Abstract
Integrated data on hepatitis B virus (HBV) patterns, HBV genotypes and mutations are lacking in human immunodeficiency virus type 1 (HIV-1) co-infected patients from Africa. This survey was conducted in 2010–2013 among 762 HIV-1-positive adults from Gabon who were predominantly treated with 3TC-based antiretroviral treatment. HBV patterns were identified using immunoassays detecting total antibody to hepatitis B core antigen (HBcAb), hepatitis B surface antigen (HBsAg), IgM HBcAb, hepatitis B e antigen (HBeAg), antibody to HBsAg (HBsAb) and an in-house real-time PCR test for HBV DNA quantification. Occult hepatitis B (OBI) was defined by the presence of isolated anti-HBc with detectable serum HBV DNA. HBV genotypes and HBV mutations were analyzed by PCR-direct sequencing method. Seventy-one (9.3%) patients tested positive for HBsAg, including one with acute hepatitis B (0.1%; 95% CI, 0.0%-0.2%), nine with HBeAg-positive chronic hepatitis B (CHB) (1.2%; 95% CI, 0.6%–2.2%), 16 with HBeAg-negative CHB (2.1%; 95% CI, 1.2%–3.3%) and 45 inactive HBV carriers (5.9%; 95% CI, 4.4%–7.8%). Sixty-one (8.0%; 95% CI, 6.2%–10.1%) patients showed OBI. Treated patients showed similar HBV DNA levels to those obtained in untreated patients, regardless of HBV patterns. Around 15.0% of OBI patients showed high (>1,000 UI/mL) viremia. The mutation M204V/I conferring resistance to 3TC was more common in HBV/A (47.4%) than in HBV/E isolates (0%) (P = .04). Our findings encouraged clinicians to promote HBV vaccination in patients with no exposure to HBV and to switch 3TC to universal TDF in those with CHB.
Introduction
Hepatitis B virus (HBV) is associated with approximately 780,000 deaths each year worlwide, mostly due to the chronic course of hepatitis B infection [1]. HBV and human immunodeficiency virus type-1 (HIV-1) share common routes of transmission. To date, 35 million individuals worldwide are HIV-1 carriers, of which 3 to 6 million have chronic hepatitis B (CHB). HBV/HIV co-infection is a huge public health problem in sub-Saharan Africa where both viruses are endemic [2]. HBV-related end-stage liver disease, such as decompensated cirrhosis and hepatocellular carcinoma (HCC), represents a growing cause of morbidity and mortality, notably in patients with low CD4+T-cell counts and high HBV DNA levels[3].
In resource-limited settings (RLS), current guidelines recommend hepatitis B surface antigen (HBsAg) screening for all HIV-1-infected patients before antiretroviral treatment (ART) initiation, and the use of first-line ART regimens containing tenofovir (TDF) in patients co-infected with HIV and HBV [4, 5]. However, HBsAg testing is not systematically performed. For instance, despite the high prevalence rates of HIV (4.2%) [6] and HBV (12.9%) in Gabon [7], data on HIV/HBV co-infections are lacking for this country. Consequently, for many years, most African HIV-HBV co-infected patients received ART regimens containing 3TC alone that were sub-optimal and “HBV-blind” [8, 9]. In addition, many African HIV-infected patients who test positive for HBsAg receive no additional testing, such as hepatitis B e antigen (HBeAg) and/or HBV DNA viral load (VL) and total antibody to hepatitis B core antigen (HBcAb), which are appropriate markers for CHB and occult hepatitis B infection (OBI), respectively [10, 11]. HBV DNA levels are useful to differentiate between patients with HBV DNA ≥ 2,000 IU/mL, indicating likely HBeAg-negative CHB and inactive HBV carriers who have HBV DNA < 2,000 IU/mL [12].
HBV genotyping and the detection of specific clinically relevant HBV mutations have been seldom studied in African patients co-infected with HIV-1. To date, HBV is classified into 10 genotypes, designated HBV/A-J, with specific geographical distributions [13, 14]. Recently, genotype A has been subdivided into four subgenotypes named HBV/A1, HBV/A2, HBV/QS-A3 (quasi-species A3) and HBV/A4 [15]. HBV/QS-A3 has been reported almost exclusively in West and Central Africa. HBV/E is also highly endemic in West and Central Africa, and has low intra-genotype diversity suggesting its recent emergence [16]. Regarding HBV mutations, the use of 3TC alone has been shown to be strongly associated with consistently high HBV viremia due to the rapid emergence of HBV mutants carrying the M204I/V mutation conferring resistance to 3TC/FTC [17, 18]. Precore (PC) and basal core promoter (BCP) mutations may represent the predominant virus species in patients with HBeAg-negative CHB in many parts of the world [19]. Escape mutations in the “a” determinant of the major hydrophilic region (MHR) of HBsAg can impair the diagnostic performance of HBsAg assays and can lead to emergence of vaccine-escape mutants (VEM) and hepatitis B immunoglobulin (HBIG) therapy failure [20].
In this study, we assessed the serological and molecular HBV patterns, including genotyping and mutational analysis, in HIV-positive patients mostly treated with 3TC-based ART regimens without TDF in Gabon.
Materials and Methods
Patients
Our cross-sectional study was conducted among HIV-1 infected patients who attended a HIV care center located at Franceville (Southeastern Gabon) between March 2010 and January 2013. Whole blood specimens were collected by venipuncture (5 ml of EDTA-anticoagulated peripheral blood). Plasma samples were then stored at -80°C. Gender, age and ART were retrieved from patients’ files. CD4+ T-cell counts, HIV-1 RNA viral load (VL) measurements and HCV serology were performed in all patients at the time of sampling [21]. The study was approved by the Ethics Committee of Gabon (N°0021/2013/SG/CNE). Written informed consent was obtained from each patient.
HBV serology
Total (IgM and IgG) HBcAb were first assessed using the Monolisa Anti-HBc PLUS immunoassay (ELISA) (Bio-Rad, Marnes-La-Coquette-France). In case of negative HBcAb results, subjects were then tested for antibody to HBsAg (HBsAb) titer with the Monolisa Anti-HBs PLUS ELISA (Bio-Rad). Subjects were considered as negative for HBsAb when titer was <10 mIU/mL. In case of positive HBcAb results, the detection of HBsAg was performed using the Monolisa HBs Ag ULTRA kit (Bio-Rad), a fully multivalent assay showing high sensitivity in the detection of HBV mutants [22]. If HBsAg result was negative, specimens were tested for HBsAb. All samples that tested positive for HBsAg were assessed for the presence of IgM HBcAb (Monolisa Anti-HBcIgM, Bio-Rad) and hepatitis B e antigen (HBeAg)/antibody to HBeAg (HBeAb) (Monolisa HBe Ag-Ab,Bio-Rad). All ELISA assays were performed according to manufacturer’s instructions.
