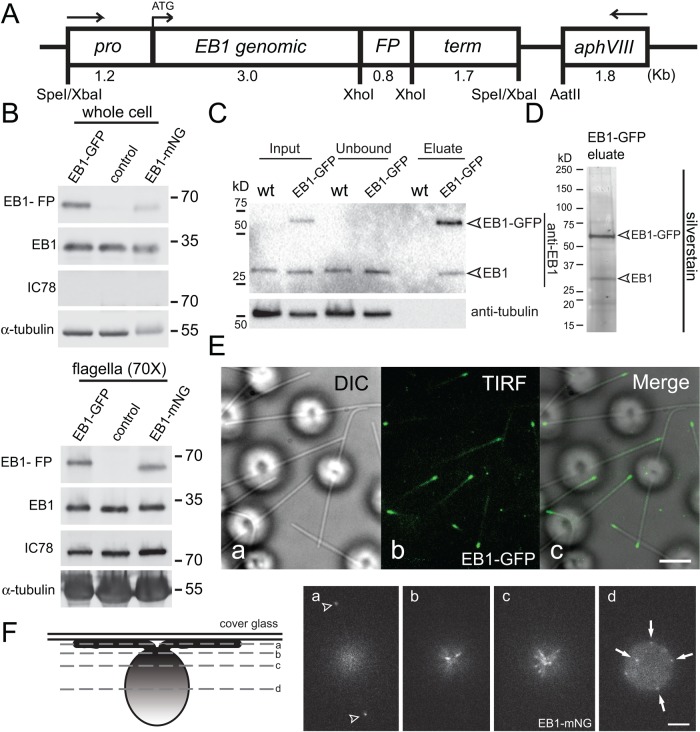
Antibody staining showed that EB1 is present in the cell bodies and at the flagellar tips of C. reinhardtii (Pedersen et al., 2003). To visualize the in vivo dynamics of EB1, we expressed EB1 fused to either GFP or the brighter NG in wild-type cells (Figure 1A; Shaner et al., 2013). Western blotting of whole cells with anti-EB1 identified two bands of ∼35 and ∼60 kDa (Figure 1B). The former was also present in untransformed control strains and represents the endogenous untagged EB1 protein; the latter represents the EB1-GFP or EB1-NG fusion proteins. For determination of subcellular distribution of EB1, isolated flagella were probed with anti-EB1 (Figure 1B). Both endogenous and FP-tagged EB1 were present in flagella of transformants. To obtain a signal of similar intensity as the whole-cell sample, ∼70 times more flagella sample (i.e., 140 flagella/cell body) had to be loaded, suggesting that ≤2% of the total EB1 is present in flagella. EB1 is known to form dimers, and we wondered whether the endogenous and transgenic EB1 interact with each other (Honnappa et al., 2005). Using an anti-GFP nanobody, we immunopurified EB1-GFP from detergent extracts of isolated flagella (Figure 1C). Endogenous EB1 remained attached to EB1-GFP after a medium-stringency salt wash (200 mM NaCl), and silver staining of the eluate showed that EB1 and EB1-GFP are the predominant proteins in the eluate indicative for the presence of EB1/EB1-GFP complexes inside flagella, putatively in the form of EB1/EB1-GFP heterodimers (Figure 1D).
FIGURE 1:
The cellular distribution of endogenous and FP-tagged EB1 is similar. (A) Schematic presentation of the EB1-FP expression vector. The sequence for either GFP or NG were integrated into the genomic DNA encompassing the EB1 gene, including its endogenous promoter (pro) and terminator (term). The selectable marker gene aphVIII was present on the same plasmid. The arrows indicate the orientation of the genes. (B) Western blot analysis of whole cells and isolated flagella of wild type (control) and strains expressing EB1-GFP or EB1-NG probed with antibodies to EB1, and as loading controls, to IC78 and α-tubulin. The flagellar samples were 70 times more concentrated than the whole-cell samples (i.e., ∼140 flagella/cell). (C) Flagellar extracts from a EB1-GFP–expressing strain and a wild-type control were incubated with anti-GFP beads and the depleted extract (Unbound), the bound fraction (Eluate), and the original extract (Input), were analyzed by SDS–PAGE and Western blotting with anti-EB1. Note that endogenous EB1 copurifies with EB1-GFP. (D) Silver staining of the eluate obtained from a strain expressing EB1-GFP by GFP affinity purification. (E) DIC (a), TIRF (b), and the corresponding merged image (c) of live EB1-GFP cells. Scale bar: 3 μm. (F) Schematic representation (left) and live images of a focal series through a EB1-NG cell. Arrowheads in a, flagellar tips; arrows in d, punctae of EB1-NG in a posterior region of the cell. Scale bar: 3 μm.
TIRF microscopy of living cells attached via their two flagella to the cover glass showed FP-tagged EB1 concentrated at the tips of flagella (Figure 1E), confirming previous observations on the endogenous EB1 based on antibody staining (Pedersen et al., 2003; Sloboda and Howard, 2007). Using steeper illumination angles allowed us to image EB1 in the cell body. EB1-NG was concentrated in the region of the basal bodies and present in a spotty distribution in more posterior regions of the cell (Figure 1F). In summary, the subcellular distribution of the endogenous EB1 is recapitulated by the FP-tagged protein, indicating that the latter is a suitable reporter for the analysis of EB1 in vivo dynamics in C. reinhardtii.
EB1-NG visualizes the dynamic properties of cytoplasmic MTs
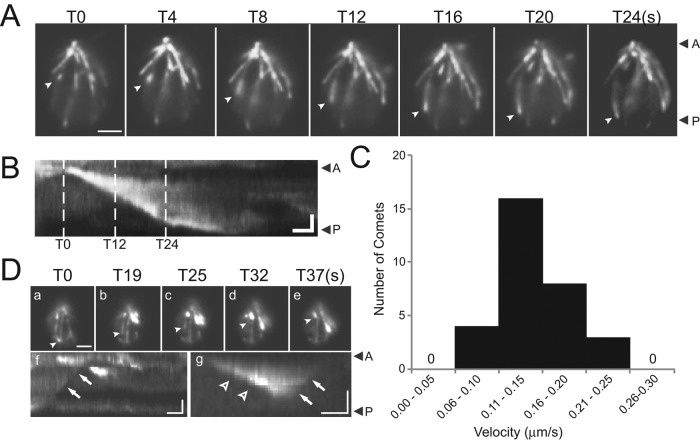
The microtubular cytoskeleton of C. reinhardtii consists of the axonemal MTs, the basal body MTs, and cytoplasmic or cortical MTs that run from their origin near the basal bodies toward the posterior end of the cell (Doonan and Grief, 1987). EB1 tracks the plus-ends of growing MTs in various cell types and thus can be used as a tool to visualize MT dynamics. EB1-NG comets were observed radiating from the basal body region and moving progressively to the posterior end of the cells, revealing the hitherto unknown dynamics of the cortical MTs in C. reinhardtii (arrowheads in Figure 2A; Supplemental Movie S1). Kymograms were used to determine the velocity of EB1-NG comets (Figure 2B); the average velocity was 0.142 μm/s (±0.04 μm/s, n = 31; Figure 2C). This corresponds to a rate of ∼8.5 μm/min, which is within the range determined for MT growth in plant (∼5 μm/min) and mammalian cells (10–20 μm/min; Mimori-Kiyosue et al., 2000; Chan et al., 2003; Salaycik et al., 2005). Near the posterior end of the cell, most EB1-NG comets became slower and weaker, but some reached the edge of the cell and continued to grow in a curve along the edge before the comet was lost. After the comets vanished, some EB1-NG remained attached to the MT and, occasionally, we observed rapid shrinkage of these signals progressing from the posterior cell region toward the anterior, indicating catastrophic depolymerization of the underlying MT (Figure 2D). We conclude that a subset of the cortical MTs of C. reinhardtii is highly dynamic and that its flagellar basal apparatus continuously nucleates MTs, similar to the mammalian centrosome. The data further establish that C. reinhardtii EB1-NG behaves similarly to EB1 in other organisms in tracking the plus-ends of growing MTs.
