Abstract
Under basal conditions, the antioxidant transcription factor NRF2 is bound to the KEAP1 protein and targeted for proteasomal degradation in the cytoplasm. In response to cellular injury or chemical treatment, NRF2 dissociates from KEAP1 and activates the transcription of protective genes and defends against injury. LH601A is a first-in-class direct inhibitor of the KEAP1-NRF2 protein-protein interaction. The purpose of this study was to determine whether LH601A activates NRF2 signaling in human kidney cells. HEK293 cells were treated with LH601A or the indirect NRF2 activator, sulforaphane (SFN) for 6 or 16 h. SFN and LH601A up-regulated NRF2 target genes HO-1 (2- to 7-fold), TRX1 (2-fold) and NQO1 mRNAs (2-fold). Both compounds also elevated HO-1 and TRX1 protein expression. Since NRF2 activation can protect tissues from injury, LH601A, a direct inhibitor of the KEAP1-NRF2 interaction may be used to defend against kidney injury and/or diseases.
Keywords: NRF2, KEAP1, nephrotoxicity, kidney, HO-1
INTRODUCTION
Nuclear factor (erythroid-derived 2)-like 2 (NRF2) is a transcription factor that mediates an important signaling pathway that prevents injury and disease. Under normal conditions, NRF2 is found in the cytoplasm bound to the Kelch-like ECH-associated protein 1 (KEAP1) and degraded. Following conditions of oxidative stress or pharmacological intervention, NRF2 dissociates from KEAP1 and translocates to the nucleus and activates the transcription of protective genes to defend against organ injury (1, 2). Examples of NRF2 cytoprotective target genes include NAD(P)H quinone oxidoreductase (NQO1), heme oxygenase-1 (HO-1), thioredoxin-1 (TRX1), glutathione reductase (GSR), and glutamate cysteine ligase (GCLC/GCLM) (3). These genes improve glutathione synthesis, detoxify reactive intermediates and metabolites, and quench reactive oxygen species.
NRF2 activators have been demonstrated to protect against acute and chronic kidney injury including cisplatin- and iron-induced nephrotoxicities (4–6), diabetic nephropathy (7, 8), and lupus-like disease (9, 10). These NRF2 activators include phytochemicals such as sulforaphane (SFN) and synthetic triterpinoids such as CDDO-Im and CDDO-Me (bardoxolone methyl). The proposed mechanism for activation of NRF2 involves covalent modification of sulfhydryl groups in critical cysteine residues of KEAP1 and subsequent prevention of the E3 ligase activity of the Cul3-KEAP1 complex (11). Recently, LH601A was identified as a direct inhibitor of the protein-protein interaction between KEAP1 and NRF2 at the ETGE motif of NRF2 using a polarization screening assay (12, 13). The advantage of a direct inhibitor of the KEAP-NRF2 interaction is a more selective mechanism of action with lower propensity for off-target properties through activation or repression of other pathways (14). The importance of avoiding off-target effects with novel NRF2 activators became apparent in the BEACON Phase III trial, which evaluated bardoxolone methyl for the treatment of chronic kidney disease in type 2 diabetic patients. The trial was terminated early due to an increased incidence of cardiac adverse events including hospitalization for heart failure (15, 16). Subsequent studies pointed to disruption of endothelin secretion as a mechanism for this adverse event (15). Supporting this hypothesis is the fact bardoxolone can decrease endothelin-1 protein expression during basal and inducible conditions in rat proximal tubule cells (15). The ability of bardoxolone to repress NFκB signaling as opposed to activation of the NRF2 pathway likely mediated disruption of endothelin secretion (15).
The purpose of this study was to determine whether LH601A stimulates NRF2 nuclear translocation and activates downstream signaling in human kidney cells. The development of specific NRF2 activators such as LH601A could create treatment options for acute and chronic kidney diseases with reduced off-target effects.
MATERIALS AND METHODS
Cell Culture
Human embryonic kidney 293 (HEK293) cells were obtained from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco's Modified Eagle Media (Life Technologies, Carlsbad, CA). Cells were cultured in a humidified 37°C incubator with 5% carbon dioxide.
Immunohistochemistry
HEK293 cells were seeded on chamber slides (Thermo Fisher Scientific, Waltham, MA) overnight and treated with 0.1% DMSO, 5 µM sulforaphane (SFN) or 100 µM LH601A for 3 h. After treatment, the cells were fixed in 4% paraformaldehyde for 5 min, then washed and blocked with 5% goat serum in 0.1% Triton-phosphate buffered saline (PBS/T) for 60 min. The cells were subsequently incubated with the primary antibody against NRF2 (H-300, sc-13032, 1:50 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C overnight. Following washing, the cells were incubated with goat anti-rabbit AlexiFluor 549 IgG (Life Technologies) for 60 min at room temperature. Nuclei were stained by DAPI. The images were analyzed using a ZERSS fluorescence inverted microscope and ProgRes capture software (GöHingen, Germany). Negative controls without primary antibody were included in the analysis (data not shown). Images were uniformly adjusted for balance, brightness and contrast using Adobe Photoshop CS3 (Adobe Systems Incorp, San Jose, CA).
