The small GTPase Rab5 regulates the early endocytic pathway of transferrin (Tfn), and Rab5 deactivation is required for Tfn recycling. Developmentally regulated GTP-binding protein 2 is required for interaction between Rab5 and RabGAP5 on endosomes and acts as a key regulator for Rab5 deactivation and Tfn recycling.
Abstract
The small GTPase Rab5 regulates the early endocytic pathway of transferrin (Tfn), and Rab5 deactivation is required for Tfn recycling. Rab5 deactivation is achieved by RabGAP5, a GTPase-activating protein, on the endosomes. Here we report that recruitment of RabGAP5 is insufficient to deactivate Rab5 and that developmentally regulated GTP-binding protein 2 (DRG2) is required for Rab5 deactivation and Tfn recycling. DRG2 was associated with phosphatidylinositol 3-phosphate–containing endosomes. It colocalized and interacted with EEA1 and Rab5 on endosomes in a phosphatidylinositol 3-kinase–dependent manner. DRG2 depletion did not affect Tfn uptake and recruitment of RabGAP5 and Rac1 to Rab5 endosomes. However, it resulted in impairment of interaction between Rab5 and RabGAP5, Rab5 deactivation on endosomes, and Tfn recycling. Ectopic expression of shRNA-resistant DRG2 rescued Tfn recycling in DRG2-depleted cells. Our results demonstrate that DRG2 is an endosomal protein and a key regulator of Rab5 deactivation and Tfn recycling.
INTRODUCTION
Intracellular vesicular trafficking contributes to diverse cellular processes, such as nutrient uptake and cell migration (Mellman, 1996). Small GTPase Rab proteins ensure the delivery of cargoes to their correct destinations by binding to various effectors, such as molecular motors and tethering factors (Stenmark, 2009). Rab5, a well-known early endosome marker, recruits early endosome antigen 1 (EEA1; Christoforidis et al., 1999a) and regulates fusion and motility of early endosomes (Nielsen et al., 1999). Rab7 plays key roles in biogenesis and maintenance of lysosomes (Bucci et al., 2000) by making endosomes competent for fusion with lysosomes (Rink et al., 2005). Rab11 is the most prominent recycling endosome marker and regulates vesicular recycling to the plasma membrane (Ullrich et al., 1996; Grant and Donaldson, 2009). As cargoes move along endocytic pathways, the transition between early and late or recycling endosomes is mediated by Rab conversion, a process in which cascades of Rab guanine nucleotide exchange factors (GEFs) and Rab GTPase-activating proteins (GAPs) act as key regulators (Hutagalung and Novick, 2011). One well-studied Rab conversion is that from Rab5 to Rab7 as endosomes mature from early endosomes to late endosomes (Rink et al., 2005). Rab5 recruits the SAND-1(Mon1)/CZZ-1 heterodimer, which in turn recruits and activates Rab7 as the endosome matures (Poteryaev et al., 2010). Activated Rab7 recruits Rab5 GAP and facilitates inactivation and removal of Rab5 (Rink et al., 2005). Defects in Rab5 GAP inhibit the Rab5-to-Rab7 conversion and result in accumulation of large vesicles containing both Rab5 and Rab7 (Chotard et al., 2010). In contrast to Rab5-to-Rab7 conversion, little is known about how Rab5-to-Rab11 conversion along the recycling pathway is controlled. However, deactivation of Rab5 seems to be an important event for cargo recycling, because defects in Rab5 deactivation interfere with recycling (Haas et al., 2005; Sun et al., 2012). In mammalian cells, RabGAP5 has been identified to have Rab5 GAP activity, and RNA interference of RabGAP5 induced a large early endosome phenotype, as seen with expression of activated Rab5 (Haas et al., 2005). In contrast, CED-10/Rac1 is recruited to recycling endosomes and deactivates Rab5 through recruitment of Rab5 GAP (TBC-2) in Caenorhabditis elegans (Sun et al., 2012), suggesting the importance of Rac1 in the control of Rab conversion along the recycling pathway. However, details of the mechanism of Rab5 deactivation during cargo recycling are largely unknown.
Developmentally regulated GTP-binding proteins (DRGs) comprise a novel class of evolutionarily conserved GTP-binding proteins and constitute a subfamily of the GTPase superfamily (Schenker et al., 1994). The DRG subfamily contains two closely related proteins, DRG1 and DRG2 (Li and Trueb, 2000). The evolutionary conservation of DRGs suggests that they may play an essential role in the control of cell growth and differentiation. DRG1 interacts with stem cell leukemia (Mahajan et al., 1996), which is a basic helix–loop–helix transcription factor involved in cell growth and differentiation. Overexpression of human DRG2 causes cell cycle arrest at the G2/M phase (Ko et al., 2004). Recently it was shown that DRG2 overexpression inhibits NF-κB function and interleukin-6 production in macrophages, as well as TH17 differentiation, and ameliorates experimental autoimmune encephalomyelitis in mice (Ko et al., 2014). However, the precise functional role of DRGs has not been discerned.
Transferrin (Tfn) and its receptor, TfnR, have been studied extensively to define mechanisms involved in receptor-mediated endocytosis (Sheff et al., 2002; Maxfield and McGraw, 2004). In this study, we unraveled an unexpected role of DRG2 in modulation of Rab5 activity and Tfn recycling. DRG2 was found to interact with Rab5 and EEA1 on endosomes in a phosphatidylinositol 3-kinase (PI3K)–dependent manner. DRG2 depletion impaired the inactivation and removal of Rab5 from Tfn-containing endosomes, causing a defect in Tfn recycling. Of interest, DRG2 depletion did not affect the recruitment of RabGAP5 and Rac1 to Rab5 endosomes but disrupted interaction between Rab5 and RabGAP5 on endosomes. These results reveal that DRG2 is associated with Rab5 endosomes and functions as a key regulator for Rab5 deactivation and Tfn recycling.
RESULTS
DRG2 colocalizes with Rab5, EEA1, and Rab11 on endosomes
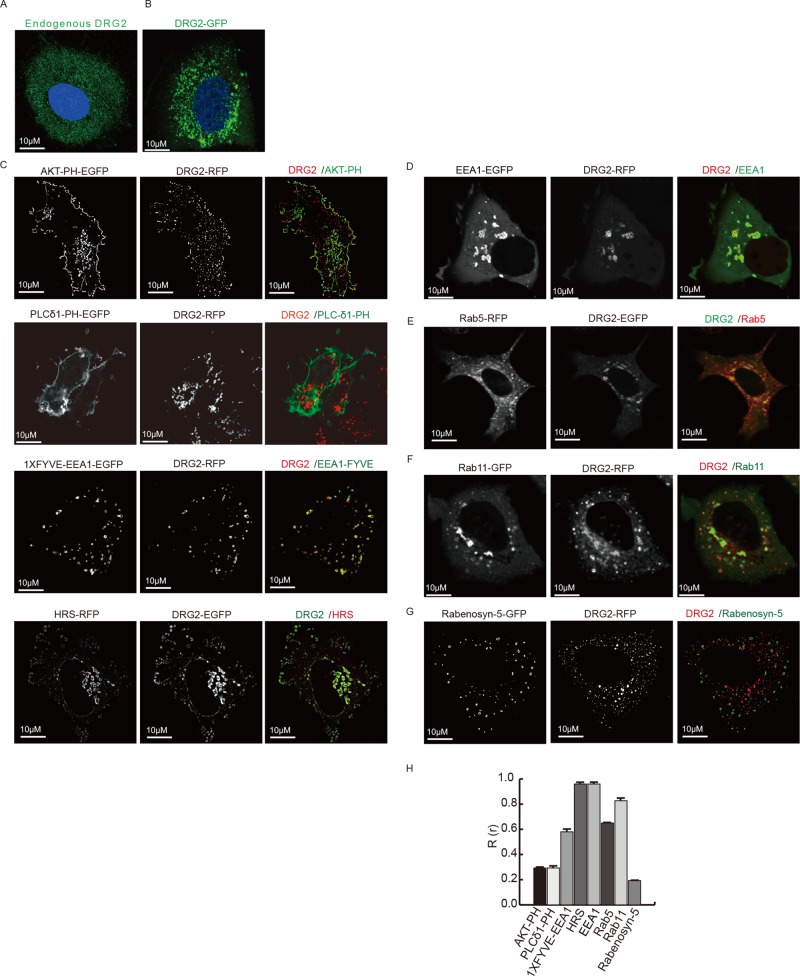
To obtain insights into the function of DRG2 in cells, we first investigated the localization of endogenous DRG2 in MCF7 cells using an anti-DRG2 antibody and immunofluorescence microscopy. DRG2 was mostly localized on peripheral, punctate, endosome-like structures (Figure 1A). Cells transfected with enhanced green fluorescent protein (EGFP)– or monomeric red fluorescent protein (mRFP)–tagged DRG2 displayed a similar distribution to that of DRG2 on the punctate structures (Figures 1B and Supplemental Figure S1A).
FIGURE 1:
DRG2 colocalizes with EEA1, Rab5, and Rab11 on endosomes. (A, B) DRG2 localizes on peripheral, punctate, endosome-like structures. (A) Endogenous DRG2 in MCF7 cells stained with anti-DRG2 antibody. (B) MCF7 cells expressing EGFP-DRG2. (C) DRG2 localizes on PI3P-containing endosomes. MCF7 cells expressing DRG2 and markers for PIs: Akt-PH for PI(3,4,5)P3/PI(3,4)P2; PLC-δ1-PH for PI(4,5)P2; and EEA1-FYVE and Hrs for PI3P. (D–G) DRG2 colocalizes with EEA1, Rab5, or Rab11 but not with Rabenosyn-5 on endosomes. MCF7 cells expressing (D) EGFP-EEA1 and mRFP-DRG2, (E) ECFP-Rab5 (red) and EGFP-DRG2, (F) EGFP-Rab11 and mRFP-DRG2, and (G) EGFP-Rabenosyn-5 and mRFP-DRG2. Representative confocal images with magnified insets of boxed areas. Blue, 4′,6-diamidino-2-phenylindole (DAPI) staining. (H) Cells from C–G were analyzed for Pearson’s colocalization coefficient (R(r)). Values are mean ± SD from three separate experiments, with 10 different cells per group per experiment. Scale bar, 10 μm.