FIGURE 2:
Fluorescent EB1 localizes to comets in the cell body. (A and B) Individual frames (A) and corresponding kymogram (B) from a recording of EB1-NG comets in the cell body. The comet marked by arrowheads moved from the flagella-bearing cell apex to its posterior, presumably tracking the tip of an elongating MT. Scale bar: 2 μm. (B) Kymogram of the comet marked in A. Dashed lines indicate time points corresponding to the frames in A. Arrowheads with A and P, anterior and posterior of the cell. Scale bar: 1 μm and 5 s. (C) Histogram depicting the distribution of the velocities of EB1-NG comets. (D, a–e) Single frames from a video depicting EB1-NG loss, presumably during catastrophic MT shortening. As described in other species, some residual EB1 remains attached to the length of the MTs in the C. reinhardtii cell body. The MT labeled by arrowheads is initially capped by EB1-NG (T0) and then retreats with time (T19–T37 in seconds). Scale bar: 2 μm. (f) Kymogram corresponding to a; the arrow indicates the trace corresponding to the EB1 signal labeled in a. (g) Kymogram showing growth and retreat of an EB1-NG comet. Arrowhead, elongation; arrows, catastrophe. Scale bars: 1 μm and 10 s. See Supplemental Movie S1.
EB1 at the flagellar tip is exchanged in an IFT-independent manner
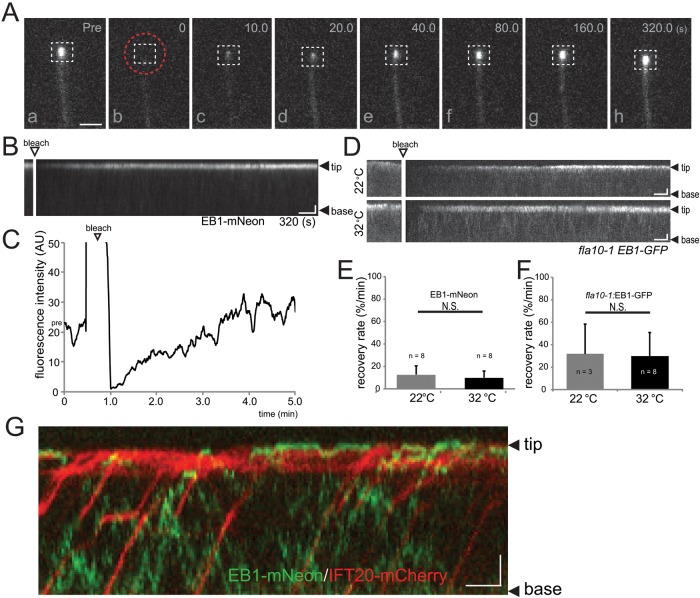
Previous data have shown that EB1 remains at the flagellar tip under various experimental conditions, including flagella shortening and inhibition of IFT. These observations could indicate that EB1 is firmly bound to the flagellar tip. To test the dynamics of flagellar EB1, we utilized fluorescence recovery after photobleaching (FRAP) analysis after bleaching either the entire flagellum (Supplemental Figure S1) or only the flagellar tip (Figure 3A; Supplemental Movie S2); both methods gave very similar results. Fluorescence recovery at the flagellar tip was apparent briefly after photobleaching, and full recovery of fluorescence was achieved in ∼3–7 min (Figure 3, B and C). Complete or near-complete recovery was also observed after repeated bleaching of the flagellar tip (Supplemental Figure S1). In conclusion, EB1 at the flagellar tip is continuously exchanged with unbleached protein from the flagellum and the cell body.
FIGURE 3:
EB1 at the flagellar tip is rapidly exchanged independent of/unaided by IFT. (A and B) Individual frames (A) and corresponding kymogram (B) from a FRAP experiment demonstrating the exchange of EB1-NG at the tips of steady-state flagella. (A) Images taken before (pre) and at various time points (0–320 s) after photobleaching of the flagellar tip using a spot laser (position indicated by the dashed red circle). The dashed white box indicates the area used for FRAP analysis. In the kymogram (B), the flagellar tip and base and the bleaching step are indicated. Scale bars: 1 μm and 20 s. (C) Quantitative analysis of a FRAP experiment. The recovery of fluorescence (in arbitrary units, a.u.) at the flagellar tip was measured after photobleaching of the entire flagellum. The signal recovers to prebleach strength in ∼3 min. Arrowhead, bleaching step. (D) Kymograms depicting recovery of EB1-GFP in flagella of fla10-1 cells maintained at 22°C and 32°C. The base of the flagella (B) and the distal tip (T) are marked. Scale bars: 2 μm and 10 s. (E and F) Mean recovery rates of wild-type (E) and fla10-1 (F) cells expressing EB1-NG and EB1-GFP, respectively. Cells were analyzed at the permissive (22°C) and restrictive (32°C) temperatures for IFT in fla10-1. Error bars indicate the SD. The differences in the rates of fla10-1 and the control strain are likely to be caused by differences in the microscope settings. (G) Merged kymogram from simultaneous imaging of EB1-NG (green) and IFT20-mCherry (red) in flagella. Note that EB1 and IFT20 move independent of each other. The base of the flagellum (base) and the distal tip (tip) are marked. Scale bars: 2 μm and 10 s.
IFT is thought to transport most flagellar proteins into and inside cilia and flagella. In C. reinhardtii, transport by IFT has been demonstrated for various axonemal proteins (Wren et al., 2013; Craft et al., 2015). To determine whether EB1 translocation to the tip and thus the recovery of EB1-FP fluorescence was IFT-dependent, we expressed EB1-FP in fla10-1, which allows one to switch off IFT by incubating cells at elevated temperatures. Because many cells moved during extended experiments, the recovery rate (percent recovery of the prebleach signal intensity/minute) instead of total recovery time was used to compare EB1-FP recovery at different conditions (Figure 3, E and F). FRAP analysis showed that there was no significant difference in the rate of EB1-FP fluorescence recovery between fla10-1 cells maintained at the permissive (22°C) or restrictive (32°C) temperature (Figure 3, D and F). Western blotting confirmed that the temperature shift was effective in abolishing IFT: at the restrictive temperature selected, IFT particle proteins were quantitatively removed from fla10-1 flagella; the levels of endogenous EB1 remained constant (Supplemental Figure S2C). Next we tested FRAP of EB1-NG at the flagellar tip of fla11-1, a temperature-sensitive mutant defective in the IFT protein IFT172, which is thought to interact with EB1 (Pedersen et al., 2005). Previous reports using antibodies did not observe EB1 at the flagellar tips of fla11-1 mutants maintained at the restrictive temperature (Pedersen et al., 2003). However, TIRF microscopy revealed that EB1-FP was present at the flagellar tips and recovered normally after photobleaching in fla11-1 cells maintained at 22°C and 32°C (Supplemental Figure S2, A and B). Western blotting of flagellar preparations from fla11-1showed that the levels of endogenous EB1 were unaffected by the temperature shift, while the levels of IFT172 were strongly reduced (Supplemental Figure S2C). We noticed that, at the permissive temperature and even more at the restrictive temperature, many fla11-1 flagella were rather short and accumulated IFT proteins at the tips (unpublished data), which could mask EB1 detection by antibodies in immunofluorescence and distort protein ratios in Western blots, putatively explaining previous findings describing the absence of EB1 from fla11-1 flagella at the restrictive temperature.
Two-color imaging revealed that EB1-NG and the IFT particle protein IFT20-mCherry move independently from each other, with IFT20 moving by IFT and EB1-FP moving by diffusion (Figure 3G). EB1-NG also moved independently of the bona fide IFT cargo mCherry-α-tubulin in steady-state and growing flagella (Supplemental Figure S3). For the entire duration of this study, transport of EB1-FP by IFT was observed only once. IFT transport of GFP-tagged EB1 and EB3 proteins was also not apparent in primary cilia of retinal pigment epithelium cells or C. elegans sensory cilia (Hao et al., 2011; Larsen et al., 2013). The data indicate that EB1-FP enters flagella and translocates to the tips independently of IFT.
Axonemal MTs bind less EB1-NG than cytoplasmic MTs
Similar to observations in other systems, C. reinhardtii EB1-NG preferably binds to the end of growing cell body MTs and vanishes from nongrowing MTs. However, EB1 is present at the tips of steady-state, growing, and even shrinking flagella (Pedersen et al., 2003), raising the question of whether the mechanisms of EB1 binding to axonemal and cytoplasmic MTs are different. We compared the intensity of the EB1-NG signal at the tips of steady-state flagella with that of the EB1-NG comets in the cell body. The latter is likely to represent the tip of a single MT, while the former contains nine A- and nine B-tubules and two central-pair MTs. The fluorescence intensity of one EB1-NG comet typically exceeded that of the total EB1-NG present at the flagellar tip (478 vs. 257 a.u. for comets and flagellar tips; SD 89.9, n = 4 and SD 74.0, n = 10, respectively). To test whether EB1-NG might be predominantly attached to the two CP MTs, we expressed EB1-NG in the central pair (CP)-deficient mutant pf18; the amounts and dynamics of EB1-NG at the tips of pf18 flagella were essentially unaltered (unpublished data). In conclusion, only small amounts of EB1 are present at the flagellar tip, suggesting that the plus-ends of axonemal MTs in steady-state flagella attract considerably less EB1 than the tips of growing singlet MTs in the cell body.