RNA Isolation and Quantitative PCR (qPCR)
HEK293 cells were seeded in 6-well plates and treated with 0.1% DMSO, 5 µM SFN or 50 or 100 µM LH601A for 6 or 16 h. Total RNA was isolated using RNABee reagent (Tel-Test Inc) according to the manufacturer’s protocol. The concentration of total RNA was quantified by UV spectrophotometry at 260/280 nm using a Nanodrop spectrophotometer 2000 (Thermo Fisher Scientific). The messenger RNA (mRNA) expression of human HO-1, NQO1, TRX1, GCLC, GCLM, and GSR was quantified by qPCR using Sybr Green as detection of amplified products in the ABI 7900HT PCR system (Applied Biosystems, Carlesbad, CA). Primer sequences have been provided in Table 1. Ct values were converted to ΔΔCt by comparing to a reference gene β-actin.
Table 1.
qPCR Primers for Human NRF2 Target Genes.
| Symbol | Forward | Reverse |
|---|---|---|
| GCLC | CCCTCGCTTCAGTACCTTAAC | GACAGCAATTGCCCATTCCA |
| GCLM | AGTGGGCACAGGTAAAACCA | CTCGTGCGCTTGAATGTCAG |
| GSR | ACCCCGATGTATCACGCAGTTA | TGTCAAAGTCTGCCTTCGTTGC |
| HO-1 | CTGCTCAACATCCAGCTCTTTG | AGTGTAAGGACCCATCGGAGA |
| NQO1 | CAAAGGACCCTTCCGGAGTAA | ACTTGGAAGCCACAGAAATGC |
| TRX1 | ACGGTGATGCTGGCAATAGG | CTGGGGTGAGCTCCACCTTA |
Western Blot Analysis
HEK293 cells were seeded in 6-well plates and treated with 0.1% DMSO, 5 µM SFN, 50 µM or 100 µM LH601A for 16 h. The cells were lysed in buffer containing 20 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1% Triton 100 and 1% protease inhibitor cocktail. Cell lysates (50 µg protein/well) were separated by SDS-PAGE electrophoresis and transferred to a nitrocellulose membrane at 4°C overnight. After blocking with 5% nonfat dry milk in 0.5% PBS/T, membranes were incubated with primary antibodies against HO-1 (ADI-SPA-895, Enzo Life Sciences, Farmingdale, NY) or TRX1 (559969, BD Pharminogen, San Jose, CA) followed by incubation with anti-rabbit or anti-mouse secondary antibody, respectively, for 1 to 2 h. The SuperSignal West Dura Chemiluminescent Substrate (Thermo Fisher Scientific) was applied to the membranes prior to detection of luminescence using a FluorChem Imager (ProteinSimple, Santa Clara, CA). Target protein band intensities were semi-quantified and normalized to β-actin levels (ab8227 antibody, Abcam).
Cell Viability Assays
HEK293 cells were seeded in 96-well plates overnight and treated with increasing concentrations of LH601A (0–100 µM) for 24 h. After treatment, cell viability was determined by the alamarBlue assay (Life Technologies) with fluorescence detection at an excitation wavelength of 570 nm and emission wavelength of 585 nm.
Statistical Analysis
Data are expressed as mean ± SE. Data were analyzed by one-way ANOVA with a Newman-Keuls post-hoc test using Prism 5.0 (GraphPad Software, Inc., San Diego, CA). Statistical significance was set at p<0.05.