Quantitative real-time HBV DNA PCR
Using the Arrow extractor (NorDiag, Biotrin International, Ireland), DNA was extracted from 240 μl of plasma, pretreated with 10 μl of proteinase K, using the Arrow Viral NA extraction kit, according to the manufacturer’s instructions. Template DNA was eluted into 60 μl of kit elution buffer. Besides clinical samples, quantification standard (AcroMetrix HBV Panel, AcroMetrix, Menica, CA, USA) was also extracted for each run with the same protocol and 1:10 diluted (from 50,000,000 IU/mL to 50 IU/mL). For amplification, we used a primers/probe set designed under the auspices of “Agence Nationale de Recherches sur le SIDA et les hépatites virales” (ANRS 12187 project) and targeting a conserved region in the HBV S gene (nucleotide (nt) positions, 379–426). All runs were performed in a 50-μl volume containing DNA extract (10 μl), Master Mix (Platinum UGD, USA) (25 μl), pure water (HyClone Pure Water, Thermo Fisher Scientific, Waltham, MA USA) (12.5 μl), forward primHBV1 (5’-GTGTCTGCGGCGTTTTATCA-3’) and reverse primHBV2 (5’-AGGCATAGCAGCAGGATGAA-3’) primers at 10 μM (1 μl each) and probe (5’FAM-TGCGGCGTTTTATCAT-MGB3’) at 5 μM (0.5 μl). Each reaction consisted of: 2 min at 50°C and 10 min at 95°C; followed by 50 cycles of 15 sec at 95°C and 1 min at 60°C each.
The lower limit of quantification (LLOQ) of our technique was 100 IU/mL and the lower limit of detection (LLOD) was 50 IU/mL. In case of detected HBV DNA below LLOQ and above or equal to the LLOD, HBV DNA PCR was repeated once and if it was detected again, the mean value for duplicates was considered in the analysis. If not, samples were allocated a value equal to half the LLOD (25 IU/mL), as undetectable specimens (i.e, not amplified (n/a) samples with no value for cycle threshold (Ct)).
HBV DNA was quantified in specimens positive for HBsAg and in those with isolated HBcAb. In addition, 50 samples negative for both HBcAb and HBsAb were randomly selected for HBV DNA detection in order to evaluate the specificity of our real-time PCR assay.
HBV genotyping and mutations by direct sequencing
Specimens positive for HBV DNA, with sufficient plasma, were studied for HBV genotyping and polymerase (pol) gene mutations in the HBV surface (S) gene. DNA extraction was done manually using the QIAamp Viral DNA Mini Kit (Qiagen, Courtaboeuf, France) and required 200 μl of plasma. HBV was amplified using a semi-nested PCR for the HBV S gene (nt positions, 56 to 911), as previously published by Olinger et al. [23]. In addition, PC and BCP regions were amplified with a nested PCR (nt positions, 1672 to 2012) that targets a fragment of 340 bp in the core (C) gene, as previously published [24].
Sequencing was performed on the 3730XL DNA Analyzer (Applied Biosystems, Vernon Hills, Illinois, USA). Pairwise alignments were performed using ClustalW (1.6) model with sequences available in GenBank, and representing all HBV genotypes. [25, 26]
Phylogenetic analyses was carried out by Bayesian inference using the Bayesian Markov chain Monte Carlo (MCMC) statistical framework implemented in MrBayes version 3.1.2 software [27, 28] under the model ‘general reversible model of evolution’ with a gamma distribution of rates across sites, run for 8 x 106 generations, with a burn-in of 25%. Calculations of Effective Sample Size (ESS) were examined with the TRACER v1.5 program (http://evolve.zoo.ox.ac.uk/) and all the ESS were greater than 1200. The tree datasets were computed using strict consensus and majority rules algorithms to evaluate the posterior probabilities of branching pattern (BPP). The final tree was visualized in the FigTree version 1.4.2 (program 2006–2014, Andrew Rambaut Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/).
Using the Mutation Reporter Tool software (http://hvdr.bioinf.wits.ac.za/mrt/), HBV resistance-associated mutations (RAMs) in the pol gene represented by V173L, L180M, A181V, A194T, S202G, M204V/I and N236T were assessed [29] [30, 31]. In addition, we looked for VEMs outside (at amino-acid (aa) position 120) and within the HBsAg immunodominant ‘a’ determinant (at aa positions 126, 129, 130, 144 and 145) [32]. Finally, A1762T/G1764A mutations in BCP region and T1858C, G1862C andG1896A mutations in PC region were analyzed [33].
Statistical analysis
Prevalence was expressed with 95% confidence intervals (95% CI). Continuous data were expressed as median value with 1st and 3rd interquartile ranges (IQR). Comparisons between groups were conducted using the Mann-Whitney test, while categorical data were expressed as percentages and compared across groups using the χ2 test. Results were 2-sided and P values < 0.05 were considered to be statistically significant.
Results
Characteristics of Study Population
A total of 762 HIV-1-positive patients were studied. Among these, 560 (73.5%) were women. Median age was 41.5 years (IQR, 34.7–49.0 years). Among the 507 treated patients, ninety-eight percent was receiving 3TC-based regimens without TDF. Median time on ART was 36 months (IQR, 13–57 months). Overall, 52.1% (264/507) of treated patients showed undetectable HIV-1 RNA VL results (Table 1).
Table 1. Main characteristics of patients co-infected with HIV-1 and HBV in Franceville, Gabon (2010–2013).
| Characteristics | Total | Male | Female |
|---|---|---|---|
| Patients, n (%) | 762 (100) | 202 (26.5) | 560 (73.5) |
| Age, median years (IQR) | 41.5 (34.7–49.0) | 45.0 (39.4–52.1) | 39.3 (33.2–48.0) |
| ART | |||
| No, n (%) | 255 (33.5) | 75 (37.1) | 180 (32.1) |
| Yes, n (%) | 507 (66.5) | 127 (62.9) | 380 (67.9) |
| Type of ART | |||
| 3TC-based regimen (without TDF), n (%) | 497 (98) | 120 (94.5) | 377 (99.2) |
| TDF-based regimen, n (%) | 10 (2) | 7 (5.5) | 3 (0.8) |
| Duration on ART, months (IQR) | 36 (13–57) | 30 (9–56) | 38 (17–57) |
| CD4 count, median cells/ml (IQR) | |||
| Untreated patients | 382 (162–618) | 355 (201–512) | 467 (245–636) |
| Treated patients | 344 (187–487) | 307 (174–462) | 352 (192–499) |
| HIV-1 RNA, median Log10 copies/mL (IQR) | |||
| Untreated patients | 4.93 (4.42–5.72) | 5.26 (4.77–6.08) | 4.86 (4.04–5.68) |
| Treated patients | < 1.24 (< 1.24–4.87) | < 1.24 (<1.24–4.86) | < 1.24 (<1.24–4.89) |
| Positive HCV serology result, n (%) | 56 (7.3) | 16 (7.9) | 40 (7.1) |
Abbreviations: ART, antiretroviral treatment; 3TC, lamivudine; TDF, tenofovir; IQR, inter quartile range.