For determining whether the growth state of flagella affects the amount of EB1-NG at the tip, cells were deflagellated by a pH shock and allowed to initiate flagellar regeneration. Cells with regenerating and steady-state flagella were mixed before imaging to allow for a direct comparison of signal strengths (Figure 4A). The EB1-NG signal at the tips of regenerating flagella was on average 2.5 times brighter than that of steady-state flagella (Figure 4B) and often extended into the flagellar shaft. FRAP analysis of growing and steady-state flagella showed similar rates of EB1-NG exchange (Supplemental Figure S2D). Using mechanical shearing, we generated cells with only one flagellum and analyzed EB1-NG distribution while such long-zero cells regrew the missing flagellum and shortened the remaining flagellum (Rosenbaum et al., 1969). EB1-NG remained attached to the tips of the longer, putatively retracting flagella, and the EB1-NG signals at the tips of growing flagella of such long-short cells were increased ∼ twofold in strength, often extending into the flagellar shaft (Figure 4, C and D). These single-cell experiments show that the tips of flagella with elongating axonemes have an increased capacity to attract EB1-FP.
FIGURE 4:
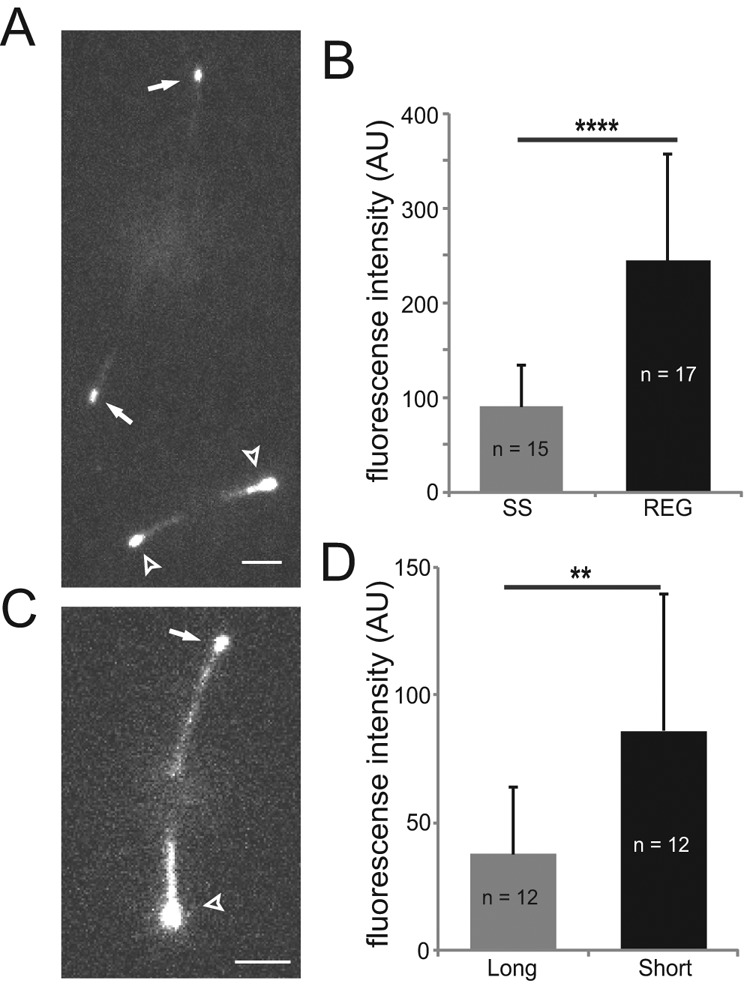
Growing flagellar tips attract more EB1. (A) TIRF image of two EB1-NG cells, one with steady-state (arrows) and one with regenerating (arrowheads) flagella. Scale bar: 3 μm. (B) Mean fluorescence intensity of EB1-NG in the tip region of steady-state (SS; n = 15) and regenerating (REG; n = 17) flagella. (243 a.u., SD 113 a.u.; n = 17 vs. 90 a.u., SD 43 a.u., n = 15); Error bars indicate SD. Significance: p ≤ 0.0001. (C) TIRF image showing EB1-NG in the flagella of a long-short cell; the long flagellum is marked by an arrow, the short one by an arrowhead. Scale bar: 3 μm. (D) Bar graph showing the mean fluorescence intensity of EB1-NG at the tips of long (n = 12) and short (n = 12) flagella of long-short cell. Error bars indicate SD. Significance: p ≤ 0.01.
Limited turnover of axonemal tubulin in steady-state flagella appears unrelated to EB1 binding
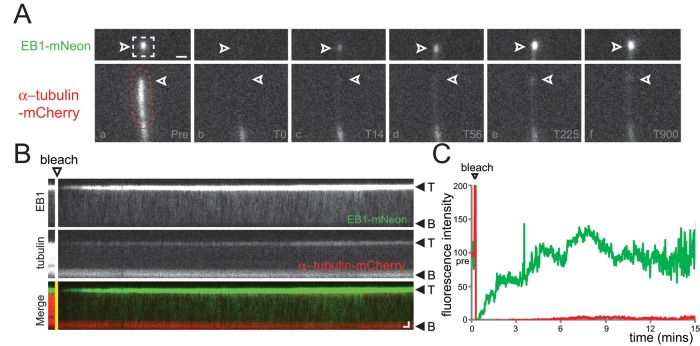
The increased presence of EB1-NG at the tips of elongating flagella suggests a causal link between tubulin polymerization and EB1-NG binding to axonemal MTs, raising the question of whether EB1 accumulation at the tips of steady-state and shortening flagella also depends on the addition of new tubulin to the axonemal MTs. Treadmilling of axonemal MTs has not been observed, and flagellar length is essentially constant within the short periods required for EB1-FP recovery (Marshall and Rosenbaum, 2001; Watanabe et al., 2004). Nevertheless, individual MTs of the axonemal bundle could shorten and reelongate without affecting the length of the entire flagellum. To address the question of how EB1 exchange and tubulin incorporation are related, we expressed EB1-NG in a strain coexpressing mCherry-α-tubulin to about ∼10% of the total α-tubulin (Figure 5 and Supplemental Figures S3 and S4). Recovery of the fluorescent signals representative for EB1 and tubulin was analyzed after photobleaching of the distal portion of the flagella. To ensure that an incorporation of mCherry-α-tubulin could be detected while continuously imaging cells by TIRF, we first analyzed growing flagella (Supplemental Figure S3). We observed dense IFT trafficking and incorporation of mCherry-α-tubulin into the elongating axoneme; the EB1 signal at the flagellar tip recovered with similar speed. In bleached steady-state flagella, however, only very little or no incorporation of mCherry-α-tubulin was observed even after prolonged observation (up to 20 min; Figure 5, A–C, Supplemental Figure S4, and Supplemental Movie S3), while EB1-NG returned to the tip at standard rates. The data reveal that only small amounts of tubulin are incorporated into the axoneme of steady-state flagella. Formally, the experiment does not exclude the possibility that bleached mCherry-α-tubulin is released from the tip and reincorporated after GDP-to-GTP exchange. However, considering the high rate of entry and diffusional mobility of FP-tagged tubulin in flagella (Craft et al., 2015), one would expect a considerable incorporation of the tagged tubulin into flagellar tips. We interpret the data to the effect that the presence of EB1 at the tips of steady-state flagella does not depend on the de novo addition of tubulin to the axoneme.
FIGURE 5:
Recovery of EB1-NG is not linked to tubulin exchange at the flagellar tip. (A–C) Gallery of individual frames (A), corresponding kymograms (B), and signal quantification (C) of a two-color FRAP experiment. (A) The distal flagellar region of a cell coexpressing EB1-NG and mCherry-α-tubulin was bleached using a laser spot that was moved along the flagellum (indicated by the dashed region), and FRAP was analyzed over several minutes (T0–T900 s). Top row, EB1-NG; bottom, mCherry-α-tubulin; arrowheads, flagellar tip. Scale bar: 1 μm. (B) Single-channel kymograms corresponding to A. The bleaching step and the orientation of the flagella are indicated. Scale bars: 1 μm and 10 s. (C) Quantification of the fluorescence intensity of EB1-NG (green) and mCherry-α-tubulin (red) corresponding to the photobleaching experiment depicted in A and B. The prebleach fluorescence intensity was set to 100 for both proteins; EB1-NG recovered rapidly and completely, while only traces of mCherry-α-tubulin were recovered even after 15 min of observation. See Supplemental Figure S4 for a similar experiment.