To determine the localization of DRG2, we stained MCF7 cells by antibodies specific for endogenous DRG2 and markers for endosomes (Rab5), Golgi, or endoplasmic reticulum (ER). Whereas limited colocalization was observed between DRG2 and Golgi (Supplemental Figure S1B), DRG2 showed significant colocalization with ER (Supplemental Figure S1C) and Rab5 (Supplemental Figure S1D). It has been reported that different organelles contain distinct sets of phosphoinositides (PIs): the plasma membrane contains phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2); Golgi membranes, phosphatidylinositol 4-phosphate; and endosomal membrane, phosphatidylinositol 3-phosphate (PI3P; Krauss and Haucke, 2007). To confirm the localization of DRG2, we cotransfected MCF7 cells with DRG2 and markers for PIs, including Akt-PH for phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3)/PI(3,4)P2, phospholipase C-δ1 (PLC-δ1)–PH for PI(4,5)P2, and EEA1-FYVE and Hrs for PI3P. Whereas DRG2 colocalized with EEA1-FYVE and Hrs on endosomes, it did not with either Akt-PH or PLC-δ1 (Figure 1C). Our results indicate that DRG2 localizes to PI3P-containing endosomes. To determine the endosome subpopulations to which DRG2 localizes, we cotransfected MCF7 cells with DRG2 and endosomal proteins such as Rabenosyn-5, EEA1, Rab5 (early endosome marker), and Rab11 (recycling endosome marker; Stenmark, 2009). Whereas DRG2 showed significant colocalization with EEA1 (Figure 1D), Rab5 (Figure 1E), and Rab11 (Figure 1F) on endosomes, it did not colocalize with Rabenosyn-5 (Figure 1G). These results demonstrate that DRG2 colocalizes with EEA1, Rab5, and Rab11 on endosomes.
DRG2 interacts with Rab5 and EEA1 on endosomes in a PI3K-dependent manner
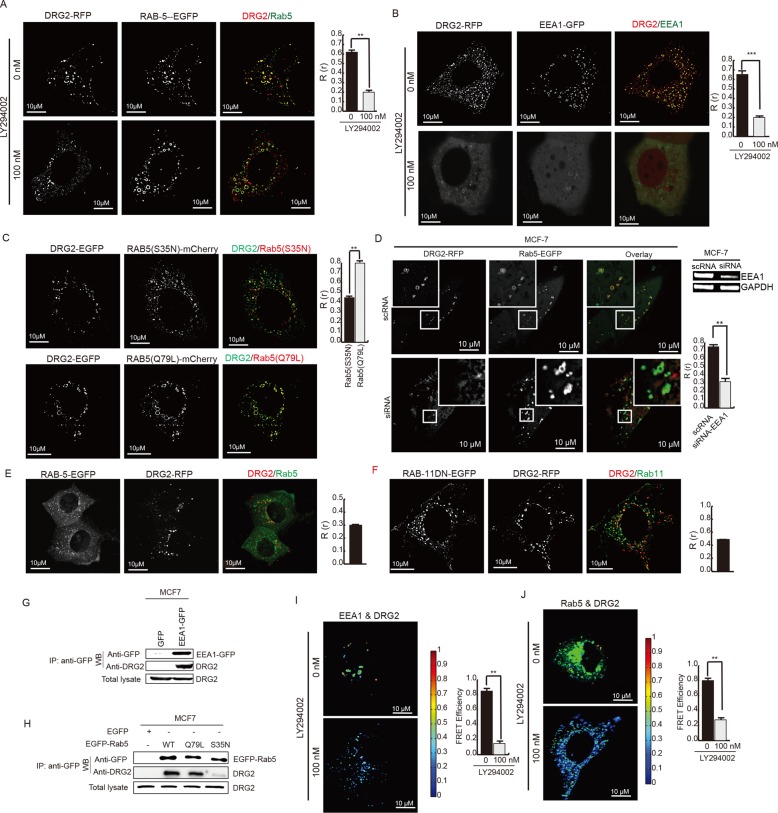
The localization of DRG2 to PI3P-containing endosomes suggested the possibility that PI3K affects the endosomal localization of DRG2. To test this, we cotransfected MCF7 cells with DRG2 and Rab5 or EEA1 and treated them with the PI3K inhibitor LY294002. As expected, inhibition of PI3K disrupted the colocalization of DRG2 with Rab5 (Figure 2A) and EEA1 (Figure 2B). The active, GTP-bound form of Rab5 induces PI3P synthesis on endosomes by interacting with PI3K (Christoforidis et al., 1999b) and mediates the recruitment of many Rab5 effectors to endosomes (Stenmark, 2009). We therefore investigated whether the activation status of Rab5 affects DRG2 recruitment to vesicular endosomes. We cotransfected MCF7 cells with DRG2 and dominant-negative (dn) Rab5(S35N) or a constitutively active Rab5(Q79L) mutant and assayed the subcellular distribution of DRG2 using confocal microscopy. DRG2 colocalized with Rab5(Q79L) but not with Rab5(S35N) mutants at vesicular endosomes (Figure 2C). These results suggest that localization of DRG2 to the Rab5-containing endosomes depends on the Rab5-PI3K pathway. EEA1 is an effector molecule of Rab5 (Christoforidis et al., 1999a). We tested whether EEA1 is involved in the recruitment of DRG2 to the Rab5-containing endosomes. We transfected MCF7 cells with small interfering RNA (siRNA) against EEA1 to inhibit the expression of EEA1. After transfection with DRG2-EGFP and Rab5-RFP, we analyzed MCF7 cells for the localization of DRG2 to the Rab5-containing endosomes. The inhibition of EEA1 blocked the localization of DRG2 to the Rab5-containg endosomes (Figure 2D), indicating that the localization of DRG2 to the Rab5-containing endosomes is dependent on the EEA1. PI3K on endosomes can be activated by growth factor signaling pathways (Tsutsumi et al., 2009). To determine whether PI3K activated by growth factors in serum affects the colocalization of DRG2 with Rab5 on endosomes, we incubated MCF7 cells cotransfected with DRG2 and wild-type Rab5 with serum-free medium. Serum deprivation inhibited the colocalization of DRG2 with Rab5 on endosomes (Figure 2E), indicating that signals from growth factors in serum may affect the localization of DRG2 to Rab5-containing endosomes. Expression of dn-Rab11(S25N) did not affect the localization of DRG2 to endosomal structures (Figure 2F). These results suggest that recruitment of DRG2 to endosomal structures is affected by the activity of Rab5 but not Rab11.
FIGURE 2:
DRG2 localizes on EEA1- and Rab5-containing endosomes in a PI3K-dependent manner and interacts with EEA1 and Rab5 on endosomes. (A, B) DRG2 colocalizes with Rab5 and EEA1 on endosomes in a PI3K-depenednt manner. MCF7 cells expressing (A) mRFP-DRG2 and EGFP-Rab5 or (B) mRFP-DRG2 and EGFP-EEA1 were incubated for 30 min in the absence or presence of 100 nM PI3K inhibitor LY294002. (C) Activation status of Rab5 affects localization of DRG2 on Rab5 endosomes. MCF7 cells expressing EGFP-DRG2 and the dn mutant Rab5(S35N) or constitutively active mutant Rab5(Q79L) fused with mCherry. (D) EEA1 inhibition blocks the localization of DRG2 on Rba5 endosomes. EEA1 was inhibited by transfecting MCF7 cells with siRNA against EEA1. MCF7 cells expressing mRFP-DRG2 and EGFP-Rab5. (E) Serum deprivation inhibits colocalization of DRG2 with Rab5 on endosomes. MCF7 cells expressing mRFP-DRG2 and EGFP-Rab5 were incubated with serum-free medium for 12 h. (F) Expression of dn mutant Rab11(S25N) does not affect localization of DRG2 to endosomes. MCF7 cells expressing mRFP-DRG2 and EGFP-Rab11(S25N). Representative confocal images with magnified insets of boxed areas. Blue, DAPI staining. Cells from A–E were analyzed for Pearson’s R(r). Values are mean ± SD from three separate experiments, with 10 different cells per group per experiment. (G, J) DRG2 interacts with EEA1 and Rab5 on endosomes in vivo. (G, H) MCF7 cells were transfected with (G) EGFP-EEA1 or (H) EGFP-Rab5(wild type), EGFP-Racb5(Q79L), or EGFP-Rab5(S35N). GFP-tagged Rab5 was immunoprecipitated from cell lysates and immunoblotted for endogenous DRG2. (I, J) MCF7 cells expressing DRG2-EGFP and (I) EEA1-mCherry or (J) Rab5-mCherry were incubated for 30 min in the absence or presence of 100 nM PI3K inhibitor LY294002 and analyzed for protein–protein interaction using FRET. The FRET efficiency percentage is depicted using discrete colors representing FRET values ranging from 0 to 10%. Values are mean ± SD from two separate experiments, with at least 10 different cells per group per experiment. **p < 0.01; ***p < 0.001. Scale bar, 10 μm.
The colocalization of DRG2 with Rab5 and EEA1 on endosomes suggests the possibility that DRG2 interacts with Rab5 and/or EEA1 on endosomes. To test this, we transfected MCF7 cells with empty pEGFP, pEGFP-EEA1, pEGFP-Rac1, pEGFP-Rab5(Q79L), or pEGFP-Rab5(S35N) and investigated the interaction between endogenous DRG2 and EEA1 or Rab5 by immunoprecipitation. As shown in Figure 2, F and G, DRG2 interacted with EEA1 and Rab5. of interest, DRG2 interacted with Rab5(Q79L) more strongly than with Rab5(S35N), indicating that DRG2 interacts with Rab5 in a Rab5 activity–dependent manner. To confirm the interaction between DRG2 and EEA1 or Rab5 on endosomes in situ, we cotransfected MCF7 cells with DRG2-mRFP and Rab5-EGFP or EEA1-EGFP. After incubation with Tfn for 20 min, cells were analyzed by fluorescence resonance energy transfer (FRET) microscopy to measure the interaction between DRG2 and Rab5 or EEA1. The FRET efficiency between DRG2-mRFP and EEA1-EGFP on endosomes was 9% (Figure 2H) and that of DRG2-mRFP and Rab5-EGFP on endosomes was 10% (Figure 2I), indicating that DRG2 interacts with Rab5 and EEA1 on endosomes. Consistent with colocalization results, treatment with the PI3K inhibitor LY294002 dramatically decreased the FRET efficiency (Figure 2, H and I), suggesting that their interactions on endosomes depend on PI3K activity.