HBV patterns among HIV-1-infected patients
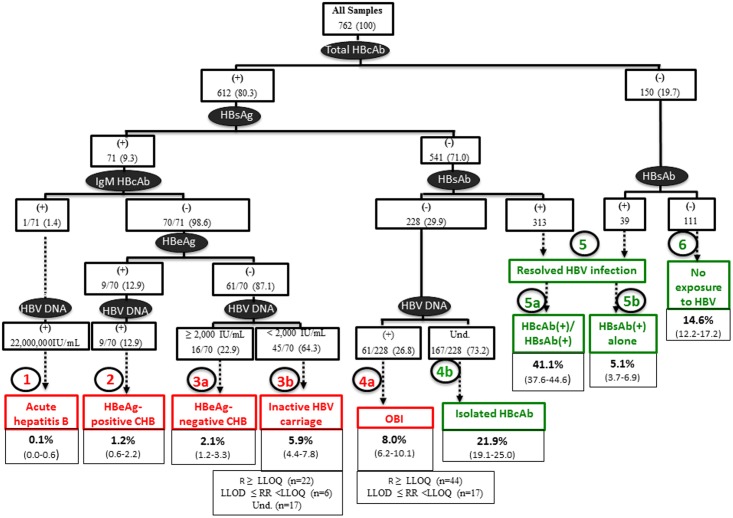
As summarized in Fig 1, nine distinct profiles were identified. First, among the 71 patients found positive for total HBcAb and HBsAg, one (0.1%) only tested positive for anti-HBc IgM (acute hepatitis B, profile 1) and had a high VL result (22,000,000 IU/mL). Second, among the 70 remaining HBc IgM-negative patients, nine (12.9%) tested positive for HBeAg with detectable HBV DNA levels (HBeAg-positive CHB, profile 2), representing 12% of the total population. Next, 61 (87.1%) tested negative for HBeAg; among which, 16 (2.1%) showed HBV DNA results ≥ 2,000 IU/mL (HBeAg-negative CHB, profile 3a) whereas 45 (5.9%) had HBV DNA values < 2,000 IU/mL (inactive HBV carriers, profile 3b). Among the 541 patients found positive for total HBcAb but negative for HBsAg, 228 tested negative for HBsAb, including 61 (8.0%) patients showing detectable DNA levels (OBI, profile 4a) versus 167 (21.9%) with undetectable HBV DNA (isolated HBcAb profile, profile 4b). Next, 313 (41.1%) individuals were positive for total HBcAb and HBsAb (resolved HBV infection, profile 5a). In the 150 patients who tested negative for total HBcAb, 39 (5.1%) had HBsAb (profile 5b) and 111 had no marker for HBV (no exposure to HBV,profile 6). HBV DNA was not amplified in 50 samples taken from patients with no exposure to HBV. No significant difference was observed between patients belonging to profiles 2, 3 and 4, according to gender, CD4 counts, HIV-1 RNA levels and HCV Ab positivity (data not shown).
Fig 1. HBV patterns defined by serological results and serum HBV DNA levels.
Infectious patterns are indicated in red boxes. Non-infectious patterns are indicated in green boxes. Overall prevalence rates are n (%) unless otherwise stated. Abbreviations: (+), positive; (-), negative; Und., undetectable; R, reactive; RR, repeatedly reactive; CHB, chronic hepatitis B; OBI, hepatitis B occult.
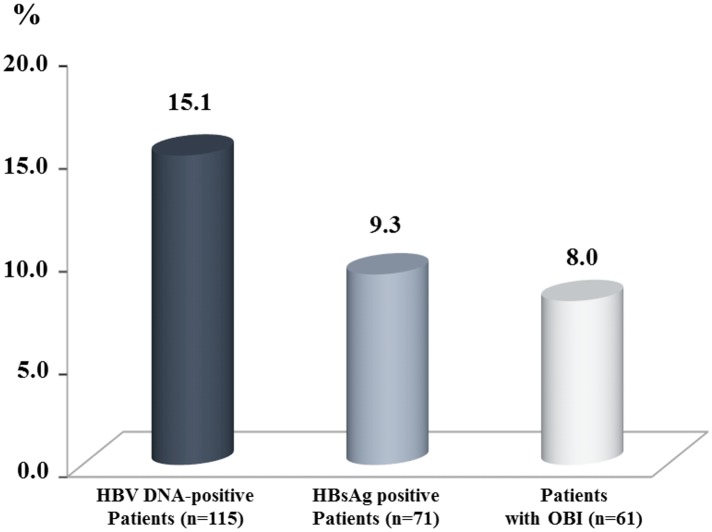
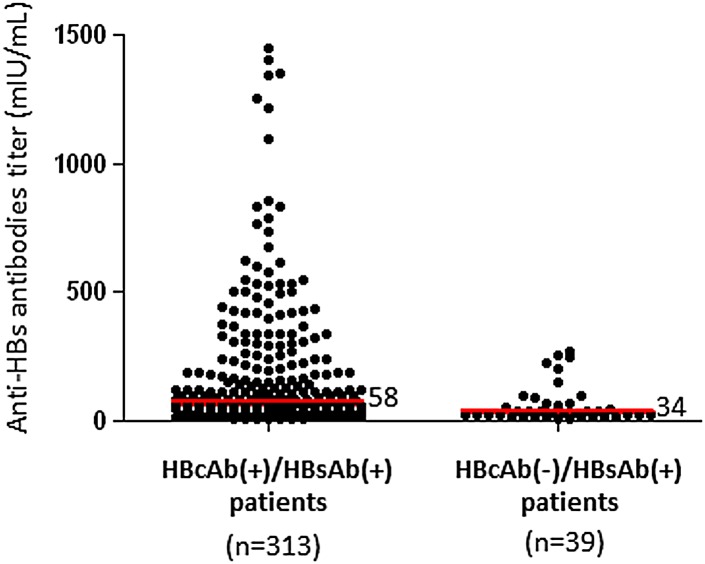
Overall, the prevalence of OBI (61/762, 8.0%) was similar to that obtained for overt (presence of circulating HBsAg) HBV infection (71/762, 9.3%) (P = .36) (Fig 2). The median anti-HBS Ab titer in patients belonging to profile 5a was significantly higher (58 mIU/mL, IQR, 30–182) than that obtained in patients belonging to profile 5b (34 mUI/mL, IQR, 19–67) (P = .0008) (Fig 3).
Fig 2. Prevalence rates of patients with detectable HBV DNA results, patients with overt (presence of circulating HBsAg) HBV infection (acute or chronic), and patients with OBI.
Fig 3. Comparison of anti-HBs antibodies levels in patients positive for both anti-HBc and anti-HBs Ab (profile 5a) and those with only anti-HBs Ab (profile 5b).