EB1-NG dwells for elongated periods of time at the flagellar tip
To characterize the diffusional behavior by which EB1 accumulates at the flagellar tip in greater detail, we used increased laser intensities, which bleached most EB1-NG particles entering the flagella within a few seconds; this prevented the accumulation of unbleached protein, enabling us to observe individual EB1-NG particles (Figure 6A; Supplemental Movie S4). The vast majority (>97%; n = 93) of the EB1-NG particles in the ciliary shaft bleached in one step, indicating the presence of a single EB1-NG (Figure 6A). In the flagellar shaft, most EB1-NG particles displayed a random back-and-forth motion with a one-dimensional (1D) diffusion coefficient of 1.06 μm2s−1 (n = 41; Figure 6B; Supplemental Movie S4). A subset (∼5%) of EB1-NG particles moved in an apparent preferred direction along the flagella, with some particles taking multiple subsequent steps in one direction (white arrows in Figure 6, Supplemental Figure S5A, and Supplemental Movies S5 and S6). Such particles were observed moving toward the flagellar tip or base; also, the displacement of the particles between frames was variable, and the runs were interrupted by one or more steps in the opposite direction. Thus these particles show characteristics typical for diffusion; however, considering the low probability of such directional runs by diffusion, we cannot exclude additional mechanisms promoting a directional movement of proteins along flagella. The latter is suggested by the parabolic distribution of the mean-square displacement over time for such particles (Supplemental Figure S5B).
FIGURE 6:
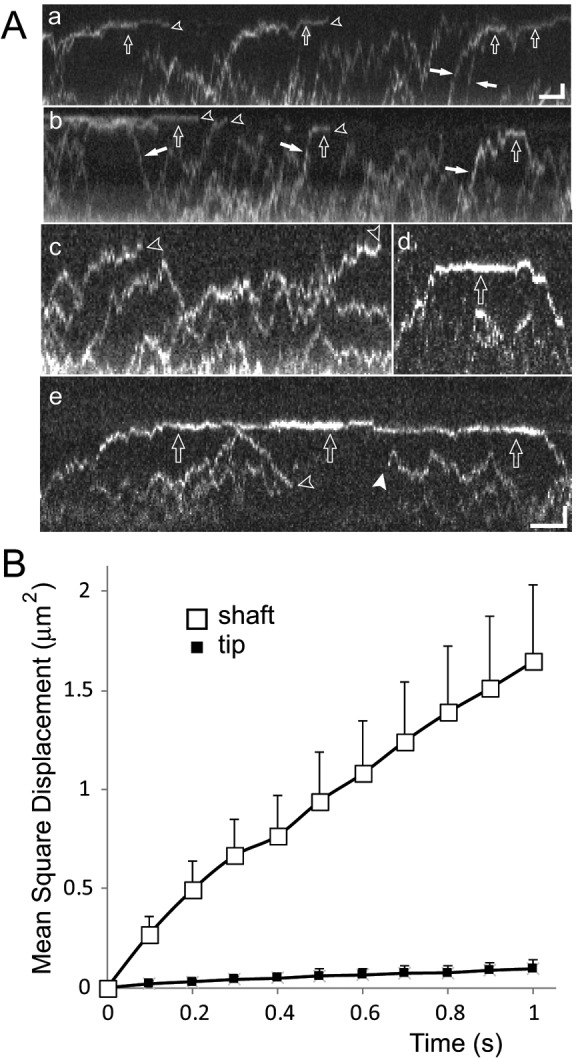
Differential mobility of EB1-NG explains its accumulation at the flagellar tip. (A) Gallery of kymograms depicting diffusion of EB1-NG in flagella. Open arrows, particles with reduced mobility near the flagellar tip; open arrowheads, bleaching events; white arrows, EB1-NG particles preferably moving in one direction along the flagellar shaft; white arrowhead, reappearance of photobleached EB1-NG as it is infrequently observed for NG-tagged proteins. Note the reduced mobility of particles approaching the tip (compare c with d and e), transiently stationary EB1-NG at the tip (a, b, d, and e), and the differences in the time EB1-NG remains trapped at the tip. Scale bars: (a and b) 1 μm and 2 s; (c–e) 1 μm and 1 s. (B) Mean-square displacement vs. time plots for EB1-NG particles moving in the flagellar shaft (open squares; n = 41) or tip segment (filled squares; n = 14).
Near the flagellar tip, EB1-NG motility was markedly reduced and interspersed with stationary periods (Figure 6A; Supplemental Movies S5 and S6). We averaged EB1-NG mobility at the tip and determined a diffusion coefficient of D = 0.063 μm2s−1 ± 0.033 μm2s−1 based on 14 trajectories of EB1-NG particles moving in the distal 1-μm segment of the flagellum (Figure 6B). EB1-NG particles became transiently trapped in the tip region, and an average resident time of 2.5 s (SD 1.6 s, n = 51) was determined for those that could be tracked from entry to exit (Figure 6A, a, d, and e). Because many particles were bleached while being trapped in the tip region (Supplemental Figure S5), the true average dwell time of EB1-NG at the tip is likely to be longer.
Simulating EB1 accumulation at the flagellar tip
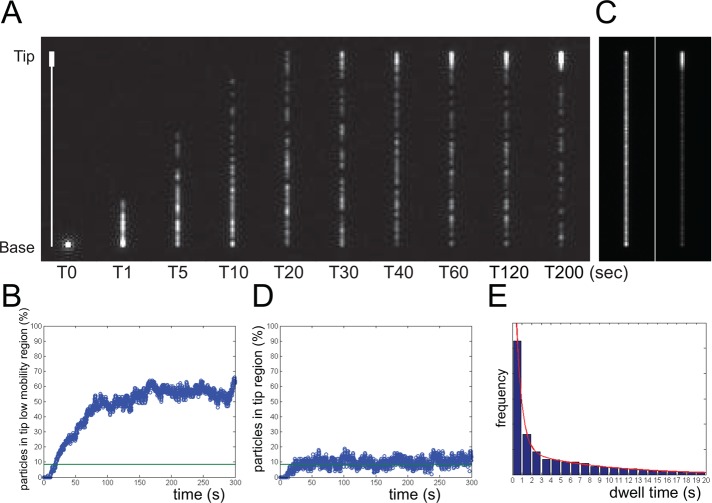
We wondered whether the observed differences in EB1-NG mobility are sufficient to explain its accumulation at the flagellar tip. We used a one-dimensional model, assuming the flagellum as a line of 12 μm in length, which is divided in a 11-μm-long proximal segment in which particles diffuse with a coefficient of 1.06 μm2s−1 and a 1-μm-long distal tip segment with a diffusion coefficient of 0.063 μm2s−1 (Figure 7A). At the beginning of the simulation, 100 particles were introduced into the proximal end of the model flagellum (Supplemental Movie S7). These parameters caused ∼55% of the particles to accumulate in the tip segment (Figure 7B), a value similar to the ∼62% (SD 6.6%, n = 14) determined for EB1-NG based on the fluorescence intensity; the somewhat higher value might reflect the omission in our measurements of the proximal portions of the flagella, which were out of the range of the TIRF excitation. In the simulation, particles remained an average of 5 s (SD 6.95) in the tip segment compared with 2.5 s in EB1-NG bleaching experiments; the latter were performed at high laser intensities, limiting the time span during which particles could be observed and thereby eliminating longer dwell times from our data.
FIGURE 7:
Modeling EB1 distribution in flagella. The flagellum was modeled as a 12-μm-long line with a 1-μm-long low-mobility region (diffusion coefficient 0.06 μm2s−1) at the tip and assuming a diffusion coefficient of 1.06 μm2s−1 for the flagellar shaft of 11 μm. One hundred particles were released at the base of the line at T0. (A) Individual frames from a simulation. The line on the left indicates the position of the flagellar shaft and the low mobility region. See corresponding Supplemental Movie S7. (B) Plot of the fraction of particles in the tip segment vs. time based on the simulation described in A. Green line, expected share of particles in 1/12 of the flagellum assuming random distribution of the particles. (C) Left, to explore to what extent the geometry of the tip will result in an accumulation of particles, we performed a simulation similar to A but using the same particle diffusion coefficient along the entire flagellar length. Note the minimal accumulation of particles at the flagellar ends in this maximum-intensity projection over ∼1000 frames. Right: A similar maximum-intensity projection but using the conditions described in A. (D) Plot of the fraction of particles in the tip segment vs. time based on the single diffusion-coefficient simulation described in C. Green line as in B. (E) Histogram showing the distribution of dwell times in the tip region based on the simulation described in A.