DRG2 is required for Tfn recycling
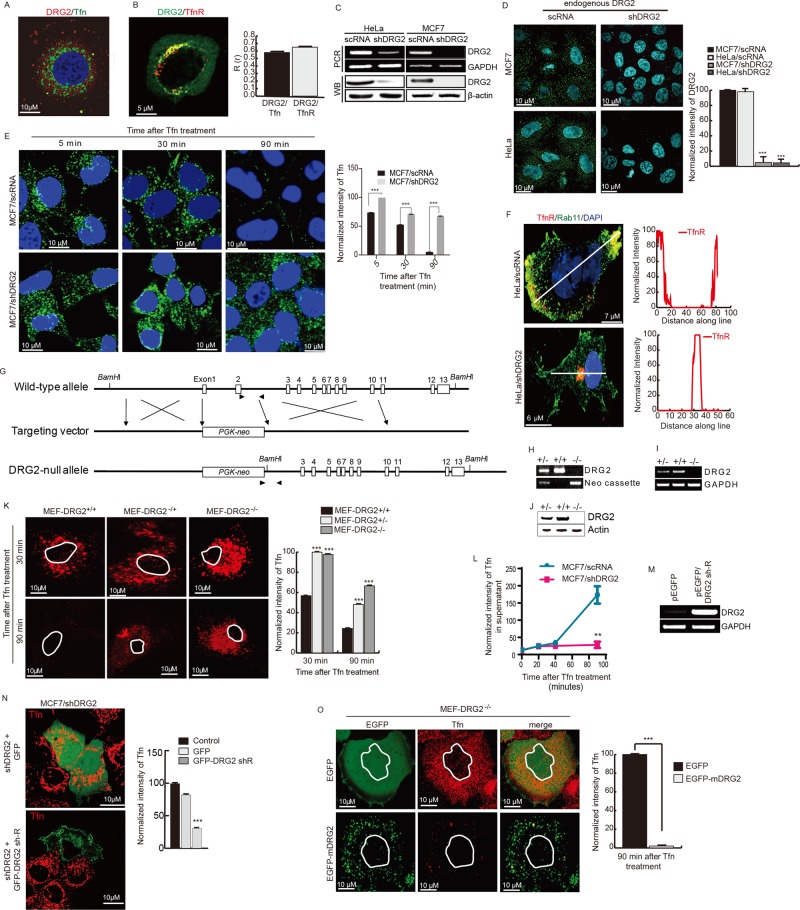
To address whether DRG2 on endosomes affects endocytic trafficking of cargoes, we used Tfn as a cargo. We found that endogenous DRG2 colocalized with Tfn and its receptor, TfnR (Figure 3, A and B, and Supplemental Figure S2, A and B). To determine the role of DRG2 in Tfn recycling, we depleted HeLa and MCF7 cells of DRG2 using short hairpin RNA (shRNA), incubated them with Alexa 633–labeled Tfn for 5, 30, and 90 min, and then investigated uptake and recycling of Tfn. Nontarget shRNA control vector–transfected cells were used as a control. The efficiency of shRNA depletion was assessed by reverse transcription PCR (RT-PCR) and Western blot analysis (Figure 3C). Down-regulation of endogenous DRG2 by shRNA was also confirmed by staining with anti-DRG2 antibody and confocal microscopy (Figure 3D and Supplemental Figure S2C). Both control and DRG2-depleted cells showed similar levels of intracellular fluorescence intensity at 5 min (Figure 3E). In control cells, internalized Tfn accumulated in the perinuclear compartment at 30 min, and then levels of intracellular Tfn decreased markedly at the 90-min time point (Figure 3E). However, in DRG2-depleted cells, internalized Tfn localized to the perinuclear compartment at 30 min, and most of the cargo remained there at 90 min (Figure 3E). DRG2 depletion also disrupted recycling of TfnR to the plasma membrane (Figure 3F and Supplemental Figure S2D). These results suggest that DRG2 deficiency does not affect the internalization of Tfn and trafficking to the perinuclear compartment but inhibits recycling of Tfn to the plasma membrane.
FIGURE 3:
DRG2 is required for Tfn recycling. (A, B) DRG2 colocalizes with Tfn and TfnR. (A) Endogenous DRG2 and Tfn–Alexa 647 (at 10 min after treatment) in MCF7 cells. (B) EGFP-DRG2 and mCherry-TfnR in HeLa cells after 10 min of Tfn treatment. Representative confocal images with magnified insets of boxed areas. Blue, DAPI staining. Graphs represent Pearson’s R(r) between DRG2 and Tfn or TfnR. Values are mean ± SD from three experiments, with 10 different cells per group per experiment. (C, D) Preparation of DRG2-depleted MCF7 and HeLa cells by using control (nontarget shRNA) or shRNA against DRG2 (shDRG2) using the pLKO vector. Depletion of DRG2 expression was confirmed by RT-PCR (C, top), Western blot (C, bottom), and fluorescence microscopy (D). (E) DRG2 depletion blocks Tfn recycling. Control and DRG2-depleted MCF7 cells were incubated with Tfn–Alexa 633 for the indicated times. Blue, DAPI staining. Graph represents the mean fluorescence intensities in arbitrary units. Values are mean ± SD from three separate experiments, with 10 different cells per group per experiment. ***p < 0.001. (F) DRG2 depletion blocks TfnR recycling. Confocal images of control and DRG2-depleted HeLa cells expressing TfnR-PAmKate and EGFP-Rab11 and incubated with Tfn for 60 min. Graphs represent linear pixel values across cells. Blue, DAPI staining. (G–K) DRG2 knockout blocks Tfn recycling. (G–J) Generation of DRG2 KO mice. (G) Schematic representation of structure of the DRG2 genomic locus and the targeting construct. Exons 1 and 2 of wild-type DRG2 were replaced by the PGK-neo cassette through homologous recombination, indicated by cross symbols. Positions of exons covering the entire coding region are indicated by open boxes. Paired arrowheads indicate the primer sets used for genotype determination. (H–J) Confirmation of DRG2 KO. (H) Genotyping. Genomic DNA from wild-type (+/+), heterozygous (+/–), and homozygous (–/–) mice was analyzed by RT-PCR using the primer sets indicated in G. Expression levels of DRG2 were analyzed by (I) RT-PCR analysis and (J) Western blot analysis. (K) MEF cells derived from wild-type (+/+), heterozygous (+/–), and homozygous (–/–) mice were incubated with Alexa 594–conjugated Tfn for indicated times. Representative confocal images. Graph represents the mean fluorescence intensities in arbitrary units. Values are mean ± SD from three separate experiments, with five different cells per group per experiment. ***p < 0.001. (L) DRG2 depletion blocks the release of internalized Tfn from MCF7 cells. After washing out of unbound Tfn–Alexa 594, Tfn released into the supernatant was analyzed at the indicated time points. n = 4. **p < 0.01. (M, N) Expression of shRNA-resistant DRG2 (DRG2 sh-R) rescues Tfn recycling. DRG2-depleted MCF7 cells were transfected with EGFP or EGFP-DRG2 sh-R. (M) Expression of DRG2 was confirmed by RT-PCR. (N) After incubation with Tfn–Alexa 594 for 90 min, images were obtained using confocal microscope. Graph shows the mean fluorescence intensities in arbitrary units. Values are mean ± SD from three experiments, with five different cells per group per experiment. ***p < 0.001. Scale bar, 10 μm. (O) MEF-DRG2−/− cells were transfected with the EGFP or EGFP-DRG2 and incubated with Cy3-conjugated Tfn for 30 and 90 min. Representative confocal images. Graph represents the mean fluorescence intensities in arbitrary units. Values are mean ± SD from three separate experiments, with five different cells per group per experiment. ***p < 0.001. Scale bar, 10 μm.
To confirm the role of DRG2 in Tfn recycling in mouse cells, we prepared mouse embryonic fibroblast (MEF) cells from wild-type, heterozygous, and homozygous DRG2-null mice (Figure 3, G– J). We treated MEF cells with Cy-3–labeled mouse Tfn for 30 and 90 min. Consistent with our observation in MCF7 cells, Tfn was not detected in MEF cells derived from wild-type mice after a 90-min incubation, whereas Tfn was observed in MEF cells derived from DRG2 KO mice at 90 min (Figure 3K), indicating that DRG2 knockout also inhibits Tfn recycling in MEF cells.
We further assessed the effect of DRG2 depletion on Tfn recycling by analyzing the release of internalized Tfn. We incubated cells with Alexa 647–labeled Tfn on ice for 30 min. After washing out unbound and surface Tfn using mild-acid buffer, we collected and analyzed the culture supernatant for fluorescence at the indicated times. Whereas a large amount of Tfn was released from control MCF7 cells at 90 min, only a small amount of Tfn was released from DRG2-depleted MCF7 cells at 90 min (Figure 3L), suggesting that recycling of Tfn is inhibited by DRG2 depletion.
To further confirm the function of DRG2 in endosomal recycling of Tfn, we monitored the rescue of this phenotype by transfecting DRG2-depleted cells with EGFP-tagged, shRNA-resistant wild-type DRG2. Expression of EGFP alone did not rescue the recycling of Tfn in DRG2-depleted cells. However, expression of shRNA-resistant DRG2 in DRG2-depleted human MCF7 and mouse MEF cells significantly decreased Tfn intensity, indicating restoration of Tfn recycling (Figure 3, M, N, and O, and Supplemental Figure S2E). These data collectively indicate that DRG2 plays an important role in the regulation of Tfn recycling.