Impact of antiretroviral treatment on HBV DNA levels according to HBV patterns
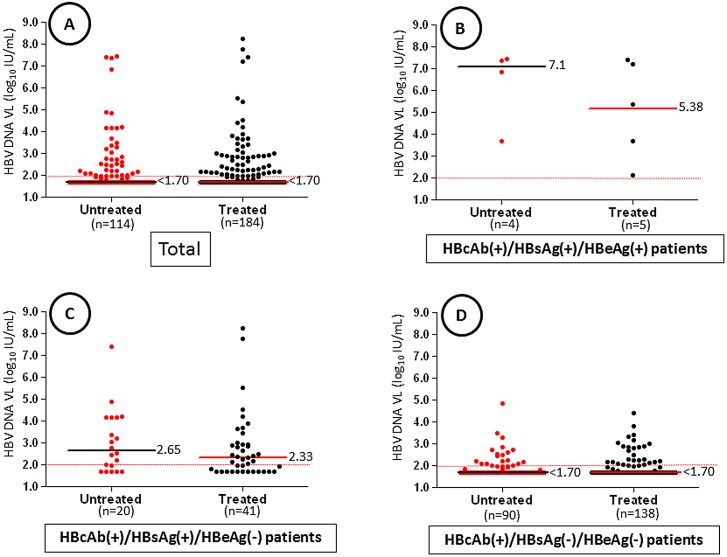
Overall, median HBV DNA levels were similar in both untreated and treated patients (P = 0.64) (Fig 4A). The lack of significance between untreated and treated patients was obtained, regardless of HBV patterns: 7.1 versus 5.38 log10 IU/mL in profile 2 (P = .41) (Fig 4B); 2.65 versus 2.33 log10 IU/mL in profile 3 (P = .34) (Fig 4C); and <1.70 versus < 1.70 log10 IU/mL in profile 4 (P = .59) (Fig 4D). Among the 61 patients with OBI, nine (14.7%) had HBV DNA level > 1,000 IU/mL, 35 (57.4%) had HBV DNA level between 100–1,000 IU/mL and 17 (27.9%) had repeatedly reactive HBV DNA results below the limit of detection.
Fig 4. Comparison of HBV DNA levels in 298 patients co-infected with HIV-1.
A) Overall comparison between patients treated with ART versus untreated patients. B) Comparison in HBsAg-positive patients with HBeAg-positive chronic hepatitis B (profile 2). C) Comparison in HBsAg-positive patients with HBeAg-negative pattern (profile 3). D) Comparison in patients with OBI or isolated anti-HBc antibodies (profile 4). Median HBV DNA levels are indicated with black bars for untreated patients and with red bars for treated patients. The red dotted line indicates the limit of detection (LOD) of the assay.
Distribution of HBV genotypes
HBV genotype was successfully analyzed in 28 samples (on 114 specimens with HBV DNA levels ≥ 50 IU/mL, 26%) from patients with HBeAg-positive CHB (n = 3), negative-HBeAg CHB (n = 6), inactive HBV carriage state (n = 9) and reactivated OBI (n = 10). PCR failed from 22 (19.3%) samples whereas 64 (56.1%) had insufficient volumes of plasma. All sequences described herein are available in the GenBank (accession numbers KM983561 to KM983588).
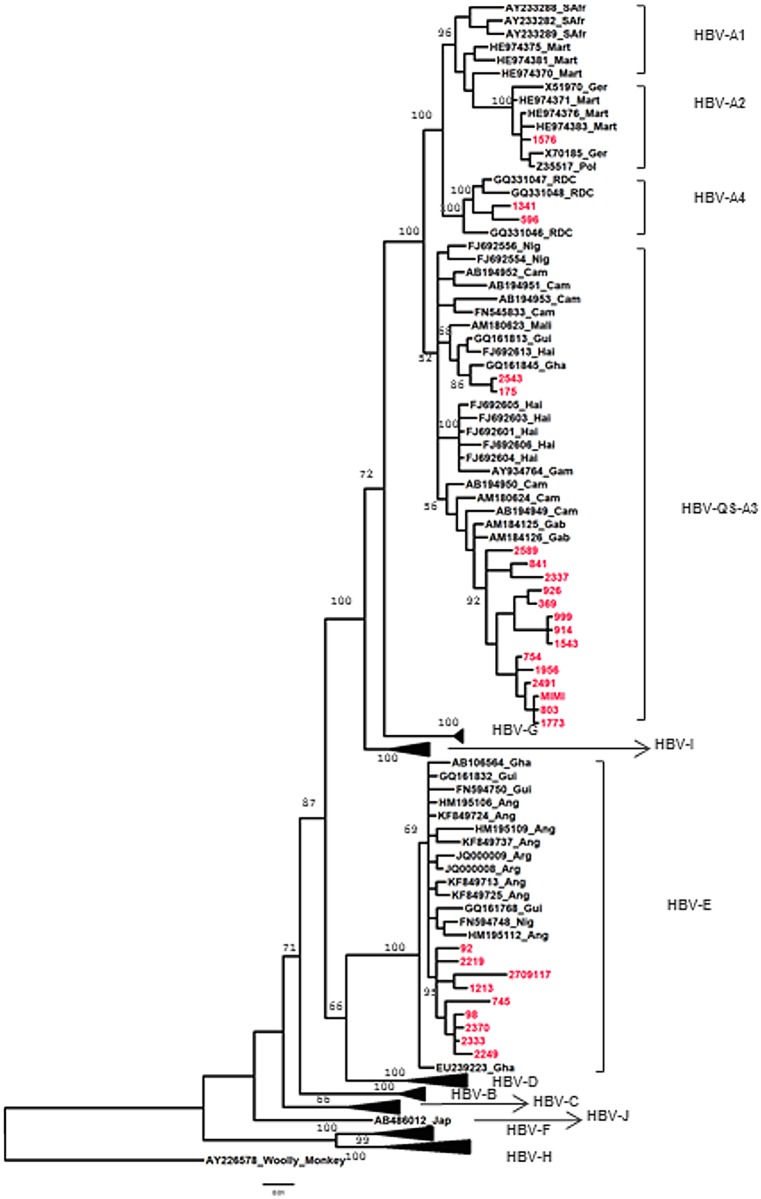
As shown in Fig 5, we revealed the presence of two HBV genotypes, including 19 sequences (67.9%) clustering with genotype HBV/A and nine (32.1%) with genotype HBV/E. For genotype A, most (n = 16) strains clustered with sub-genotype QS-A3, and were closely related to isolates from Cameroon and Gabon. Two additional strains were classified as sub-genotype A4 and one clustered with sub-genotype A2. The nine HBV/E strains were closely related to isolates from Angola and Ghana. Genotype HBV/A was more frequently encountered in females (14/19, 73.7%), whereas HBV/E was found in a majority of males (7/9, 77.8%) (P = .03).
Fig 5. Phylogenetic analysis of a 765 bp HBV-S gene region from different HBV isolates, with Woolly Monkey strain (AY226578) as the root.
The phylogenetic tree was inferred by the Bayesian method in the GTR model, with gamma-distributed rates at sites, 8 million generations and effective sample sizes greater than 1200. BPP (Branching Posterior Probability) are shown along the main nodes. HBV sequences obtained during this study are represented in red.