We wondered to what extent EB1 accumulated at the tip simply because the direction into which particles can travel is restricted. In simulations assuming the same diffusion coefficient (1.06 μm2s−1) along the entire length of the flagellum, the concentration of particles in the distal segment was only very slightly elevated compared with the flagellar shaft (Figure 7, C and D). In summary, the simple model essentially recapitulates the experimental results on EB1-NG. We conclude that the distinct motilities of EB1 in the flagellar shaft and tip segment are sufficient to explain its accumulation at the flagellar tip without the need of motor-driven transport.
DISCUSSION
EB1 transiently binds to the flagellar tip with long dwell times
In vivo imaging was used to analyze the behavior of the MT plus-end tracking protein EB1 in flagella of C. reinhardtii. Similar to observations on singlet MTs, EB1-NG transiently attaches to the flagellar tip, most likely binding to the distal portions of axonemal MTs. However, we observed several features distinguishing EB1-NG behavior at the flagellar tip from that of EB1 at the end of growing cytoplasmic MTs. Compared with the latter, the tips of steady-state flagella accumulate only small amounts of EB1, which were only slightly elevated in growing flagella, indicative of a comparatively limited number of axonemal binding sites for EB1. Chlamydomonas flagella elongate with a maximum rate of ∼400 nm/min compared with rates of 8 μm/min and more determined for cytoplasmic singlet MTs (Srayko et al., 2005; Bhogaraju et al., 2014). Assuming a similar GTPase activity of tubulin in cytoplasmic and flagellar MTs, the slow growth rate of the latter will minimize the size of any GTP/GDP+Pi tubulin zone, putatively restricting EB1 binding. EB1 has been used as an indicator for the presence of GTP-tubulin, and the presence of EB1 at the tips of steady-state flagella raises the question of whether axonemal MTs permanently maintain a GTP cap (Seetapun et al., 2012). In C. reinhardtii, tubulin turnover at the axonemal tip is a rather slow process: It takes dozens of minutes before hemagglutinin-tagged tubulin introduced into unlabeled flagella using sexual cell fusion becomes incorporated to detectable levels (Marshall and Rosenbaum, 2001; Lechtreck et al., 2013b). Similarly, we showed that little or no recovery of fluorescence occurs after photobleaching of mCherry-α-tubulin in steady-state flagella. Also, one would expect that the EB1 signal is lost or diminished during flagellar shortening, when the axoneme depolymerizes and any GTP-tubulin maintained at the tip should be lost. EB1, however, remains attached to the tips of shortening flagella and displays unaltered exchange rates in FRAP experiments (Pedersen et al., 2003; this study). EB1 binding to the tips of nongrowing flagella is therefore unlikely to indicate the presence of GTP-tubulin. To solve this conundrum, we propose that the tip of the axonemal MT has a lattice conformation allowing for EB1 binding independent of the GTP status of tubulin. Indeed, EB1 binding is not strictly linked to the GTP state of tubulin: The EB1 comets observed on growing singlet MTs exceed the presumed GTP/GDP+Pi cap in length, suggesting a delay between GTP hydrolysis and the conformational changes in the lattice that will abolish EB1 binding (Maurer et al., 2014). The tips of axonemal MTs are crowned by cap structures that surround the MTs and insert plugs into the MT lumen (Dentler and Rosenbaum, 1977). These cap structures could generate a microtubular lattice to which EB1 can bind. Notably, these cap structures are maintained during flagellar shortening and therefore could preserve the EB1-binding sites as they track depolymerizing axonemal MTs. Compared with the transient binding sites at the end of rapidly growing singlet MTs, which are short-lived due to the chemical instability of GTP-tubulin in the lattice, the stable binding sites proposed here for axonemal MTs should result in different EB1 exchange characteristics. Indeed, EB1-NG was trapped for seconds in the distal flagella segment compared with mean dwell times of just 0.05 s determined in vitro for EB1 at the tips of growing singlet MTs or along GTP-γ-S MTs (Bieling et al., 2008; Chen et al., 2014). The possibility that EB1 binds to nontubulin tip proteins cannot be excluded, but the observation that growing flagella bind more EB1 links its binding to the conformation of the axonemal MT lattice.
In C. elegans sensory cilia, tubulin exchange at the microtubule tips of the middle and distal segments is clearly detectable after a few minutes (Hao et al., 2011). EB1-GFP localizes to these ciliary microtubule ends but does not recover to detectable levels after photobleaching, and movement of EB1-GFP in cilia was not observed. EB1 in C. elegans cilia could firmly reside at the sites of de novo tubulin incorporation. This contrasts our observations in C. reinhardtii, which show rapid EB1-GFP recovery but only little incorporation of tubulin in steady-state cilia. It is unknown whether the elaborate tip structures observed in many motile cilia are present in C. elegans sensory cilia or other primary cilia. Additional studies are required to determine whether (motile and nonmotile) cilia have principally different tip structures, tubulin exchange rates, and EB1 dynamics.
EB1 moves into and inside flagella by diffusion
In imaging experiments, EB1-NG did not comigrate with the IFT-B complex protein IFT20, and FRAP of EB1-NG at the flagellar tip was not affected when IFT was switched off using conditional mutants. IFT-like transport of EB1-FP was observed only once, likely representing an unusual event that might have been caused, for example, by clumping with a genuine IFT cargo. We conclude that EB1 enters and moves inside C. reinhardtii flagella independently of IFT. The transition zone at the flagellar base is thought to function as a diffusional barrier for large cytoplasmic proteins (Kee et al., 2012; Breslow et al., 2013). Despite the predicted molecular weight of ∼95–120 kDa for EB1-FP (hetero-)dimers, the Stokes radii of their globular entities are below the estimated size-exclusion limit, suggesting that EB1 can freely diffuse from the cell body into the flagellum. Previously, we showed that GFP-tagged tubulin dimers (∼130 kDa) diffuse apparently freely into C. reinhardtii flagella (Craft et al., 2015). Tubulin, however, is also a cargo of IFT, and vast amounts of tubulin are transported via IFT during flagellar growth. The elongation of the axonemal MTs will remove soluble tubulin from the flagellar matrix, generating a diffusional current and resulting in the net entry of tubulin from the cell body into the flagellum. However, diffusion alone is apparently insufficient to supply enough tubulin for flagellar growth. We propose that the different modes of transport observed for tubulin and EB1 reflect differences in the subcellular distribution of these proteins: estimates suggest that the concentration of soluble EB1 in the flagellum is similar to that in the cell body (see Materials and Methods). Binding sites at the flagellar tips and the end of growing cytoplasmic MTs will then locally accumulate EB1. In contrast, the tubulin concentration in the flagellar matrix, in particular the matrix of growing flagella, is likely to exceed that in the cell body cytoplasm (Craft et al., 2015). This suggests that IFT functions in concentrating soluble tubulin inside the flagellar matrix above cell body levels; a high tubulin concentration could be necessary to promote an efficient elongation of the axoneme.
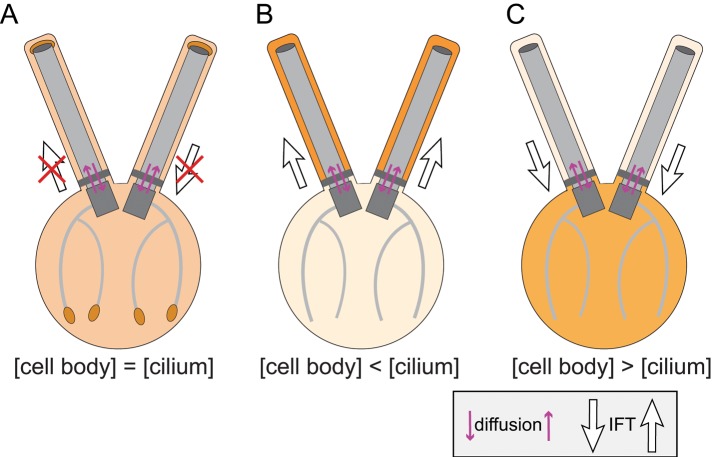
Flagellar assembly and maintenance requires a mix of diffusion and motor-driven protein transport. It is of interest to determine which flagellar proteins move by diffusion, by IFT, or a combination of both, and whether rules exist allowing one to predict which mode of transport will be used by a particular protein (Figure 8). One prediction would be that proteins with elevated concentrations in the flagellar matrix versus the cell body cytoplasm will be transported by IFT, while proteins that are in a diffusional equilibrium between cell and flagellum will not use IFT; a prerequisite is that the proteins in question can efficiently enter and exit flagella by diffusion. Similarly, small cell body proteins, which might leak into flagella by diffusion, might depend on IFT to be removed from flagella. The membrane-associated protein phospholipase D, for example, is retained to >99% in the cell body under wild-type conditions but accumulates progressively in flagella when IFT is abolished (Lechtreck et al., 2013a). In conclusion, IFT could function in moving proteins that pass freely in and out of flagella through the transition zone and against cellular concentration gradients.