DRG2 depletion inhibits the dissociation of Rab5 from Tfn-containing endosomes
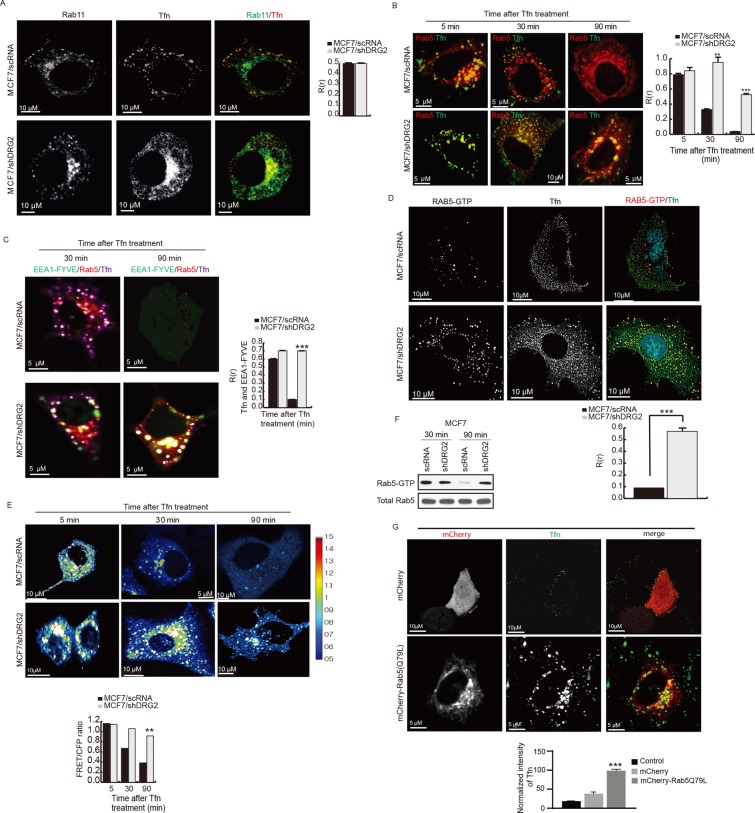
Rab11 is known to associate with recycling endosomes and be required for Tfn recycling (Takahashi et al., 2012). Thus we asked whether DRG2 depletion inhibits the recruitment of Rab11 to Tfn-containing endosomes. In both control and DRG2-depleted MCF7 cells, Rab11 and internalized Tfn colocalized to vesicular structures (Figure 4A), indicating that DRG2 depletion does not affect the recruitment of Rab11 to Tfn-containing endosomes.
FIGURE 4:
DRG2 depletion inhibits the inactivation of Rab5 on endosomes. (A) DRG2 depletion does not affect the recruitment of Rab11 to Tfn-containing endosome. Representative confocal images of control and DRG2-depleted MCF7 cells expressing EGFP-Rab11 and incubated with Tfn-Alexa 647 for 30 min. Blue, DAPI staining. Graph represents Pearson’s R(r) between Rab11 and Tfn. (B) DRG2 depletion leads to defects in dissociation of Rab5 from Tfn-containing endosomes. Confocal images of control and DRG2-depleted MCF7 cells expressing mRFP-Rab5 and incubated with Tfn-Alexa 647 for the indicated times. Graph represents Pearson’s R(r) between Rab5 and Tfn. (C–E) DRG2-depleted cells show sustained Rab5 activity on Tfn-containing endosomes. (C) Sustained colocalization of Rab5 and FYVE-EEA1 on Tfn-containing endosomes in DRG2-depleted cells. Confocal images of control and DRG2-depleted MCF7 cells expressing mRFP-Rab5 and EGFP-FYVE-EEA1 and incubated with Tfn–Alexa 647 for 30 and 90 min. Graph represents Pearson’s R(r). (D) Detection of active GTP-bound Rab5 on Tfn-containing endosomes in DRG2-depleted MCF7 cells. Control and DRG2-depleted MCF7 cells were incubated with Tfn–Alexa 647 for 30 min. Cells were stained with anti–Rab5-GTP antibody. Graph represents Pearson’s R(r). (E) FRET analysis for detection of GTP-bound Rab5 in living cells. FRET images were obtained from control and DRG2-depleted MCF7 cells expressing Raichu-Rab5 and incubated with Tfn for the indicated times. FRET is presented as pseudocolor images. Graph shows FRET/CFP ratio. (F) GTP-loaded Rab5 pull-down assay. GTP-loaded Rab5 was pulled down from lysates of control and DRG2-depleted MCF7 cells expressing mRFP-Rab5 and incubated with Tfn for 30 min. (G) Expression of Rab5 (Q79L) mimics the phenotypes of DRG2-depleted cells. Confocal images of MCF7 cells expressing mCherry or mCherry-Rab5 (Q79L) and incubated with Tfn–Alexa 647 for 30 min. Graph represents the mean fluorescence intensities in arbitrary units. All values are mean ± SD from three experiments, with 10 different cells per group per experiment. **p < 0.01; ***p < 0.001. Scale bar, 10 μm.
Deactivation of Rab5 and subsequent dissociation of Rab5 from the endosome is necessary for proper recycling of cargoes (Hutagalung and Novick, 2011). We next investigated whether in DRG2-depleted cells, Rab5 fails to dissociate from Tfn-containing endosomes. To test this, we treated control and DRG2-depleted MCF7 cells overexpressing Rab5-mRFP with Tfn–Alexa 647 and analyzed them for colocalization of Rab5 and Tfn. In both control and DRG2-depleted MCF7 cells, a significant fraction of Tfn internalized for 5 min colocalized with Rab5 (Figure 4B and Supplemental Figure S3A), suggesting that DRG2 does not affect the initial recruitment of Rab5 to Tfn-containing early endosomes. However, at 30 min, whereas <40% of Tfn in control cells colocalized with Rab5, most of the Tfn in DRG2-depleted cells still colocalized with Rab5 (Figure 4B and Supplemental Figure S3A). In addition, >50% of Tfn in DRG2-depleted cells colocalized with Rab5 in perinuclear compartment, even after 90 min (Figure 4B and Supplemental Figure S3A). Thus our results suggest that DRG2 is required for dissociation of Rab5 from Tfn-containing endosomes.
It has been reported that inhibition of recycling of Tfn leads to lysosomal degradation of TfnR (Navaroli et al., 2012). To determine whether blocking of Tfn recycling from recycling endosome to the plasma membrane directs Tfn to lysosomes, we transfected control and DRG2-depleted MCF7 cells with mGFP-LAMP1. After Tfn–Alexa 647 treatment for 30 min, cells were analyzed for colocalization of Tfn and LAMP1. Of interest, in DRG2-depleted cells, Tfn colocalized with LAMP1 (Supplemental Figure S4A). However, as shown in Figure 3E, the Tfn level did not decrease. Thus, although the compartments that accumulate Tfn upon DRG2 depletion resemble late endosomes morphologically, they represent abnormal early endosomes that are unable to degrade Tfn. Our data are consistent with the accumulation of Tfn-positive, multivesicular endosome–like organelles upon inhibition of early endosomal recycling (Bright et al., 2001).
Inactivation of Rab5 is required for the Rab5/Rab7 transition (Rink et al., 2005; Chotard et al., 2010). Defects in Rab5 deactivation lead to failure in progression from early to late endosomes (Rink et al., 2005). Thus we predict that failure in Rab5 deactivation induced by DRG2 depletion may prevent the degradation of epidermal growth factor receptor (EGFR), a cargo destined for degradation. First, we transfected cells with EGFR and treated them with Tfn and EGF and determined whether DRG2 depletion affects the sorting of these cargoes into different endosome subpopulations. As shown in Supplemental Figure S4B, we could not observe any defects in the separation of these cargoes into different endosomes in DRG2-depleted cells. We next determined the fate of EGFR in control and DRG2-depleted cells. Consistent with our hypothesis, we found that, whereas the intensity of EGFR decreased dramatically in control MCF7 cells, it did not decrease in DRG2-depleted MCF7 cells (Supplemental Figure S4, C and D).
DRG2 depletion inhibits Rab5 deactivation on endosomes
Deactivation of the early-acting GTPase RAB5 is required to allow proper functioning of recycling endosomes associated with later-acting GTPases (Hutagalung et al., 2011). Defects in Rab5 deactivation or inhibition of Rab5 GAP by siRNA interfere with cargo recycling (Haas et al., 2005; Sun et al., 2012). Thus we speculated that inhibition of Tfn recycling in DRG2-depleted cells could result from a defect in Rab5 deactivation. To address this hypothesis, we investigated whether DRG2 deficiency prevents the deactivation of Rab5 on endosomes. We applied several criteria to assess the state of Rab5 activation in endosomes. First, we determined the PI3P level of Tfn-containing endosomes. Rab5-GTP recruits PI3K to endosomes (Christoforidis et al., 1999b). Thus the level of the lipid product of PI3K, PI3P, on endosomes can be used to monitor the state of activation of Rab5. Here we used the FYVE domain of EEA1, which can bind to PI3P (Patki et al., 1998) to detect endosomal Rab5-GTP levels. In both control and DRG2-depleted MCF7 cells, most Tfn colocalized with EEA1-FYVE at 30 min after Tfn treatment (Figure 4C and Supplemental Figure S3B). However, the size of PI3P-containing endosomes in DRG2-depleted MCF7 cells was greater than that in control MCF7 cells (Figure 4C and Supplemental Figure S3B). In addition, at 90 min after Tfn treatment, whereas most of the Tfn was still colocalized with EEA1-FYVE in DRG2-depleted cells, it was hard to detect EEA1-FYVE–positive endosomes in control cells (Figure 4C and Supplemental Figure S3B). This suggests that Rab5 activity in DRG2-depleted cells is higher than that in the control cells. Second, we directly stained Rab5-GTP using anti–Rab5-GTP antibody and determined Rab5 activity on the Tfn-containing endosomes at 30 min after Tfn treatment. As shown in Figure 4D, whereas the Rab5-GTP level was extremely low in control cells, it was high in DRG2-depleted cells. Third, we used ratiometric FRET microscopy to visualize active, GTP-loaded Rab5 in living cells. Here we used Raichu-Rab5 (which consists of yellow fluorescent protein [YFP], the Rab5-binding domain of EEA1, cyan fluorescent protein [CFP], and Rab5a; Kitano et al., 2008) as an intramolecular biosensor. Active, GTP-bound Rab5 would increase in FRET between CFP and YFP. In control cells, very distinct FRET signals were detected in the endosomal compartment at an early time point (5 min) after Tfn treatment and subsequently decreased after 30 min. In contrast, in DRG2-depleted cells, FRET signals detected at an early time point (5 min) were maintained in endosomal structures until 90 min after Tfn treatment (Figure 4E), indicating that DRG2 depletion leads to defects in Rab5 deactivation in endosomes. Fourth, we assessed whether DRG2 depletion influences Rab5 GTP loading, using a pull-down approach with an antibody that selectively binds GTP-loaded Rab5. Of importance, whereas the level of GTP-bound Rab5 in control cells decreased significantly at 90 min after Tfn treatment, the level of GTP-bound Rab5 in DRG2-depleted cells was maintained at 90 min (Figure 4F). Collectively these results indicate that DRG2 deficiency prevents the deactivation of Rab5.