Distribution of specific HBV mutations according to HBV genotype
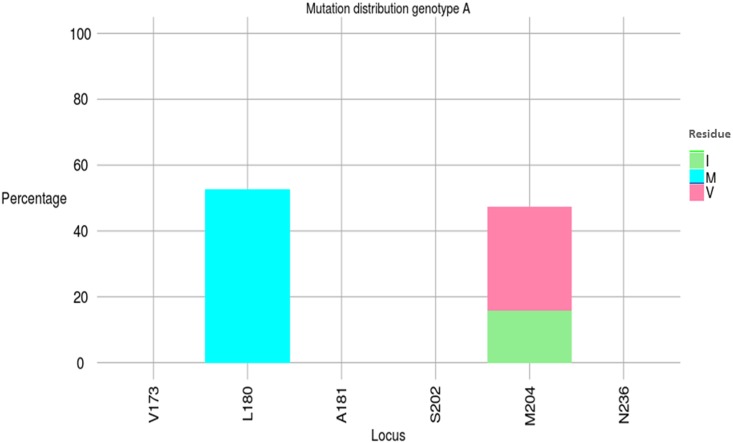
Regarding RAMs, the mutation M204V/I, associated with the compensatory mutation L180M, was significantly more common in genotype HBV/A (9/19, 47.4%) than in genotype HBV/E (0/9, 0%) (Yate’s corrected χ2, P = .04) (Fig 6, Table 2). The median duration on 3TC-based ART was not significantly different between patients harboring HBV/A strains (21 months; IQR, 0–55.5) and those carrying HBV/E strains (5.5 months; IQR, 0–49.5) (P = .66). Among the 9 patients harboring HBV/A variants with RAMs, six were treated with 3TC-based ART whereas three were untreated.
Fig 6. Mutation distribution graphs for the polymerase region.
Sequence alignments were submitted to Mutation Reporter Tool (http://hvdr.bioinf.wits.ac.za/mrt/) to produce graphs for genotype A. None mutation found in nine sequences from genotype E. Letters preceding each locus on the X-axis show the reference motifs. To facilitate comparison, conserved loci were not suppressed.
Table 2. Specific HBV mutations according to HBV patterns and genotypes.
| ID | Sex | Age (yrs) | CD4 (/mL) | HIV-1 RNA (log) | ART duration (months) | HBV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBV DNA (log) | Pattern | GT | Viral mutations | |||||||||
| RAMs | VEM | BCP | PC | |||||||||
| 175 | M | 51 | 54 | 6.02 | 0 | 7.45 | 2 | A | WT | WT | A1762T/G1764A | WT |
| 329 | F | 40 | 499 | 3.19 | 60* | 7.39 | 2 | A | L180M, M204V | WT | WT | WT |
| 2219 | M | 58 | 94 | 4.66 | 0 | 7.37 | 2 | E | WT | WT | WT | WT |
| 2709117 | M | 45 | 10 | 5.52 | 0 | 7.40 | 3a | E | WT | WT | WT | G1896A |
| MIMI | F | 50 | 618 | <1.24 | 4* | 5.52 | 3a | A | WT | T126I | WT | G1896A |
| 841 | F | 46 | 752 | <1.24 | 98* | 4.52 | 3a | A | WT | WT | WT | WT |
| 2337 | M | 42 | 34 | 6.45 | 0 | 4.17 | 3a | A | WT | WT | WT | WT |
| 2370 | F | 53 | 426 | 4.44 | 0 | 4.15 | 3a | E | WT | WT | A1762T/G1764A | G1896A |
| 1213 | M | 40 | 666 | 3.42 | 67* | 3.44 | 3a | E | WT | WT | nd | nd |
| 2249 | M | 46 | 37 | 4.40 | 0 | 3.06 | 3b | E | WT | P120T | WT | WT |
| 1341 | F | 36 | 618 | <1.24 | 34* | 3.02 | 3b | A | WT | WT | nd | nd |
| 2543 | M | 40 | 266 | <1.24 | 6** | 2.94 | 3b | A | WT | WT | nd | nd |
| 1543 | F | 49 | 318 | 5.21 | 30* | 2.79 | 3b | A | L180M, M204V | WT | nd | nd |
| 914 | F | 38 | 618 | 3.89 | 12* | 2.52 | 3b | A | L180M, M204V | WT | nd | nd |
| 2589 | F | 34 | 405 | 4.0 | 4** | 2.47 | 3b | A | WT | WT | nd | nd |
| 596 | F | 31 | 227 | <1.24 | 53* | 2.43 | 3b | A | WT | WT | WT | WT |
| 2333 | M | 46 | 372 | <1.24 | 11* | 2.39 | 3b | E | WT | WT | A1762T/G1764A | G1896A |
| 999 | F | 33 | 970 | <1.24 | 58* | 2.03 | 3b | A | L180M, M204V | WT | WT | WT |
| 1956 | F | 30 | 258 | nd | 0 | 4.84 | 4a | A | L180M, M204V | WT | nd | nd |
| 98 | F | 50 | 207 | 6.41 | 85* | 4.41 | 4a | E | WT | WT | A1762T/G1764A | G1896A |
| 1773 | F | 38 | 533 | <1.24 | 21* | 3.82 | 4a | A | L180M, M204I | WT | nd | nd |
| 1576 | M | 35 | 533 | 3.09 | 0 | 3.50 | 4a | A | WT | WT | WT | G1896A |
| 803 | M | 44 | 208 | 5.59 | 46* | 2.80 | 4a | A | L180M/M204I | WT | nd | nd |
| 2491 | F | 29 | 277 | 4.90 | 0 | 2.73 | 4a | A | L180M/M204I | WT | nd | nd |
| 926 | F | 70 | 488 | 3.56 | 0 | 2.72 | 4a | A | L180M/M204V | WT | nd | nd |
| 754 | F | 37 | 22 | 5.07 | 81* | 2.69 | 4a | A | WT | WT | nd | nd |
| 92 | M | 46 | 316 | 3.84 | 19** | 2.63 | 4a | E | WT | WT | nd | nd |
| 745 | M | 40 | 547 | <1.24 | 32* | 2.63 | 4a | E | WT | WT | nd | nd |
*3TC-based ART;
**TDF-based ART.
Abbreviations: ART, antiretroviral treatment; F, female; M, male; GT, genotype; BCP, basal core promoter; PC, precore; WT, wild type; nd, not done.
Regarding influence of mutations in the polymerase on the S-gene, no mutation implicated in vaccine escape was found in all genotypes.
The BCP/PC region was successfully sequenced in 14 strains (failed PCR in five samples and insufficient volumes in nine). One HBV/A strain (1/8, 12.5%) had BCP mutations whereas three (3/6, 50%) HBV/E strains showed the A1762T and G1764A double mutation (P = .35) (Table 2). The classical PC G1896A mutation was found in two (2/8, 14%) and four (4/6, 66.7%) HBV/A and HBV/E isolates, respectively (P = .31). All 14 sequences are available in the GenBank (accession numbers KT724763 to KT724776).
Discussion
Our study provided a comprehensive picture of HBV patterns identified in a large group of HIV-1-positive patients in Gabon who were mostly receiving 3TC-containing ART without TDF. We found herein prevalence rates of HBeAg-positive CHB, HBeAg-negative CHB, inactive HBV carriage and OBI of 1.2%, 2.1%, 5.9% and 8.0%, respectively. In our relatively old population, HBsAg-positive patients in the HBeAg-negative phase were approximately 6 times more frequent than those who were in the HBeAg-positive window, likely reflecting the natural history of HBV infection in sub-Saharan Africa where most individuals are infected at childhood by horizontal transmission [34].