FIGURE 8:
The role of diffusion and IFT in flagellar protein transport. The saturation of the orange background color indicates protein concentration, with dark colors specifying high concentrations. (A) Proteins with similar concentrations in the cell body cytoplasm and the ciliary matrix might not require IFT for transport. An example is EB1, which accumulates locally by being captured onto microtubule plus-ends. (B) Proteins in which the concentration in the ciliary matrix exceeds that in the cell body cytoplasm require IFT to be concentrated in the ciliary compartment. An example is tubulin, which enters cilia by diffusion and by IFT (Craft et al., 2015). IFT of tubulin and the tubulin concentration in the ciliary matrix are elevated during ciliary growth, presumably to allow for an efficient elongation of the axoneme. (C) Many proteins are abundant in the cell body but efficiently excluded from flagella. If such proteins are able to enter cilia by diffusion, IFT might function as a scavenger, exporting such proteins from cilia. An example in phospholipase D, which is retained to > 98% in the cell body of C. reinhardtii and removed from cilia in an IFT- and BBSome-dependent manner (Lechtreck et al., 2013a). In summary, IFT functions in moving proteins against concentration gradients into and out of cilia. While this is likely to apply to small proteins, which are able to diffuse through the transition zone, larger proteins might depend on IFT to pass through the transition zone.
A diffusion-to-capture mechanism is sufficient to accumulate proteins in flagella
Simulations in model flagella consisting of two regions with distinct mobile behavior essentially recapitulated our in vivo observations with respect to the amount of the EB1-NG protein trapped in the low-mobility region at the tip and the dwell time of individual particles in this region. In the simulated kinetic, however, 35% of the particles required just 38 s to enter the tip region, which is faster than the experimental data on EB1-NG. The simulation commences with an empty tip region, whereas bleached molecules have to exit the tips in order for unbleached EB1-NG to replace them in the FRAP experiments. The rate-limiting step of FRAP is clearly the slow exchange of photobleached molecules at the tip with unbleached molecules rather than supply of unbleached EB1-NG by diffusion. For EB1, the local concentration in the low-mobility region is ∼18 times higher than in the rest of the flagellum and cytoplasm. The simulations support our in vivo observations that a diffusion-to-capture mechanism is sufficient to explain the accumulation of EB1 at the flagellar tip.
In the case of EB1, the binding sites in the tip region are permanent, but it is worthwhile to consider situations in which the interaction between a diffusing protein and its flagellar binding sites are regulated. On the activation of such binding sites, the protein will rapidly accumulate by diffusion and, when binding is abolished, the released protein will exit the flagellum until the equilibrium concentration is reestablished. Such a mechanism appears to drive the light-regulated import and export of membrane-associated signaling proteins such as arrestin from the cilia-like outer segment of rods (Calvert et al., 2006). Regulated binding to the ciliary tip is also characteristic for the hedgehog signaling protein Gli; kinesin Kif7 is dispensable for Gli transport but maintains a Gli-binding compartment at the ciliary tip (He et al., 2014). Diffusion and regulated capture could explain the delivery of building blocks into growing cilia and the transient accumulation of signaling proteins in cilia without evoking IFT involvement.
MATERIALS AND METHODS
Strains and culture conditions
C. reinhardtii was maintained in batch cultures in a modified M (minimal media) at 21°C with a light/dark cycle of 14:10 h. For flagellar isolation and in vivo TIRF microscopy experiments, cultures were aerated and supplemented with 0.5% CO2. The following strains were used in this study: wild-type (CC-620 and CC-621), fla10-1 (CC-1919), fla11-1 (CC-1920), ift20-1 IFT20-mCherry (Lechtreck et al., 2009), and mCherry-α-tubulin in CC-620 (Craft et al., 2015).
Transgenetic strain generation
For expression of fluorescent protein–tagged EB1, a 6-kb genomic DNA fragment, encompassing EB1 including 1.2 and 1.7 kb of the 5′ and 3′ flanking sequence, was amplified by PCR using a Chlamydomonas BAC clone as a template and primers (gcacacggtctagattcgcactgccgtgagc) and (gtctagaccaggcatcggaagtggcttcggagcc), each containing an XbaI site. The PCR fragment was digested with XbaI and ligated into the complementary SpeI site in the pGEM-T Easy vector also containing a paromomycin (PMM)-resistant cassette to create the plasmid pCrEB1 (Zhu et al., 2013). For tagging of the C-terminus of CrEB1, a 2.8-kB 3′ CrEB1 genomic DNA was amplified from the BAC clone using the primer pair (gcaagaccggtgacatgaagcacagcg) and (ccagagcgactgacccaggcatcg) and TA cloned into pGEM-T vector (Promega). A QuikChange site-directed mutagenesis kit (Stratagene, San Diego, CA) was used to convert the sequence before the stop codon into a XhoI site into which a GFP-encoding fragment derived by PCR from pKL3-GFP was inserted (Lechtreck et al., 2009). The tagged DNA was reamplified to add a KpnI site through an antisense primer (taggtaccccagagcgactgacccaggc). After digestion with KpnI, the tagged PCR fragment was inserted in pCrEB1, replacing the corresponding untagged fragment in pCrEB1 to create the plasmid pCrEB-GFP. For generation of the EB1-NG derivate, NG DNA was custom synthesized based on the NG protein sequence, using the Chlamydomonas codon bias, and amplified from the pBR25-CrNG plasmid using the primer pair (ctcgagatggtgtccaagg) and (ctcgagcttgtacagctcgtcc) with added XhoI sites (Craft et al., 2015). The XhoI-digested NG fragment replaced the GFP fragment in the pCrEB-GFP to create pCrEB-NG. An aliquot of these genomic constructs was transformed into Chlamydomonas cells using the glass bead method (Kindle, 1990), and positive transformants were selected on TAP plates containing 10 μg/ml PMM. The PMM-resistant clones were screened for fluorescence using a Nikon Eclipse wide-field microscope and a CoolSNAP-ES CCD camera. The fla10-1 EB1-GFP and fla11 EB1-GFP were generated and selected similarly. The IFT20-mCherry EB1-NG strain was generated by transforming IFT20-mCherry cells (Lechtreck et al., 2009) with the EB1-NG plasmid via electroporation. Positive transformants were selected on TAP media plates containing 10 μg/ml PMM, and resistant clones were screened via TIRF microscopy. The mCherry-α-tubulin EB1-NG coexpressing strain was generated by transforming EB1-mNeon cells with pBR25-mCherry-α-tubulin. The plasmid was constructed by PCR amplification of the mCherry gene from the pKL3-IFT20-mCherry construct described by Lechtreck et al. (2009). The amplified mCherry gene was digested with XhoI and BamHI and inserted into the pBR25-sfGFP-α-tubulin expression vector, replacing the sfGFP fragment (Rasala et al., 2013; Craft et al., 2015). Positive transformants were selected on TAP plates containing 10 μg/ml zeocin in constant light and identified by TIRF microscopy.
Flagellar isolation and Western blotting
For preparation of whole-cell samples, the cell pellet from a 5-ml late–log phase TAP liquid culture was resuspended with 50 μl 10 mM HEPES buffer, followed by the addition of 100 μl 5X SDS–PAGE sample buffer and boiling for 5 min. After 2 μl 1.7 mg/ml phenylmethylsulfonyl fluoride was added, the insoluble remnants were removed by centrifugation, and the supernatant was processed for SDS–PAGE and Western blotting. Flagellar samples were prepared as previously described (Yang et al., 2008). The following antibodies were used for analysis: rabbit polyclonal anti-EB1 (1:5000; Pedersen et al., 2003), mouse monoclonal anti-IC78 (1:5000; King and Witman, 1990), and mouse monoclonal anti α-tubulin (1:5000; Sigma). Western blots were developed using anti-mouse and anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Invitrogen), and chemiluminescence images were captured and documented using a UVP Autochemi Bioimaging System (Cambridge, UK).