Constitutively active Rab5 Q79L mimics the DRG2 knockdown phenotype
Defects in Rab5 deactivation interfere with cargo recycling (Haas et al., 2005; Sun et al., 2012), and, thus, expression of constitutively active Rab5(Q79L) should have the same effect on Tfn trafficking as DRG2 depletion. To test this, we analyzed MCF7 cells transfected with mCherry-Rab5 CA(Q79L) for Tfn trafficking. Similar to DRG2 depletion, endocytosed Tfn accumulated in the perinuclear compartment and colocalized with Rab5 CA(Q79L) after a 90-min treatment (Figure 4G). These data clearly suggest that DRG2 depletion leads to defects in Rab5 deactivation and thus inhibits recycling of Tfn.
DRG2 deficiency does not interfere with the recruitment of Rab5 GAP to Rab5-containing endosomes
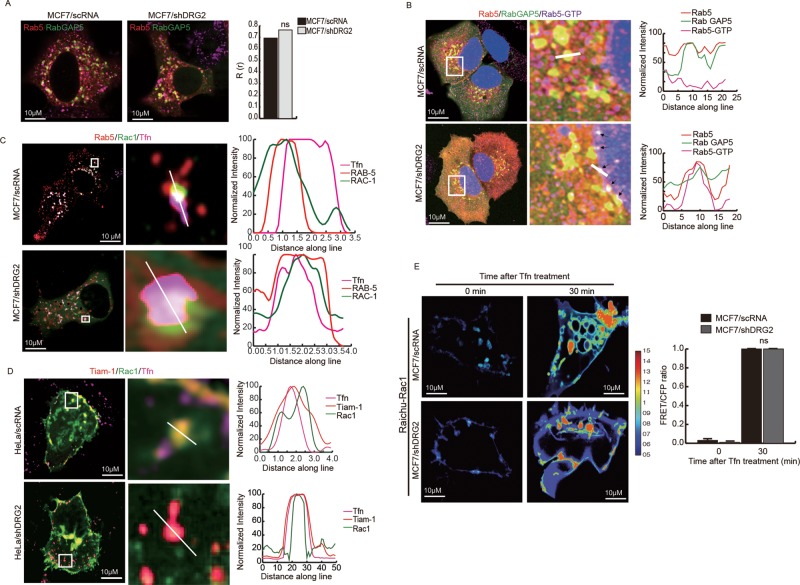
Two GAPs, RabGAP5 (Haas et al., 2005) and RN-Tre (Lanzetti et al., 2000, 2004), have been reported to have GAP activity toward Rab5. Thus we investigated whether DRG2 deficiency leads to defects in the recruitment of these two GAPs to Rab5-containing endosomes. To determine the effect of DRG2 deficiency on the recruitment of RabGAP5 to Tfn-containing endosomes, we cotransfected control and DRG2-depleted MCF7 cells with mRFP-Rab5a and EGFP-RabGAP5 or EGFP-RN-Tre and incubated them with Alexa 647–labeled Tfn for 30 min. Unexpectedly, most of the RN-Tre localized on the plasma membrane and did not colocalize with Rab5 and Tfn in both control and DRG2-depleted cells (Supplemental Figure S5). These results indicate that RN-Tre does not act as a GAP against Rab5 on endosomes in these cells. However, RabGAP5 showed significant colocalization with Rab5 on endosomes in both control and DRG2-depleted cells (Figure 5A and Supplemental Figure S6A). These results suggest that RabGAP5 may act as a GAP against Rab5, and DRG2 depletion does not affect RabGAP5 recruitment to recycling endosomes.
FIGURE 5:
DRG2 depletion does not affect the recruitment of Rab5GAP, Rac1, and Tiam-1 to endosomes. (A) DRG2 depletion does not affect the recruitment of RabGAP5 to Rab5-containing endosomes. Control and DRG2-depleted MCF7 cells cotransfected with mRFP-Rab5 and EGFP-Rab GAP5 were incubated with Tfn for 30 min. Representative confocal images. Graph shows Pearson’s R(r) between Rab5 and Rab GAP5. Values are mean ± SD from three experiments, with five cells per group per experiment. ns, not significant. (B) Detection of active GTP-bound Rab5 on RabGAP5-containing endosomes in DRG2-depleted cells. Control and DRG2-depleted MCF7 cells were cotransfected with mRFP-Rab5 and EGFP-RabGAP5. After incubation with Tfn for 30 min, cells were stained with anti-Rab5 GTP antibody, and images were obtained using a confocal microscope. Blue, DAPI staining. Representative images with magnified insets of boxed areas. Graphs show linear pixel values across endosomes. (C, D) DRG2 deficiency does not affect recruitment of Rac1 and Tiam-1 to endosomes. Control and DRG2-depleted MCF7 cells were cotransfected with (C) EGFP-Rac1 and mRFP-Rab5 or (D) mCherryR-TIAM-1 and EGFP-Rac1 and incubated with Alexa 647–conjugated Tfn for 30 min. Representative confocal images. Graphs show linear pixel values across endosomes. (E) FRET analysis for detection of GTP-bound Rac1 in living cells. FRET images were obtained from control and DRG2-depleted MCF7 cells expressing Raichu-Rac1 and incubated with Tfn for the indicated times. FRET is presented as pseudocolor images. Graph shows FRET/CFP ratio. Values are mean ± SD from three experiments, with 10 different cells per group per experiment. ns, not significant. Scale bar, 10 μm.
We next tested whether colocalization of RabGAP5 with Rab5 on endosomes is sufficient to inactivate Rab5. We stained HeLa cells coexpressing EGFP-RabGAP5 and mRFP-Rab5 with anti–Rab5-GTP antibody. Most Rab5 on the endosome containing RabGAP5 was not stained with anti-Rab5-GTP antibody in control cells (yellow spots in Figure 5B and Supplemental Figure S6B). However, in DRG2-depleted cells, most Rab5 was stained with anti–Rab5-GTP (white spots in Figure 5B and Supplemental Figure S6B). These data indicate that the colocalization of Rab5 and RabGAP5 is not sufficient for inactivation of Rab5 and that DRG2 is required for inactivation of Rab5 on endosomes.
DRG2 deficiency does not affect the recruitment of Rac1 and Tiam-1 to endosomes
Rac1 is recruited to recycling endosomes, where it mediates deactivation of Rab5 (Sun et al., 2012). To determine whether DRG2 deficiency affects the recruitment of Rac1 to Rab5-containing endosomes, we cotransfected control and DRG2-depleted MCF7 cells with EGFP-Rac1 and mRFP-Rab5a and incubated them with Alexa 647–labeled Tfn for the indicated times. DRG2 deficiency did not disrupt the recruitment of Rac1 to Rab5-containing endosomes (Figure 5C and Supplemental Figure S6C). Our result indicates that DRG2 is not involved in the recruitment of Rac1 to Rab5-containing endosomes.
Rac1 GEF Tiam-1 activates Rac1 in endosomes (Palamidessi et al., 2008). To determine whether DRG2 depletion affects the recruitment of Tiam-1 to Rac1-containing endosomes, we cotransfected control and DRG2-depleted MCF7 cells with Tiam-1 and Rac1 and incubated them with Alexa 594–labeled Tfn for the indicated time. There was no significant difference in recruitment of Tiam-1 to Rac1-containing endosomes between control and DRG2-depleted MCF7 cells (Figure 5D and Supplemental Figure S6D), indicating that DRG2 depletion does not affect the recruitment of TIAM to Rac1-containing vesicles.
Next we determined Rac1 activity on endosomes by FRET assay. Control and DRG2-depleted MCF7 cells were transfected with the Raichu-Rac1 construct, and the FRET signal was examined at 10 min after Tfn treatment using FRET microscopy. Both control and DRG2-depleted cells showed a strong FRET signal on endosomes (Figure 5E). Collectively these results suggest that DRG2 depletion does not affect Rac1 activity on tubular structures.
DRG2 depletion impairs the interaction between Rab5 and RabGAP5 on endosomes
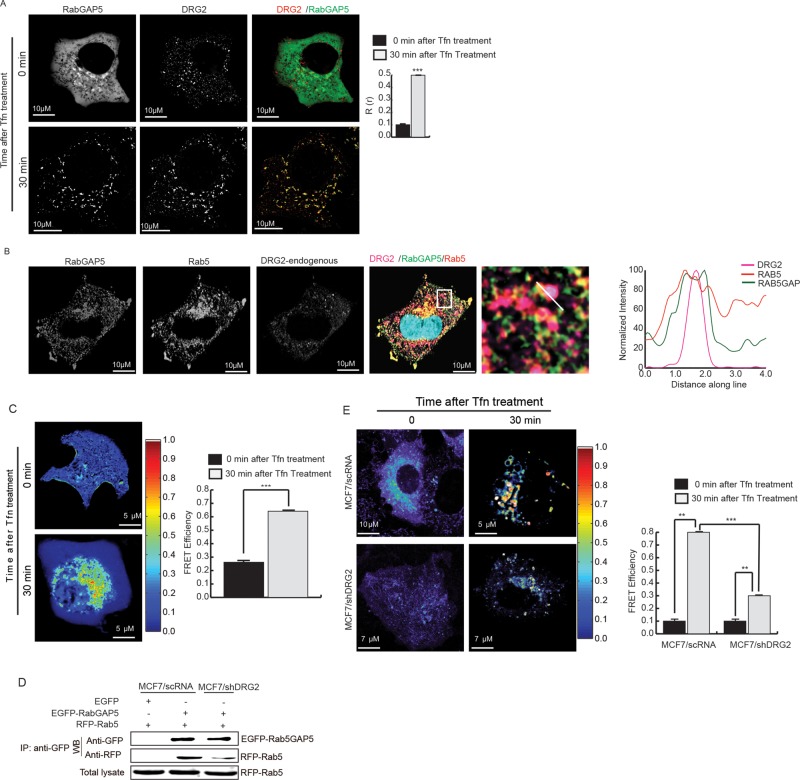
How does DRG2 deactivate Rab5 on endosomes? It is unlikely that DRG2 on endosomes acts as GAP for Rab5, because DRG2 did not show any GAP activity against Rab5 in an in vitro assay (Supplemental Figure S7). RabGAP5 interacts with Rab5 and inactivates it (Haas et al., 2005). Thus we determined whether DRG2 affect the interaction between Rab5 and RabGAP5 on endosomes. We first tested whether DRG2 colocalizes with RabGAP5 on endosomes. We cotransfected MCF7 cells with mRFP-DRG2 and EGFP-RabGAP5 and examined colocalization before and after Tfn treatment. DRG2 did not colocalize with RabGAP5 in the absence of Tfn stimulation. However, at 30 min after Tfn treatment, DRG2 colocalized with RabGAP5 on endosomes (Figure 6A). When we observed MCF7 cells expressing DRG2, Rab5, and RabGAP5, we found that these three proteins colocalized on endosomes (Figure 6B). Next we determined interaction between DRG2 and RabGAP5 on endosomes in MCF7 cells expressing mRFP-DRG2 and EGFP-RabGAP5 by FRET assay. FRET assay showed that DRG2 did not interact with RabGAP5 in the absence of Tfn stimulation but interacted with it on endosomes at 30 min after Tfn treatment (Figure 6C). Colocalization of DRG2 with Rab5 and RabGAP5 (Figure 6B) and interactions of DRG2 with Rab5 (Figure 2F) and RabGAP5 (Figure 6C) on endosomes suggest the possibility that DRG2 may mediate interaction between Rab5 and RabGAP5. To determine the role of DRG2 in the interaction between Rab5 and RabGAP5, we cotransfected control and DRG2-depleted MCF7 cells with EGFP-RabGAP5 and mRFP-Rab5 and examined their interaction before and after Tfn treatment by immunoprecipitation and FRET assay. In immunoprecipitation analysis, Rab5 interacted with RabGAP5 more strongly in control cells than in DRG2-depleted cells (Figure 6D). Before Tfn treatment, both control and DRG2-depleted cells showed FRET efficiencies of <1% (Figure 6E). However, at 30 min after Tfn treatment, whereas high FRET efficiency (8%) was detected from endosomes in control cells (Figure 6E, top), FRET efficiency on endosomes was low (2.5%) in DRG2-depleted cells compared with control cells (Figure 6E, bottom). These results suggest that DRG2 interacts with RabGAP5 on endosomes and is required for the interaction between Rab5 and RabGAP5 on endosomes.