RESULTS
LH601A Stimulates NRF2 Expression and Nuclear Translocation in HEK293 Cells
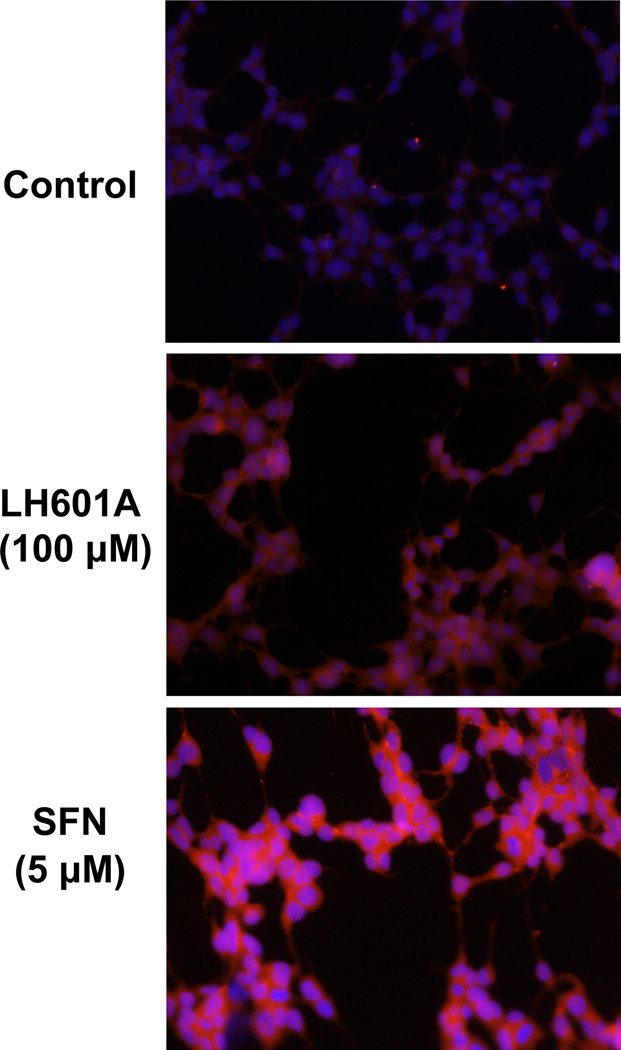
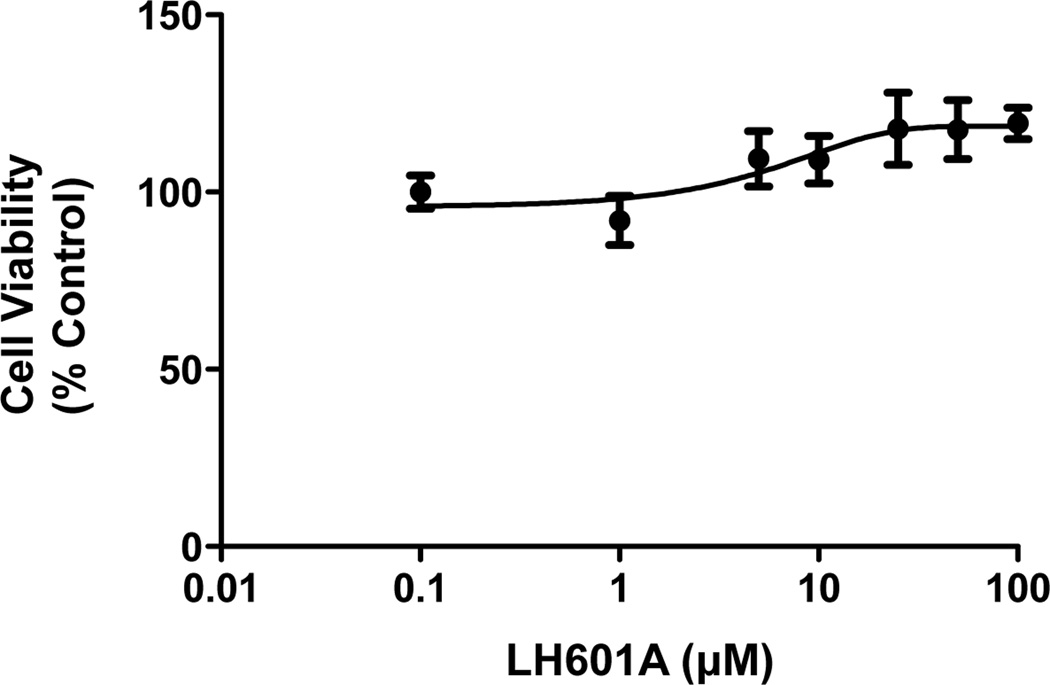
Indirect immunofluorescent staining was used to assess the relative expression and localization of NRF2 protein in HEK293 cells in response to LH601A and SFN. Cells exposed to LH601A (100 µM) and SFN (5 µM) had increased NRF2 protein staining (red staining) and nuclear accumulation (co-localization with blue DAPI staining) at 3 h compared to control cells (Fig. 1). The NRF2 staining signal was stronger with SFN than LH601A. LH601A (0–100 µM) did not alter cell viability in HEK293 cells (Fig. 2).
Fig. 1. NRF2 Expression and Nuclear Translocation.
HEK-293 cells were treated with vehicle, LH601A, and SFN for 3 h, fixed, and stained for NRF2 expression (red staining) and nuclear localization (blue staining). Images were acquired at 20X magnification.
Fig. 2. Cell Viability.
HEK-293 cells were treated with LH601A (0.1–100 µM) and cell viability was assessed at 24 h. Data are presented as mean ± SE (n=4–6).