Immunoprecipitation of EB1-GFP
Isolated flagella from the EB1-GFP–expressing strain and a control strain were resuspended in HMEK (30 mM HEPES, 5 mM MgSO4, 25 mM KCL, and 0.5 mM EGTA) supplemented with protease inhibitor cocktail (Sigma-Aldrich). Flagella were lysed by adding an equal volume of HMEK plus 200 mM NaCl and 0.5%NP-40 for 30 min on ice, and the axonemes were removed by centrifugation (20,000 × g, 4°C for 10 min). The membrane+matrix fraction was incubated with GFP-nAB agarose slurry (GFP-nAB, Allele Biotechnology) equilibrated with binding buffer (HMEK, 150 mM NaCl) followed by nutation for 1 h at 4°C. The slurry was washed three times with binding buffer (HMEK, 150 mM NaCl); this was followed by three washes with wash buffer (HMEK, 200 mM NaCl). The bound proteins were eluated with 1 M glycine (pH 2.5) and were analyzed by SDS–PAGE followed by Western blotting or silver stain (Bio-Rad Technologies).
Flagellar regeneration and long-short cell generation
For obtaining cells with regenerating flagella, cells grown in M media were deflagellated by a pH shock, pelleted via centrifugation, and resuspended in fresh M media (Lefebvre, 1995). Cells were allowed to regrow flagella at room temperature under constant light with gentle agitation. To delay the regrowth of flagella, we placed cells on ice until needed. Long-short cells were generated by passing cells in M media repeatedly (∼4–6 times) through a 26 G × ½ in. needle attached to a 1-ml syringe. This method resulted in a small percentage of long-short cells that were identified by microscopy.
In vivo microscopy
A Nikon Eclipse Ti-U inverted microscope equipped with a 60×/1.49 numerical aperture (NA) TIRF objective and a through-the-objective TIRF illumination system with 75-mW, 561-nm and 40-mW, 488-nm diode lasers (Spectraphysics) was utilized for in vivo imaging experiments (Lechtreck, 2013). The excitation light was filtered with a Nikon GFP/mCherry TIRF filter cube, and the two-color emission light was separated by using a splitting device (Photometrics DualView2). Photobleaching of flagella was accomplished using two approaches. For bleaching the entire flagellum, the 488-nm laser-emission intensity was increased to ∼10% for 5–30 s. For bleaching a specific area of the flagellum, a focused 488-nm laser beam passing through the specimen in epifluoresence mode was used for <2 s. Increased laser intensities were used to image individual EB1-FP molecules. For in vivo imaging, 8–10 μl of cells was placed in on a 24 × 60 mm no. 1.5 coverslip and allowed to settle for ∼1–3 min. Then a 22 × 22 mm no. 1.5 coverslip containing an equal volume of 10 mM HEPES and 6.25 mM EGTA (pH 7.4) was placed on top of the large cover glass to form an observation chamber. Cells were imaged at room temperature (∼24°C) or after incubation at 32°C, using an objective heater (Bioptechs). Images were recorded and documented at 1–31 frames/s using the iXon X3 DU897 EMCCD camera (Andor) and Elements software package (Nikon). ImageJ (National Institutes of Health) with the LOCI Bio-formats Importer (University of Wisconsin) and Multiple Kymogram (European Molecular Biology Laboratory) plug-ins was used to generate movies and kymograms and to retrieve single frames from the Element ND2 files. Kymograms, individual frames for time-lapse series, and movies were cropped and adjusted for brightness and contrast in ImageJ and Photoshop (Adobe). All figures were assembled using Illustrator (Adobe). Movies were cropped, adjusted for brightness and contrast, rotated, and converted to 8-bit format in ImageJ. The corrected movies were exported as AVI files, and QuickTimePro was used for scene selection.
FRAP and fluorescence intensity analysis
For determination of the fluorescence intensity, videos were opened in ImageJ, and the flagellar tip region or another region of interest (ROI) was selected using the Rectangle tool. The fluorescence intensity inside the selected region was determined using the Plot Z-axis tool, and the data were exported into Excel. The fluorescence intensity in the ROI was corrected for the background fluorescence using ROI of the same size. For FRAP analysis, videos were imported into ImageJ, an ROI was selected with the Rectangle tool, and the fluorescence of the ROI was determined from the Plot Z-axis tool. After background subtraction in Excel, the highest intensity value before the bleaching event was set to 100%, and the recovery of fluorescence (as percentage of the prebleached value) was calculated. In a subset of movies, the fluorescence lost during the experiment was calculated using the unbleached flagellar tip of the same cell as an internal control.
Estimation of the cellular distribution of EB1 and tubulin
Western blotting indicated a ratio of 70:1 for EB1 in the cell body versus flagella. The cell body has a volume of ∼250 μm3 compared with 0.75 μm3 for a 12-μm-long flagellum. However, the volume of freely accessible cytoplasm and flagellar matrix is likely to be considerably lower. We used ∼20% cytoplasm for the cell body, with its numerous cell organelles and vesicles that exclude tubulin, and 50% for the flagellum, in which a considerable volume is occupied by the axoneme. Then the cell body cytoplasm is ∼66 times larger than that of the two flagella. This suggests that EB1 has a similar concentration in the cell body and the flagellum. The concentrations of tubulin have been estimated earlier. In brief, the two flagella contain ∼20% of the cell's total tubulin, corresponding to an ∼12 times higher concentration in the flagellar compartment compared with the accessible cytoplasm (∼20% of the total cell volume). In steady-state flagella, ∼10% of the tubulin is soluble; the share of soluble tubulin in the C. reinhardtii cell body is unknown but has been estimated to be ∼40% or more in other cells. Assuming that ∼60% of the cell body tubulin is polymerized, the concentration of soluble tubulin in the flagellar matrix is twice that of the cell body cytoplasm during steady-state and approximately four times higher during flagellar growth.
Calculation of EB1-FP diffusion coefficient
To calculate the diffusion coefficient of EB1-NG in flagella, we utilized the same methods described for DRC4-GFP and GFP-α-tubulin (Wren et al., 2013; Craft et al., 2015). In short, 11 movies were selected that illustrated a high number of diffusing particles, and specific trajectories were identified from those movies using the ImageJ plug-in Mosaic Particle Tracker (Sbalzarini and Koumoutsakos, 2005). For each trajectory identified, the ratio of total distance traveled versus end-to-end distance was calculated; particles were excluded if this ratio was greater than 2.0, as these particles were likely not diffusing but rather undergoing a type of directed transport. The mean-square displacement versus time was calculated using the remaining 41 trajectories.
Similarly, EB1-NG was analyzed diffusing with a markedly reduced motility within the most distal 1-μm segment, noted as the flagellar tip. From 14 trajectories analyzed, a diffusion coefficient of 0.063 μm2s–1 was determined.
To analyze those particles that displayed a directionally biased translocation along the cilia shaft, we determined the diffusion coefficient from 14 such trajectories. In this case, trajectories were excluded if the resulting ratio of total distance traveled to end-to-end distance was < 4.0; the excluded particles were likely undergoing typical diffusion instead of directed translocation.
Simulations
The simulations of one-dimensional diffusion of EB1 were written in Python. One hundred particles were initialized to a position at the base of the flagellum (x = 0). At each time step, the position of each particle was updated by selecting a random step, dx, from a Gaussian distribution with SD, 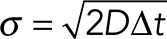 , where D is the diffusion constant and Δt is the time step. For our simulations Δt = 0.1 s. D is position dependent:
, where D is the diffusion constant and Δt is the time step. For our simulations Δt = 0.1 s. D is position dependent:
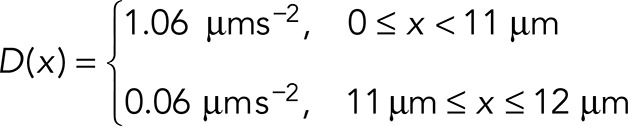 |
At the base (x = 0) and at the tip (x = 12), reflecting boundary conditions were used. That is, if the new position x + dx was greater than 12 μm or less than 0 μm, the new position was set to x = 12 or 0 μm, respectively. For each time step, the number of particles in the tip region (11 μm ≤ x ≤ 12 μm) was counted. Each time a particle entered the tip, a counter was started to keep track of how long it spent at the tip. When a particle left the tip, the dwell time was added to a list, and the counter was reset to zero. The list was then used to generate the histogram of tip dwell times. Simulations using a D(x) of 1.06 μms–2 for the entire flagellum and simulations in which the low-mobility region is moved from the tip down into the flagellar shaft were generated similarly.