FIGURE 6:
DRG2 depletion impairs the interaction between Rab5 and RabGAP5 on endosomes. (A) DRG2 colocalizes with RabGAP5 on endosomes. MCF7 cells were cotransfected with mRFP-DRG2 and EGFP-RabGAP5 and incubated with Tfn for the indicated times. Representative confocal images with magnified insets of boxed area. Graph represents Pearson’s R(r). (B) Endogenous DRG2 colocalizes with Rab5 and RabGAP5 on endosomes. MCF7 cells expressing mRFP-Rab5 and EGFP-RabGAP5 were incubated with Tfn for 30 min and stained with antiDRG2 antibody. Images were obtained using a confocal microscope. Blue, DAPI staining. Representative images with a magnified inset of boxed area. Graphs show linear pixel values across endosomes. (C) DRG2 interacts with RabGAP5 on endosomes. MCF7 cells expressing mRFP-DRG2 and ERFP-RabGAP5 were incubated with Tfn for indicated times and analyzed for protein–protein interaction using FRET. The FRET efficiency percentage is depicted using discrete colors representing FRET values ranging from 0 to 1. (D, E) DRG2 depletion inhibits interaction between Rab5 and RabGAP5 on endosomes. Control and DRG2-depleted MCF7 cells were cotransfected with mRFP-Rab5 and EGFP-RabGAP5 and analyzed for protein–protein interaction using (D) immunoprecipitation and (E) FRET analysis. The FRET efficiency percentage is depicted using discrete colors representing FRET values ranging from 0 to 1. All values are mean ± SD from three experiments, with 10 different cells per group per experiment. **p < 0.01; ***p < 0.001. Scale bar, 10 μm.
DISCUSSION
Rab5 regulates fusion and motility of early endosomes. Inactivation and removal of Rab5 from early endosome is required for recycling of cargoes (Haas et al., 2005; Sun et al., 2012). However, only a few molecules that regulate Rab5 activity during cargo recycling pathway have been identified, and thus the underlying mechanisms are not well understood. Here we identified DRG2 as a key regulator of Rab5 inactivation and Tfn recycling. DRG2 was found to colocalize with Rab5 and EEA1 on endosomes. DRG2 did not have GAP activity toward Rab5. However, in DRG2-depleted cells, Rab5 was not inactivated and failed to dissociate from Tfn-containing endosomes. DRG2 depletion did not affect Tfn uptake but perturbed Tfn transport from endosomes to the plasma membrane. Of importance, ectopic expression of DRG2 rescued Rab5 deactivation and Tfn recycling in DRG2-depleted cells. This is the first study to show that DRG2 is required for inactivation of Rab5 and Tfn recycling.
DRG2’s localization to Rab5 endosomes raised the question of how DRG2 is associated with those endosomes. We showed that DRG2 localized PI3P-containing endosomes, and PI3K inhibitors blocked localization of DRG2 to Rab5 endosomes. PI3K is recruited to endosomes by Rab5-GTP and induces localized synthesis of PI3P (Christoforidis et al., 1999b). Our results indicate that localization of DRG2 to Rab5 endosomes depends on PI3K and PI3P. Numerous protein domains have been identified that promote association with PI head groups (Kutateladze, 2010). In particular, PI3P can be recognized by FYVE, PH, and PX domains (Lindmo and Stenmark, 2006; Kutateladze, 2010). However, DRG2 does not contain any of these domains, indicating that DRG2 associates with endosomes by interacting with proteins on Rab5 endosomes in a PI3K-dependent manner. More than 20 proteins have been reported to interact with Rab5 and be involved in regulation of endosomal fusion and trafficking (Christoforidis et al., 1999a). Consistently, we found that DRG2 interacted with Rab5, indicating that interaction between DRG2 and Rab5 mediates the localization of DRG2 to Rab5 endosomes. Rab5 recruits PI3Ks in a GTP-dependent manner, resulting in localized synthesis of PI3P (Christoforidis et al., 1999b). In turn, EEA1 (Simonsen et al., 1998) and Rabenosyn-5 (Nielsen et al., 2000), which are Rab5 effectors, bind PI3P through the interaction of a FYVE domain in a PI3K-dependent manner. In this study, we also found that, whereas DRG2 did not colocalize with Rabenosyn-5 on endosomes, it colocalized and interacted with EEA1 on endosomes. In addition, colocalization and interaction of DRG2 with EEA1 on endosomes were disrupted by PI3K inhibitor. Our results suggest the possibility that DRG2 localizes to Rab5 endosomes through interaction with EEA1 as well as with Rab5. Considering that PI3P on endosomes is synthesized by PI3K, which is recruited by Rab5-GTP (Christoforidis et al., 1999b), endosomal localization of DRG2 should depend on the activation status of Rab5. Consistently, DRG2’s localization to the Rab5 endosomes was affected by GTP- or GDP-bound Rab5. In addition, it was affected by serum deprivation, indicating that PI3K activated by signal pathways as well as Rab5 may be involved in the recruitment of DRG2 to the endosomes. Further work is necessary to elucidate the mechanisms for the endosomal localization of DRG2.
Recruitment of GAP for Rab5 has been reported to facilitate the inactivation of Rab5 in Rab5-to-Rab7 conversion (Rink et al., 2005; Chotard et al., 2010). Failure to inactivate Rab5 in DRG2-depleted cells raised the question of whether DRG2 is required for the recruitment of Rab5 GAPs to Rab5-containing endosomes. RabGAP5 (Haas et al., 2005) and RN-Tre (Lanzetti et al., 2000, 2004; have been reported to have GAP activity toward Rab5 on endosomes. However, there is controversy regarding their Rab5 GAP activity. Itoh et al. (2006) reported that RabGAP5 does not show GAP activity toward Rab5 in in vitro GAP activity assay. It has been reported that RN-Tre does not interact with Rab5 in a yeast two-hybrid system (Haas et al., 2005) and localizes to focal adhesions (Palamidessi et al., 2013). In our study, we found that, whereas RabGAP5 colocalized with Rab5 on endosomes, RN-Tre mostly localized to plasma membranes and did not colocalize with Rab5 on endosomes. Our results indicate that RabGAP5 but not RN-Tre may act as a GAP toward Rab5 on endosomes in our cell system. DRG2 depletion did not inhibit the recruitment of RabGAP5 to Rab5 endosomes. However, recruitment of RabGAP5 was not sufficient for inactivation of Rab5 on endosomes, since active GTP-bound Rab5 was detected in RabGAP5-containing endosomes in DRG2-depleted cells. Sun et al. (2012) suggested the importance of recruitment of Rac1 to the recycling endosomes for Rab5 inactivation. Thus we tested whether DRG2 depletion affects the recruitment of Rac1 to the endosomes. DRG2 depletion did not affect the recruitment of Rac1 to the Tfn-containing endosomes. In addition, DRG2 depletion did not affect Rac1 activity on endosomes and the recruitment of Tiam-1, a Rac1 GEF (Palamidessi et al., 2008), to the Tfn-containing endosomes. Our results indicate that DRG2 does not affect the recruitment of RabGAP5, Rac1, and Tiam-1 to the Rab5-containing endosomes. It has been reported that binding of a GAP to Rab is necessary to inactivate the Rab (Pfeffer, 2001) and RabGAP5 interacts with Rab5 and inactivates it (Haas et al., 2005). Even though RabGAP5 colocalizes with Rab5 on endosomes, the impairment of interaction between these two proteins would result in failure in inactivation of Rab5. In our study, we found that DRG2 depletion disrupted the interaction between Rab5 and RabGAP5 on endosomes. Collectively our results indicate that even though DRG2 does not affect the recruitment of RabGAP5 to Rab5 endosomes, it is required for interaction between Rab5 and RabGAP5 and thus for deactivation of Rab5 on endosomes. It is likely that DRG2 directly mediates the interaction between these two proteins, since DRG2 was found to interact with Rab5 and RabGAP5 on endosomes by FRET assay. Further work is necessary to elucidate the detailed mechanisms of how DRG2 regulates the interaction between Rab5 and RabGAP5.
In conclusion, this study identifies DRG2 as a key molecule that regulates Rab5 activity on endosomes and Tfn recycling. DRG2 interacted with EEA1 and Rab5 and localized to Rab5 endosomes. DRG2 did not affect the recruitment of RabGAP5 to Rab5 endosomes. DRG2 interacted with RabGAP5 on endosomes, and DRG2 depletion impaired interaction between Rab5 and RabGAP5, Rab5 deactivation, and Tfn recycling. Dissecting how DRG2 on the endosomes regulates interaction between Rab5 and RabGAP5 and thus Rab5 activity will provide new insights into regulation of cargo recycling.