As recommended by WHO, our survey supports the universal use of TDF because no significant difference was obtained in HBV DNA levels between patients receiving 3TC-containing regimens versus untreated individuals, regardless of HBV patterns. When considering HBV/A genotypes only, we observed a high (~50%) prevalence of M204V/I and L180M mutations. Our data are consistent with those obtained in Malawi reporting HBV DNA VL rebounds coinciding with the rapid emergence of major resistance and compensatory mutations coexisting on the same HBV genomes [9]. In another study conducted in Cameroon, RAMs were detected in 60% of HIV/HBV co-infected patients at month 24 [18]. Importantly, we identified in our survey three natural M204V/I mutations in 3TC-naive patients, as already described in other settings [17]. For these three patients, we were not able to identify any previous 3TC-based treatment. Taken together, these findings highlight the urgency to abandon the use of 3TC as the sole HBV active agent and to switch to TDF in HIV/HBV co-infected patients.
Consistently with other studies conducted either in Africa [10, 35] or Asia [36], HBcAb was a reliable surrogate marker of OBI. Unexpectedly, among our patients with HBcAb ‘alone’, ~20% (44/228) showed HBV DNA levels above 100 IU/mL. Previous studies assessing HBV viremia in subjects with HBcAb alone have reported widely divergent results. For instance, Launay and colleagues reported HBV viremia exceeding 104 copies/mL in six of ten patients with OBI [37]. In contrast, Khamduang et al. obtained three (1.5%) specimens with HBV DNA levels between 101 and 1,000 IU/mL among 200 Thai HIV-positive pregnant women with OBI [36]. It is likely that high HBV DNA levels in OBI are more common during routine HIV care in RLS where unplanned 3TC-based ART interruptions are highly prevalent (>40% in our population, as previously documented) [6], and are recognized to be associated with severe HBV reactivations [38–40].
Our study revealed a highly complex molecular virology of HBV during HIV co-infection. First, a dual QS-A3 and E HBV genotypic presence was confirmed in HIV-positive patients [7, 41], suggesting that mixed infections and/or recombination events are possible in Gabon. Second, we describe for the first time in the S gene, in this country, the presence of sub-genotypes A2 and A4, highlighting the high degree of HBV/A genetic diversity in Africa. However HBV full-genome sequences are required in our country to identify recombinant A/E strains and confirm the circulation of HBV sub-genotype A2 and A4, there origin and how they integrated this country. Third, the M204V/I RAM were more common in HBV/A strains. Fourth, PC/BCP mutants were also prevalent, notably in HBV/E genotypes. Our findings suggest that HBV genotypes should be further monitored, at least in African research laboratories, because they may display specific mutational S, P and C patterns that can differentially impact on HBV treatment and prevention.
Our study has limitations, including its cross-sectional design and the absence of alanine aminotransferase (ALT) values. One important limitation was represented by the loss of significant information on HBV genotypes and mutations since we were able to analyze only 26% of HBV DNA-positive patients. It was mostly due to the unavailability of relevant plasma specimens. It made difficult the identification of the most common mutations in HBeAg-negative patients in our setting. Another strong limitation of our study was represented by OBI assessment. At the time of our survey, OBI was defined by the presence of isolated anti-HBc with detectable (<200 IU/mL) serum HBV DNA. To date, according to the last statements (Taormina definition), the diagnosis of OBI requires detection of DNA positivity in at least two different genomic regions (to avoid the overestimation of OBI due to DNA false positivity), with a sensitivity of detection of <10 copies of HBV DNA per reaction (to prevent the underestimation of OBI). However, our assay constituted a unique opportunity to quantify HBV DNA in a RLS like ours because it was specific, cheap (~25–30 USD per a test) and could be performed on an open platform that was already used for HIV-1 RNA monitoring. On the other hand, the bases for the failure of HBsAg detection, despite high (>103−104 copies/mL) HBV DNA levels and no apparent immune escape mutations, remain obscure. To explain this striking pattern, different hypothesis (such as the significant impact of HBV/E on the sensitivity of HBsAg assays, or the presence of HBV surface gene mutations located outside the sequenced region) have been previously put forward by others (Launay et al, 2011). Finally, the clinical relevance of HBV DNA levels, HBV genotypes and HBV mutations among HIV/HBV co-infected patients was not investigated according to HBV-related liver disease.
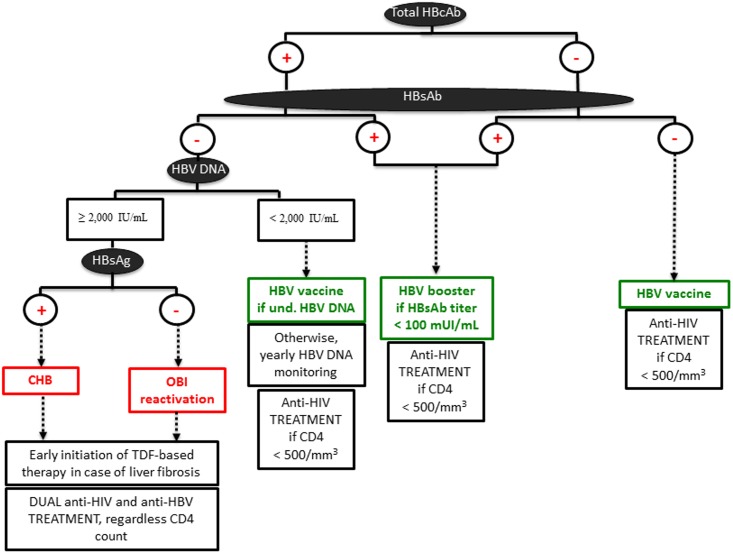
In conclusion, besides HCV [21] and HDV [7, 41], which are also highly prevalent in Gabon, we propose a new standardized diagnostic procedure for HBV screening in HIV-positive people from sub-Saharan Africa, before ART initiation (Fig 7). In this algorithm, initial testing should be done using total HBcAb, and HBsAb allowing HBV vaccine to be further implemented in patients with no exposure to HBV [42]. In addition, a booster vaccine dose may be offered in HBsAb(+) and HBcAb(+)/HBsAb(+) patients showing low (<100 IU/mL) HBsAb levels. Gabon introduced hepatitis B in its universal infant Expanded Programme of Immunization (EPI) in 2004 [43], indicating that patients in the present study were likely not immunized due to HBV vaccine but through HBV resolved infections. Patients who are found positive only for HBcAb should have an initial assessment for HBV DNA VL. If HBV DNA level is < 2,000 IU/mL, a simple VL follow-up is required. HBV vaccine should be promoted in case of undetectable VL results. In contrast, if HBV DNA VL result is ≥ 2,000 IU/mL, HBsAg should be done to differentiate patients with CHB and those with OBI activation. A dual, TDF-based, anti-HIV and anti-HBV therapy will be then initiated in these patients in case of liver fibrosis. Patients treated with 3TC will switch to TDF since prior exposure and/or resistance to 3TC does not compromise TDF-based regimens efficiency [44].