The movies were created by generating a point-spread function (PSF) at the position of each particle at each time step. In each frame, each particle was assumed to emit 500 photons corrupted by Poisson noise. For generating the image of a particle, an ideal pupil function, P(μ,ν), was generated with radius NA/λ, where the NA was chosen to be 1.2; (μ,ν) is the position in the pupil plane and has units of 1/length. The ideal pupil is defined as
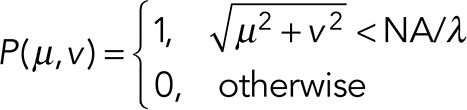 |
The ideal pupil function was then multiplied by 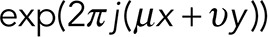 , where (x, y) is the position of the particle. The image of the particle is then
, where (x, y) is the position of the particle. The image of the particle is then
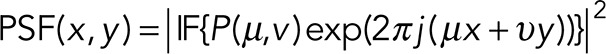 |
where IF{} denotes the Fourier transform. In this way, it is straightforward to generate subpixel particle movements. The PSFs generated by each particle are added to generate the image for each frame. A background of 20 photons is added to each pixel, and then Poisson noise is added to the image to generate the final image of each frame. The pixel size is 110 nm, and the number of pixels used to generate each image is 128 × 128.
Supplementary Material
Acknowledgments
We thank Lotte Pedersen and Joel Rosenbaum for the gift of antibodies to C. reinhardtii EB1. This work was support by the Marquette University Startup program to P.Y., a Marquette University teaching assistantship to Y.L., and grants from the National Science Foundation (NSF 1350654) to P. K. and the National Institutes of Health (GM110413) to K.F.L.
Abbreviations used:
- 1D
one-dimensional
- EB1
end-binding protein 1
- FP
fluorescent protein
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- IFT
intraflagellar transport
- MT
microtubules
- NA
numerical aperture
- NG
mNeonGreen
- PMM
paromomycin
- PSF
point-spread function
- ROI
region of interest
- TIRF
total internal reflection fluorescence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0608) on December 2, 2015.
REFERENCES
- Akhmanova A, Steinmetz MO. Microtubule +TIPS at a glance. J Cell Sci. 2010;123:3415–3419. doi: 10.1242/jcs.062414. [DOI] [PubMed] [Google Scholar]
- Bhogaraju S, Weber K, Engel BD, Lechtreck KF, Lorentzen E. Getting tubulin to the tip of the cilium: one IFT train, many different tubulin cargo-binding sites. Bioessays. 2014;36:463–467. doi: 10.1002/bies.201400007. [DOI] [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr, Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Chan J, Calder GM, Doonan JH, Lloyd CW. EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat Cell Biol. 2003;5:967–971. doi: 10.1038/ncb1057. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rolls MM, Hancock WO. An EB1-kinesin complex is sufficient to steer microtubule growth in vitro. Curr Biol. 2014;24:316–321. doi: 10.1016/j.cub.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol. 2015;208:223–237. doi: 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Dickinson DJ, Wood CC, Goldstein B, Slep KC. Crescerin uses a TOG domain array to regulate microtubules in the primary cilium. Mol Biol Cell. 2015;26:4248–4264. doi: 10.1091/mbc.E15-08-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler WL, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. III. Structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J Cell Biol. 1977;74:747–759. doi: 10.1083/jcb.74.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan JJ, Grief C. Microtubule cycle in Chlamydomonas reinhardtii: an immunofluorescence study. Cell Motil Cytoskel. 1987;7:381–392. [Google Scholar]
- Gray RS, Abitua PB, Wlodarczyk BJ, Szabo-Rogers HL, Blanchard O, Lee I, Weiss GS, Liu KJ, Marcotte EM, Wallingford JB, Finnell RH. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat Cell Biol. 2009;11:1225–1232. doi: 10.1038/ncb1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Thein M, Brust-Mascher I, Civelekoglu-Scholey G, Lu Y, Acar S, Prevo B, Shaham S, Scholey JM. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol. 2011;13:790–798. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Jr., Kapoor TM, Anderson KV. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, John CM, Kostrewa D, Winkler FK, Steinmetz MO. Structural insights into the EB1-APC interaction. EMBO J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Witman GB. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J Biol Chem. 1990;265:19807–19811. [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Grigoriev I, Akhmanova A, Pedersen LB. Analysis of microtubule plus-end-tracking proteins in cilia. Methods Enzymol. 2013;524:105–122. doi: 10.1016/B978-0-12-397945-2.00007-X. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF. In vivo imaging of IFT in Chlamydomonas flagella. Methods Enzymol. 2013;524:265–284. doi: 10.1016/B978-0-12-397945-2.00015-9. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, Witman GB. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013a;201:249–261. doi: 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Gould TJ, Witman GB. Flagellar central pair assembly in Chlamydomonas reinhardtii. Cilia. 2013b;2:15. doi: 10.1186/2046-2530-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA. Flagellar amputation and regeneration in Chlamydomonas. Methods Cell Biol. 1995;47:3–7. doi: 10.1016/s0091-679x(08)60782-7. [DOI] [PubMed] [Google Scholar]
- Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, Warren P, Nicastro D. Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+ binding proteins, and stable protofilaments. J Biol Chem. 2014;289:17427–17444. doi: 10.1074/jbc.M114.568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPγS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci USA. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr Biol. 2014;24:372–384. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Geimer S, Sloboda RD, Rosenbaum JL. The microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr Biol. 2003;13:1969–1974. doi: 10.1016/j.cub.2003.10.058. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Piao T, Luo M, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Barrera DJ, Ng J, Plucinak TM, Rosenberg JN, Weeks DP, Oyler GA, Peterson TC, Haerizadeh F, Mayfield SP. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013;74:545–556. doi: 10.1111/tpj.12165. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Salaycik KJ, Fagerstrom CJ, Murthy K, Tulu US, Wadsworth P. Quantification of microtubule nucleation, growth and dynamics in wound-edge cells. J Cell Sci. 2005;118:4113–4122. doi: 10.1242/jcs.02531. [DOI] [PubMed] [Google Scholar]
- Satish Tammana TV, Tammana D, Diener DR, Rosenbaum J. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J Cell Sci. 2013;126:5018–5029. doi: 10.1242/jcs.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Schroder JM, Larsen J, Komarova Y, Akhmanova A, Thorsteinsson RI, Grigoriev I, Manguso R, Christensen ST, Pedersen SF, Geimer S, Pedersen LB. EB1 and EB3 promote cilia biogenesis by several centrosome-related mechanisms. J Cell Sci. 2011;124:2539–2551. doi: 10.1242/jcs.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetapun D, Castle BT, McIntyre AJ, Tran PT, Odde DJ. Estimating the microtubule GTP cap size in vivo. Curr Biol. 2012;22:1681–1687. doi: 10.1016/j.cub.2012.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen I, Veprintsev D, Akhmanova A, Steinmetz MO. End binding proteins are obligatory dimers. PloS One. 2013;8:e74448. doi: 10.1371/journal.pone.0074448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skube SB, Chaverri JM, Goodson HV. Effect of GFP tags on the localization of EB1 and EB1 fragments in vivo. Cytoskeleton (Hoboken) 2010;67:1–12. doi: 10.1002/cm.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloboda RD, Howard L. Localization of EB1, IFT polypeptides, and kinesin-2 in Chlamydomonas flagellar axonemes via immunogold scanning electron microscopy. Cell Motil Cytoskel. 2007;64:446–460. doi: 10.1002/cm.20195. [DOI] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Vasudevan KK, Jiang YY, Lechtreck KF, Kushida Y, Alford LM, Sale WS, Hennessey T, Gaertig J. Kinesin-13 regulates the quantity and quality of tubulin inside cilia. Mol Biol Cell. 2015;26:478–494. doi: 10.1091/mbc.E14-09-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Hayashi M, Yagi T, Kamiya R. Turnover of actin in Chlamydomonas flagella detected by fluorescence recovery after photobleaching (FRAP) Cell Struct Funct. 2004;29:67–72. doi: 10.1247/csf.29.67. [DOI] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF. A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol. 2013;23:2463–2471. doi: 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Owen HA, Yang P. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J Cell Biol. 2008;180:403–415. doi: 10.1083/jcb.200705069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liu Y, Sivadas P, Gupta A, Yang P. Molecular tools for studying the radial spoke. Methods Enzymol. 2013;524:19–36. doi: 10.1016/B978-0-12-397945-2.00002-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.