MATERIALS AND METHODS
Mice and cell culture
C57BL/6 mice were purchased from Orient Bio (Sungnam, Korea). Animal experiments were approved by the Animal Care and Use Committee of the University of Ulsan. MEFs were prepared from day 12.5 embryos derived from crosses between DRG2+/− mice as described previously (Conner, 2001). In this study, MEFs were used within passage 5 to avoid replicative senescence. MCF7 was maintained in RPMI supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). HeLa cell was maintained in DMEM supplemented with 10% FBS (Invitrogen). LY294002 (Biomol, Hamburg, Germany) was used to inhibit PI3K activity at 100 nM. Holo–human transferrin (T0665) was purchased from Sigma-Aldrich (St. Louis, MO).
Generation of DRG2 KO mice
DRG2 KO mice were generated by Macrogen (Seoul, Korea). Briefly, a targeting vector was designed to replace DRG2 exons 1 and 2 with a PGK-neo cassette using 5′ 2.6-kb and 3′ 7.3-kb arm fragments of DRG2 ligated into the pPNT vector (Figure 3G). The targeting vector was electroporated into 129/Sv embryonic stem cells, and six clones of recombination-positive embryonic stem cells were selected by Southern blot analysis using probes to the 3′ flanking regions of DRG2. Embryonic stem cell clones were injected into blastocysts, and these were then transferred into pseudopregnant ICR female mice to generate chimeric male mice. Chimeric male mice were mated with C57BL/6 female mice, and germline transmission of the mutant allele was confirmed by Southern blot analysis of tail DNA from F1 offspring. The DRG2- knockout (KO) mouse strain was backcrossed with the C57BL/6 strain and maintained for more than nine generations by cross-mating between heterozygous (+/–) female mice and WT male mice. PCR-based genotyping analysis of tail-tissue genomic DNA was performed for DRG2 and PGK-neo using the primers shown in Supplemental Table S1. When heterozygous females mated with heterozygous males, only wild-type and heterozygous pups were born, without homozygous mutants.
Plasmids, siRNA, shRNAs, and transfections
pcDNA6-V5/hDRG2 (human DRG2) and pcDNA6-V5/mDRG2 (mouse DRG2) constructs were described previously (Ko et al., 2014). The following constructs were generated with PCR primers (Supplemental Table S1): pmRFPC3-hDRG2, pEGFPN1-hDRG2, and pEGFPC1-RabGAP5. A plasmid encoding shRNA-resistant DRG2, pEGFPN1-shRNAR-DRG2, was generated with PCR primers (Supplemental Table S1) corresponding to the sequence of DRG2 shRNA #3 (DRG2-shRNA, TRC0000047195; Sigma-Aldrich) using the pcDNA6-V5/hDRG2 plasmid as a template. Plasmid constructs pmRFPC3-Rab5a, mCherry-Rab5 CA (Q79L), mCherry-Rab5 DN (S34N), pEGFP-EEA1, pEGFPC1-Rab11wt/DN(S25N), pTfnR-PAmKate, pTfnR-PAmcherry1, pcDNA3-EGFP-Rac1(WT/Q61L/T17N), pIRESneo-EGFP-α-tubulin, LAMP1-GFP HRS-RFP, Rabenosyn-5-GFP, and pEGFP-EGFR were purchased from Addgene (Cambridge, MA). pEGFP-EEA1 FYVE and Akt-PH-GFP were kindly provided by Tamas Balla (National Institutes of Health, Bethesda, MD). Raichu Rab5 was kindly provided by Michiyuki Matsuda (Kyoto University, Kyoto, Japan). Raichu Rac1 and Halo-tagTiam-1 mCherry were kindly provided by Maxime Dahan (Institut de Biologie de l’Ecole Normale Superieure, Paris, France). pECFP-C2 Rab5a was kindly provided by Emilia Galperin (University of Kentucky, Lexington, KY). Plasmid construct pLKO-DRG2-shRNA containing shRNA against human DRG2 (DRG2-shRNA, TRC0000047195) and nontarget shRNA control vector (MISSION pLKO.1-puro nonmammalian shRNA control plasmid DNA, SHC002) were purchased from Sigma-Aldrich. PLC-δ-PH-EGFP was kindly provided by Pann-Ghill Suh (Ulsan National Institute of Science and Technology, Ulsan, Korea). RN-Tre-GFP was kindly provided by Letizia Lanzetti (Istituto di Candiolo, Torino, Italy).
To generate DRG2-depleted cells, cells were transfected with pLKO-DRG2-shRNA and selected by puromycin (P9620; Sigma-Aldrich). To transiently inhibit EEA1 expression, cells were transfected with siRNA against EEA1 (3′-AAGTTTCAGATTCTTTACAAA-5′; Nazarewicz et al., 2011) or scrambled siRNA as a control (Santa Cruz Biotechnology, Santa Cruz, CA). Cells were transfected with the various plasmid constructs using TurboFect (Thermo Scientific, Schwerte, Germany).
Coimmunoprecipitation, SDS–PAGE, and immunoblotting
Cell extracts were incubated with 1.5 μg of anti-V5 (V8012; Sigma-Aldrich) or anti-GFP (G10362; Invitrogen) antibodies and 10 μl of Dynabeads (Invitrogen). Proteins were resolved by SDS–PAGE, transferred onto Hybond-P membranes (Amersham Biosciences, Piscataway, NJ), and probed with appropriate dilutions of the following antibodies: anti-human DRG2 (GW22260A; Sigma-Aldrich), anti-GFP (sc-9996; Santa Cruz Biotechnology), anti-mRFP (632496; Clontech, Palo Alto, CA), anti-Rab5 GTP (26911; NewEast Biosciences, Malvern, PA), anti-V5 (20-783-70389; GenWay, San Diego, CA), and anti–β-actin (A2228; Sigma-Aldrich). Immunoreactivity was detected using a GE HealthCare Image Quant system.
Semiquantitative RT-PCR
Semiquantitative RT-PCR was performed using Taq polymerase (Solgent, Daejeon, Korea). PCR primer pairs are listed in Supplemental Table S1.
GTPase activity assay
Purified recombinant proteins glutathione S-transferase (GST)–Rab5 and GST-DRG2 were purchased from Bioclone (San Diego, CA). A 200-ng amount of recombinant Rab5 was incubated with or without 40 ng of recombinant DRG2 in a buffer containing 20 mM Tris (pH 8.0), 1 mM EDTA, 2 mM dithiothreitol (DTT), and 0.25 μCi of [α-32P]GTP for 5 min at room temperature. After addition of 5 mM MgCl2 (final concentration) and a further incubation step at 37°C for 20 min, the reaction was stopped by adding 5× stop solution (0.2% SDS, 2 mM EDTA, 2 mM DTT, 0.5 mM GTP, 0.5 mM GDP). Samples were chromatographed on polyethyleneimine-cellulose thin-layer plates in 0.5 M KH2PO4 (pH 3.5), and radioactivity was visualized by autoradiography on x-ray film (Fisher Scientific, King of Prussia, PA).
Confocal microscopy
Cells were transfected with the various plasmid constructs for 24 h. Before the Tfn-internalization assay, Tfn present in serum was removed by two washes with DMEM. Cells were then incubated in DMEM–4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) containing 100 ng/ml EGF (E9644; Sigma-Aldrich; for EGFR degradation assay, 150 μg/ml cycloheximide was added 1 h before imaging) and/or 20 μg/ml Alexa Fluor 594–, 633–, or 647–conjugated human Tfn (T-13343, T-23362, and T23366; Molecular Probes, Eugene, OR) or Cy3-conjugated mouse Tfn (015-160-050; Jackson Immunoresearch, West Grove, PA) on ice for 30 min. Unbound EGF and Tfn were removed by washing three times with ice-cold DMEM-HEPES. Cell surface Tfn was removed by a mild-acid wash for 2 min on ice. After incubation for the indicated times at 37°C, cells were washed three times with ice-cold phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (Sigma-Aldrich). Endogenous DRG2 was stained using chicken anti-DRG2 antibody (GW22260A; Sigma-Aldrich) and goat anti-chicken immunoglobulin G (IgG) conjugated with fluorescein isothiocyanate (A16055; Invitrogen)/Alexa Fluor 647 goat anti-chicken IgG (A21449; Invitrogen). Early endosomes, ER, and Golgi were stained using anti-PDI antibody (ab3672; Abcam) and anti–58K Golgi protein (ab27043; Abcam, Boston, MA), respectively.
Confocal images were acquired in sequential mode using a 100× Plan Apochromat numerical aperture 1.4 oil objective and the appropriate filter combination on an Olympus 1000/1200 laser-scanning confocal system or Zeiss LSM 700/780 at the Ulsan National Institute of Science and Technology Bio-imaging Center (Ulsan, Korea). All images were saved as TIFF files, and their contrast was adjusted with ImageJ (version 1.19 m; National Institutes of Health, Bethesda, MD). Pixel-by-pixel colocalization analysis was performed using ImageJ (plug-in JACoP; Bolte and Cordelieres, 2006). Images were analyzed with MetaMorph 7 software (Universal Imaging) and Imaris Bitplane-7.7.2 at the Ulsan National Institute of Science and Technology Bio-imaging Center.
Flow cytometry
Cells were transfected with pEGFP-EGFR or pEGFP-N1 for 24 h. Cells were incubated with 100 ng/ml EGF on ice for 30 min, and unbound EGF was removed by washing with ice-cold DMEM-HEPES (150 μg/ml cycloheximide was added before EGF treatment). After incubation for the indicated times at 37°C, flow cytometric analysis was performed with a FACSCanto II (BD Biosciences, San Jose, CA).
Fluorometric analysis for Tfn released from cells
Cells were incubated with 20 μg/ml Alexa Fluor 594–conjugated human Tfn (T-13343; Molecular Probes) on ice for 30 min. Unbound Tfn was removed by washing three times with ice-cold PBS. After incubation for the indicated times at 37°C, Tfn concentration in the culture supernatant was determined based on its fluorescence by fluorometry.