Fig 7. Suggested diagnostic strategy for HBV screening in resource-limited settings.
Acknowledgments
We are indebted to all patients who participated to this study.
Special thanks to Serge Sadeuh Mba for phylogenetic support (Centre Pasteur du Cameroun).
Data Availability
All HBV sequences are now deposited in the databank and we have modified the acknowledgments section of our manuscript accordingly. Due to ethical restrictions for the protection of patient privacy, our data are available upon request to BIVIGOU-MBOUMBA Berthold (bivigou.berthold@gmail.com), and ROUET François (franrouet@yahoo.fr).
Funding Statement
The Centre International de Recherches Médicales de Franceville (CIRMF), Gabon, is funded by the Gabonese Government, Total Gabon and the French Foreign Ministry. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Organization WH. Hepatitis B. Fact sheet N°204 from world health organization media center. Available online on december the 25 of 2014, http://www.who.int/mediacentre/factsheets/fs204/fr/. 2014.
- 2.Parvez MK. HBV and HIV co-infection: Impact on liver pathobiology and therapeutic approaches. World journal of hepatology. 2015;7(1):121–6. 10.4254/wjh.v7.i1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kourtis AP, Bulterys M, Hu DJ, Jamieson DJ. HIV-HBV coinfection—a global challenge. N Engl J Med. 2012;366(19):1749–52. Epub 2012/05/11. 10.1056/NEJMp1201796 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkins E, Nelson M, Agarwal K, Awoyemi D, Barnes E, Bhagani S, et al. British HIV Association guidelines for the management of hepatitis viruses in adults infected with HIV 2013. HIV Med. 2013;14 Suppl 4:1–71. 10.1111/hiv.12106 . [DOI] [PubMed] [Google Scholar]
- 5.Lemoine M, Eholie S, Lacombe K. Reducing the neglected burden of viral hepatitis in Africa: Strategies for a global approach. J Hepatol. 2014. Epub 2014/12/03. 10.1016/j.jhep.2014.10.008 . [DOI] [PubMed] [Google Scholar]
- 6.Liegeois F, Vella C, Eymard-Duvernay S, Sica J, Makosso L, Mouinga-Ondeme A, et al. Virological failure rates and HIV-1 drug resistance patterns in patients on first-line antiretroviral treatment in semirural and rural Gabon. J Int AIDS Soc. 2012;15(2):17985 10.7448/IAS.15.2.17985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makuwa M, Mintsa-Ndong A, Souquiere S, Nkoghe D, Leroy EM, Kazanji M. Prevalence and molecular diversity of hepatitis B virus and hepatitis delta virus in urban and rural populations in northern Gabon in central Africa. J Clin Microbiol. 2009;47(7):2265–8. 10.1128/JCM.02012-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamers RL, Zaaijer HL, Wallis CL, Siwale M, Ive P, Botes ME, et al. HIV-HBV co-infection in southern Africa and the effect of lamivudine versus tenofovir-containing cART on HBV outcomes. Jaids. 2013. Epub 2013/07/31. 10.1097/QAI.0b013e3182a60f7d . [DOI] [PubMed] [Google Scholar]
- 9.Aoudjane S, Chaponda M, Gonzalez Del Castillo AA, O'Connor J, Noguera M, Beloukas A, et al. Hepatitis B virus (HBV) sub-genotype A1 infection is characterized by high replication levels and rapid emergence of drug resistance in HIV-positive adults receiving first-line antiretroviral therapy (ART) in Malawi. Clin Infect Dis. 2014. Epub 2014/08/08. 10.1093/cid/ciu630 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attia KA, Eholie S, Messou E, Danel C, Polneau S, Chenal H, et al. Prevalence and virological profiles of hepatitis B infection in human immunodeficiency virus patients. World journal of hepatology. 2012;4(7):218–23. 10.4254/wjh.v4.i7.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell TG, Makondo E, Martinson NA, Kramvis A. Hepatitis B virus infection in human immunodeficiency virus infected southern African adults: occult or overt—that is the question. PLoS One. 2012;7(10):e45750 10.1371/journal.pone.0045750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azmi AN, Tan SS, Mohamed R. Practical approach in hepatitis B e antigen-negative individuals to identify treatment candidates. World J Gastroenterol. 2014;20(34):12045–55. 10.3748/wjg.v20.i34.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochwoto M, Chauhan R, Gopalakrishnan D, Chen CY, Ng'ang'a Z, Okoth F, et al. Genotyping and molecular characterization of hepatitis B virus in liver disease patients in Kenya. Infect Genet Evol. 2013;20:103–10. 10.1016/j.meegid.2013.08.013 . [DOI] [PubMed] [Google Scholar]
- 14.Forbi JC, Ben-Ayed Y, Xia GL, Vaughan G, Drobeniuc J, Switzer WM, et al. Disparate distribution of hepatitis B virus genotypes in four sub-Saharan African countries. J Clin Virol. 2013;58(1):59–66. 10.1016/j.jcv.2013.06.028 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pourkarim MR, Amini-Bavil-Olyaee S, Kurbanov F, Van Ranst M, Tacke F. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J Gastroenterol. 2014;20(23):7152–68. 10.3748/wjg.v20.i23.7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foupouapouognigni Y, Mba SA, Betsem a Betsem E, Rousset D, Froment A, Gessain A, et al. Hepatitis B and C virus infections in the three Pygmy groups in Cameroon. J Clin Microbiol. 2011;49(2):737–40. Epub 2010/11/26. JCM.01475-10 [pii] 10.1128/JCM.01475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirandola S, Campagnolo D, Bortoletto G, Franceschini L, Marcolongo M, Alberti A. Large-scale survey of naturally occurring HBV polymerase mutations associated with anti-HBV drug resistance in untreated patients with chronic hepatitis B. J Viral Hepat. 2011;18(7):e212–6. 10.1111/j.1365-2893.2011.01435.x . [DOI] [PubMed] [Google Scholar]
- 18.Kouanfack C, Aghokeng AF, Mondain AM, Bourgeois A, Kenfack A, Mpoudi-Ngole E, et al. Lamivudine-resistant HBV infection in HIV-positive patients receiving antiretroviral therapy in a public routine clinic in Cameroon. Antivir Ther. 2011. Epub 2012/02/01. 10.3851/IMP1911 . [DOI] [PubMed] [Google Scholar]
- 19.Lazarevic I. Clinical implications of hepatitis B virus mutations: recent advances. World J Gastroenterol. 2014;20(24):7653–64. 10.3748/wjg.v20.i24.7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q, Wang Y. Comprehensive analysis of the prevalence of hepatitis B virus escape mutations in the major hydrophilic region of surface antigen. J Med Virol. 2012;84(2):198–206. 10.1002/jmv.23183 . [DOI] [PubMed] [Google Scholar]