GTP-loaded Rab5 pull-down assay
Dynabeads were coated with anti–Rab5-GTP mouse monoclonal antibody (26911; NewEast Biosciences). Beads were then incubated with protein extract from mRFP-Rab5–transfected cells for 1 h at 15°C. Precipitated active Rab5 was detected by immunoblotting using anti-DsRed polyclonal antibody (632496; Clontech).
Ratiometric FRET microscopy to detect Rab5 and Rac1 activity
We used pRaichu-Rac1 and pRaichu-Rab5 as FRET probes for Rab5 and Rac1 GTPases, respectively. Raichu-Rac1 consists of YFP, CRIB of PAK1, Rac1, CFP (amino acids 1–237), and the CAAX box of Ki-Ras (Itoh et al., 2002). Raichu-Rab5 consists of Venus, the amino-terminal Rab5-binding domain of EEA1 (amino acid residues 36–218), SECFP, and Rab5a (Kitano et al., 2008). FRET analysis for Rac1 and Rab5 was conducted as described previously (Aoki and Matsuda, 2009). Briefly, control and DRG2-depleted MCF7 cells were transfected with pRaichu-Rac1 or pRaichu-Rab5. Images were captured using the ratiometric FRET module in the FluoView program of the LSM FV1000 confocal microscope (Olympus, Tokyo, Japan). CFP and YFP images were processed by ImageJ and MetaMorph software (Universal Imaging, West Chester, PA). After background subtraction, FRET ratio images were generated using MetaMorph software and visualized in intensity-modulated display mode. In this display mode, eight colors from red to blue are used to represent the FRET ratio (YFP/CFP), with the intensity of each color indicating the mean intensity of YFP and CFP.
Sensitized emission FRET to detect protein interaction
EGFP and mRFP were used as a donor and acceptor FRET pair for all FRET measurements. Cells were maintained in phenol red–free medium during imaging. FRET images were corrected for cross-talk between donor and acceptor as previously described (Elangovan et al., 2003). FRET images were analyzed using the Olympus FluoView 4.2 Program FRET module. Donor and acceptor fluorescence signals were detected in the 501–533- and 565–651-nm spectral regions, and images were acquired with zoom 4 and line average 8 (scan speed 7). Ten donor (a), acceptor (d), and FRET (b and c) images were acquired for each donor- and acceptor-only sample, whereas 13–15 e, f, and g images were acquired for the double-labeled samples.
Several cells were imaged by using donor only (EGFP), acceptor only (mRFP), and donor and acceptor colabeled cells under the same experimental conditions. By calculating the correction factor based on the pixel-by-pixel intensity of single-labeled cells (EGFP/mRFP) and then applying these values as a correction factor to the appropriate matching pixels of the double-labeled cells (EGFP and mRFP combination: EGFP-EEA-1 and mRFP-DRG2, EGFP-RABGAP5 and mRFP-Rab5), we obtain
precision FRET (PFRET) = f − DSBT − ASBT
where f is the uncorrected FRET, ASBT is the acceptor spectral bleedthrough, and DBST is the donor spectral bleedthrough signal acquired by single-labeled cells. The donor bleedthrough signal in the FRET channels for all of the pixel elements of the whole image is determined by the equation
DSBT signal = (b/a)e
where a is the donor channel image with donor excitation in single-labeled donor specimens, b is the acceptor channel image with donor excitation in single-labeled donor specimens, and e is the donor channel image with donor excitation in double-labeled donor and acceptor specimens.
The acceptor bleedthrough signal in the FRET channels for all the pixel elements of the whole image is determined by the equation
ASBT signal = (c/d)g
where c is the acceptor channel image with donor excitation in single-labeled acceptor specimens, d is the acceptor channel image with acceptor excitation in single-labeled acceptor specimens, and g is the acceptor channel image with acceptor excitation in double-labeled donor and acceptor specimens. This equation not only removes the spectral bleedthrough but also nullifies the effect of the variation in fluorescence protein expression levels.
The FRET efficiency is calculated by using the formula
En = 1 − IDA/[IDA + PFRET × (ψdd/ψaa) × Qd]
where ψdd/ψaa = (photomultiplier tubes [PMT] gain of donor channel/PMT gain of acceptor channel) × (spectral sensitivity of donor channel/spectral sensitivity of acceptor channel) and Qd is the donor quantum yield.
We represented all FRET images by using false-color scoring for sensitized FRET intensities over the arbitrary range from 0 (dark blue) to 1 (white).
Statistical analysis
Unpaired Student’s t tests (two-tailed) were used to determine the significance of differences between groups. p < 0.05 is considered significant.
Supplementary Material
Acknowledgments
We thank Pann-Ghill Suh (Ulsan National Institute of Science and Technology, Ulsan, Korea), Letizia Lanzetti (Istituto di Candiolo, Torino, Italy), Tamas Balla (National Institutes of Health, Bethesda, MD), Michiyuki Matsuda (Kyoto University, Kyoto, Japan), Maxime Dahan (Institut de Biologie de l’Ecole Normale Superieure, Paris, France), Emilia Galperin (University of Kentucky, Lexington, KY), and Steve Caplan (University of Nebraska, Omaha, NE) for providing plasmid constructs used in this study. This work was supported by Korea Research Foundation Grants funded by the Korean Government (MOEHRD; 2014005655, 2014R1A6A1030318, HI14C2434).
Abbreviations used:
- DRG2
developmentally regulated GTP-binding protein
- EEA1
early endosome antigen 1
- EGFP
enhanced green fluorescent protein
- EGFR
epidermal growth factor receptor
- FRET
fluorescence resonance energy transfer
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- MEF
mouse embryonic fibroblast
- mRFP
monomeric red fluorescent protein
- MVE
multivesicular endosome
- PI3K
phosphatidylinositol 3-kinase
- PI3P
phosphatidylinositol 3-phosphate
- shRNA
small hairpin or short hairpin RNA
- siRNA
small interfering RNA
- Tfn
transferrin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0558) on November 18, 2015.
REFERENCES
- Aoki K, Matsuda M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat Protoc. 2009;4:1623–1631. doi: 10.1038/nprot.2009.175. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Bright NA, Lindsay MR, Stewart A, Luzio JP. The relationship between lumenal and limiting membranes in swollen late endocytic compartments formed after wortmannin treatment or sucrose accumulation. Traffic. 2001;2:631–642. doi: 10.1034/j.1600-0854.2001.20906.x. [DOI] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L, Mishra AK, Sylvain MA, Tuck S, Lambright DG, Rocheleau CE. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2285–2296. doi: 10.1091/mbc.E09-11-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999a;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao LY, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999b;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Conner DA. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb2302s51. Chapter 23, Unit 23.2. [DOI] [PubMed] [Google Scholar]
- Elangovan M, Wallrabe H, Chen Y, Day RN, Barroso M, Periasamy A. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29:58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Biol Cell. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of Rac and Cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Satoh M, Kanno E, Fukuda M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 2006;11:1023–1037. doi: 10.1111/j.1365-2443.2006.00997.x. [DOI] [PubMed] [Google Scholar]
- Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:U241-U215. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- Ko MS, Kim HJ, Kim HK, Yoon NA, Lee UH, Lee SC, Chung DK, Lee BJ, Suh JH, Cho WJ, Park JW. Developmentally regulated GTP-binding protein 2 ameliorates EAE by suppressing the development of TH17 cells. Clin Immunol. 2014;150:225–235. doi: 10.1016/j.clim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Ko MS, Lee UH, Kim SI, Kim HJ, Park JJ, Cha SJ, Kim SB, Song H, Chung DK, Han IS, et al. Overexpression of DRG2 suppresses the growth of Jurkat T cells but does not induce apoptosis. Arch Biochem Biophys. 2004;422:137–144. doi: 10.1016/j.abb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- Krauss M, Haucke V. Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep. 2007;8:241–246. doi: 10.1038/sj.embor.7400919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429:309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- Li B, Trueb B. DRG represents a family of two closely related GTP-binding proteins. Biochim Biophys Acta. 2000;1491:196–204. doi: 10.1016/s0167-4781(00)00025-7. [DOI] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- Mahajan MA, Park ST, Sun XH. Association of a novel GTP binding protein, DRG, with TAL oncogenic proteins. Oncogene. 1996;12:2343–2350. [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Navaroli DM, Bellvé KD, Standley C, Lifshitz LM, Cardia J, Lambright D, Leonard D, Fogarty KE, Corvera S. Rabenosyn-5 defines the fate of the transferrin receptor following clathrin-mediated endocytosis. Proc Natl Acad Sci USA. 2012;109:E471–E480. doi: 10.1073/pnas.1115495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarewicz RR, Salazar G, Patrushev N, San Martin A, Hilenski L, Xiong S, Alexander RW. Early endosomal antigen 1 (EEA1) is an obligate scaffold for angiotensin II-induced, PKC-alpha-dependent Akt activation in endosomes. J Biol Chem. 2011;286:2886–2895. doi: 10.1074/jbc.M110.141499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- Palamidessi A, Frittoli E, Ducano N, Offenhauser N, Sigismund S, Kajiho H, Parazzoli D, Oldani A, Gobbi M, Serini G, et al. The GTPase-activating protein RN-tre controls focal adhesion turnover and cell migration. Curr Biol. 2013;23:2355–2364. doi: 10.1016/j.cub.2013.09.060. [DOI] [PubMed] [Google Scholar]
- Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional PtdIns(3)P-binding motif. Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Schenker T, Lach C, Kessler B, Calderara S, Trueb B. A novel GTP-binding protein which is selectively repressed in SV40-transformed fibroblasts. J Biol Chem. 1994;269:25447–25453. [PubMed] [Google Scholar]
- Sheff D, Pelletier L, O’Connell CB, Warren G, Mellman I. Transferrin receptor recycling in the absence of perinuclear recycling endosomes. J Cell Biol. 2002;156:797–804. doi: 10.1083/jcb.20111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sun L, Liu O, Desai J, Karbassi F, Sylvain MA, Shi A, Zhou Z, Rocheleau CE, Grant BD. CED-10/Rac1 regulates endocytic recycling through the RAB-5 GAP TBC-2. PLoS Gene. 2012;8:e1002785. doi: 10.1371/journal.pgen.1002785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin HW, Katoh Y, Nakayama K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci. 2012;125:4049–4057. doi: 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- Tsutsumi K, Fujioka Y, Tsuda M, Kawaguchi H, Ohba Y. Visualization of Ras-PI3K interaction in the endosome using BiFC. Cell Signal. 2009;21:1672–1679. doi: 10.1016/j.cellsig.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.