- 21.Rouet F, Deleplancque L, Mboumba BB, Sica J, Mouinga-Ondeme A, Liegeois F, et al. Usefulness of a Fourth Generation ELISA Assay for the Reliable Identification of HCV Infection in HIV-Positive Adults from Gabon (Central Africa). PLoS One. 2015;10(1):e0116975 Epub 2015/01/27. 10.1371/journal.pone.0116975 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Servant-Delmas A, Mercier-Darty M, Ly TD, Wind F, Alloui C, Sureau C, et al. Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J Clin Virol. 2012;53(4):338–45. 10.1016/j.jcv.2012.01.003 . [DOI] [PubMed] [Google Scholar]
- 23.Olinger CM, Venard V, Njayou M, Oyefolu AOB, Maiga I, Kemp AJ, et al. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections and recombinations. J Gen Virol. 2006;87 Part 5:1163–73. 0002371553–0012. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Aoyama K, Ohno N, Iwata K, Akahane Y, Baba K, et al. The precore/core promoter mutant (T1762A1764) of hepatitis B virus: clinical significance and an easy method for detection. J Gen Virol. 1995;76 (Pt 12):3159–64. . [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronquist F, Larget B, Huelsenbeck JP, Kadane JB, Simon D, van der Mark P. Comment on "Phylogenetic MCMC algorithms are misleading on mixtures of trees". Science. 2006;312(5772):367; author reply 10.1126/science.1123622 . [DOI] [PubMed] [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4. . [DOI] [PubMed] [Google Scholar]
- 29.Makondo E, Bell TG, Kramvis A. Genotyping and molecular characterization of hepatitis B virus from human immunodeficiency virus-infected individuals in southern Africa. PLoS One. 2012;7(9):e46345 Epub 2012/10/03. 10.1371/journal.pone.0046345 PONE-D-12-16469 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menendez-Arias L, Alvarez M, Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol. 2014;8:1–9. 10.1016/j.coviro.2014.04.005 . [DOI] [PubMed] [Google Scholar]
- 31.Kim do Y, Chang HY, Lim SM, Kim SU, Park JY, Kim JK, et al. Quasispecies and pre-existing drug-resistant mutations of hepatitis B virus in patients with chronic hepatitis B. Gut and liver. 2013;7(3):329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian T, Yan H, Shen L, Wang F, Zhang S, Cao Y, et al. Change in hepatitis B virus large surface antigen variant prevalence 13 years after implementation of a universal vaccination program in China. J Virol. 2013;87(22):12196–206. 10.1128/JVI.02127-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannoun C, Soderstrom A, Norkrans G, Lindh M. Phylogeny of African complete genomes reveals a West African genotype A subtype of hepatitis B virus and relatedness between Somali and Asian A1 sequences. J Gen Virol. 2005;86(Pt 8):2163–7. 10.1099/vir.0.80972-0 . [DOI] [PubMed] [Google Scholar]
- 34.Rouet F, Chaix ML, Inwoley A, Anaky MF, Fassinou P, Kpozehouen A, et al. Frequent occurrence of chronic hepatitis B virus infection among west African HIV type-1—Infected children. Clin Infect Dis. 2008;46(3):361–6. 0002522212–0002. 10.1086/525531 [DOI] [PubMed] [Google Scholar]
- 35.Amponsah-Dacosta E, Lebelo RL, Rakgole JN, Selabe SG, Gededzha MP, Mayaphi SH, et al. Hepatitis B virus infection in post-vaccination South Africa: Occult HBV infection and circulating surface gene variants. J Clin Virol. 2015;63:12–7. 10.1016/j.jcv.2014.11.032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khamduang W, Ngo-Giang-Huong N, Gaudy-Graffin C, Jourdain G, Suwankornsakul W, Jarupanich T, et al. Prevalence, Risk Factors, and Impact of Isolated Antibody to Hepatitis B Core Antigen and Occult Hepatitis B Virus Infection in HIV-1-Infected Pregnant Women. Clinical Infectious Diseases. 2013;56(12):1704–12. CCC:000319466600004. 10.1093/cid/cit166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Launay O, Masurel J, Servant-Delmas A, Basse-Guerineau AL, Meritet JF, Laperche S, et al. High levels of serum hepatitis B virus DNA in patients with 'anti-HBc alone': role of HBsAg mutants. J Viral Hepat. 2011;18(10):721–9. 10.1111/j.1365-2893.2011.01482.x . [DOI] [PubMed] [Google Scholar]
- 38.Bagaglio S, Porrino L, Lazzarin A, Morsica G. Molecular characterization of occult and overt hepatitis B (HBV) infection in an HIV-infected person with reactivation of HBV after antiretroviral treatment interruption. Infection. 2010;38(5):417–21. 10.1007/s15010-010-0032-1 . [DOI] [PubMed] [Google Scholar]
- 39.Costantini A, Marinelli K, Biagioni G, Monachetti A, Ferreri ML, Butini L, et al. Molecular analysis of hepatitis B virus (HBV) in an HIV co-infected patient with reactivation of occult HBV infection following discontinuation of lamivudine-including antiretroviral therapy. BMC Infect Dis. 2011;11:310 10.1186/1471-2334-11-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inuzuka T, Ueda Y, Morimura H, Fujii Y, Umeda M, Kou T, et al. Reactivation from occult HBV carrier status is characterized by low genetic heterogeneity with the wild-type or G1896A variant prevalence. J Hepatol. 2014;61(3):492–501. 10.1016/j.jhep.2014.04.033 . [DOI] [PubMed] [Google Scholar]
- 41.Makuwa M, Caron M, Souquiere S, Malonga Mouelet G, Mahe A, Kazanji M. Prevalence and genetic diversity of hepatitis B and delta viruses in pregnant women in gabon: Molecular evidence that hepatitis delta virus clade 8 originates from and is endemic in central Africa. J Clin Microbiol. 2008;46(2):754–6. 0002531003–0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaiklang K, Wipasa J, Chaiwarith R, Praparattanapan J, Supparatpinyo K. Comparison of immunogenicity and safety of four doses and four double doses vs. standard doses of hepatitis B vaccination in HIV-infected adults: a randomized, controlled trial. PLoS One. 2013;8(11):e80409 10.1371/journal.pone.0080409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ategbo S, Ngoungou EB, Koko J, Vierin Y, Zang N, C. E., Moussavou M, A. Immunization coverage of children aged 0 to 5 years in Libreville (Gabon). Sante. 2010;20:215–9. [DOI] [PubMed] [Google Scholar]
- 44.Price H, Dunn D, Pillay D, Bani-Sadr F, de Vries-Sluijs T, Jain MK, et al. Suppression of HBV by tenofovir in HBV/HIV coinfected patients: a systematic review and meta-analysis. PLoS One. 2013;8(7):e68152 10.1371/journal.pone.0068152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All HBV sequences are now deposited in the databank and we have modified the acknowledgments section of our manuscript accordingly. Due to ethical restrictions for the protection of patient privacy, our data are available upon request to BIVIGOU-MBOUMBA Berthold (bivigou.berthold@gmail.com), and ROUET François (franrouet@yahoo.fr).