LH601A Induces NRF2 Target Genes in HEK293 Cells
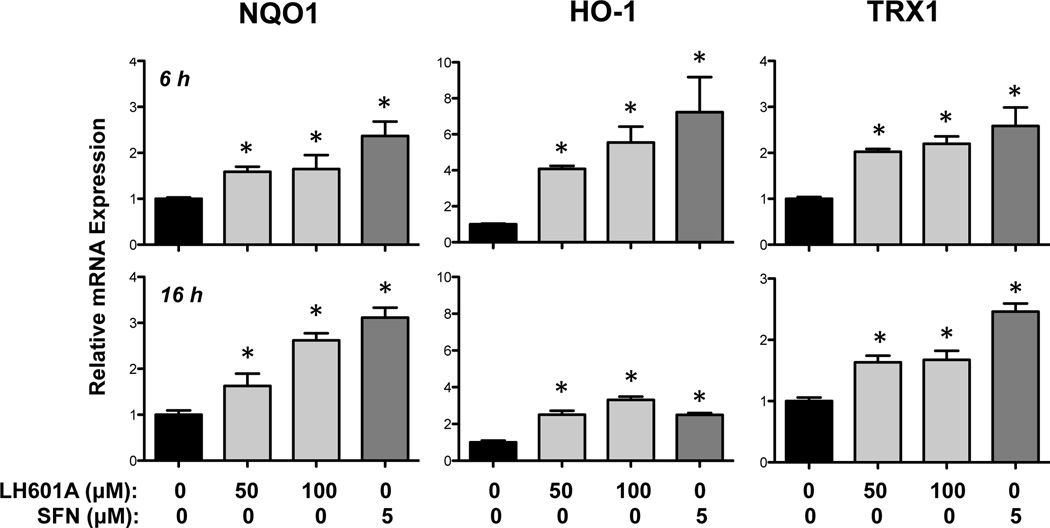
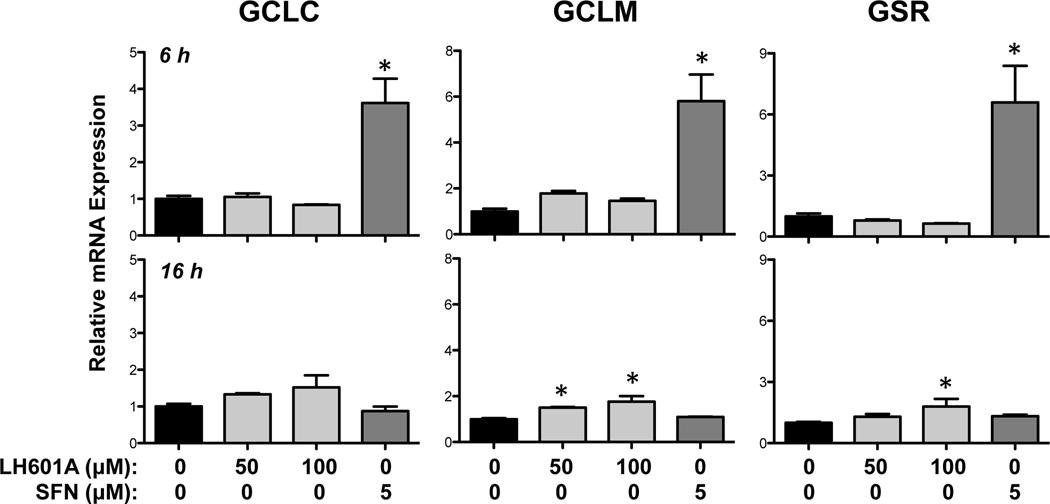
The ability of LH601A (50 and 100 µM) to enhance expression of NRF2 target genes was assessed at 6 and 16 h (Figs. 3 and 4). LH601A and SFN increased NQO1 and TRX1 mRNAs between 2- and 3-fold at both 6 and 16 h (Fig. 3). Both NRF2 activators enhanced HO-1 mRNA expression between 4- and 7-fold at 6 h, with lesser fold-changes at 16 h. Interestingly, LH601A had little to no effect on the regulation of glutathione-related genes whereas SFN enhanced GCLC, GCLM, and GSR mRNAs between 3- and 6-fold at 6 h (Fig. 4).
Fig. 3. Regulation of NRF2 Target Genes NQO1, HO-1, and TRX1.
Messenger RNA expression was quantified by qPCR in HEK-293 cells at 6 and 16 h after exposure to vehicle, LH601A, and SFN. Data are presented as mean ± SE (n=3–9). Asterisks (*) represent statistically significant differences (p < 0.05) compared to vehicle controls.
Fig. 4. Regulation of NRF2 Target Genes GCLC, GCLM, and GSR.
Messenger RNA expression was quantified by qPCR in HEK-293 cells at 6 and 16 h after exposure to vehicle, LH601A, and SFN. Data are presented as mean ± SE (n=3–9). Asterisks (*) represent statistically significant differences (p < 0.05) compared to vehicle controls.
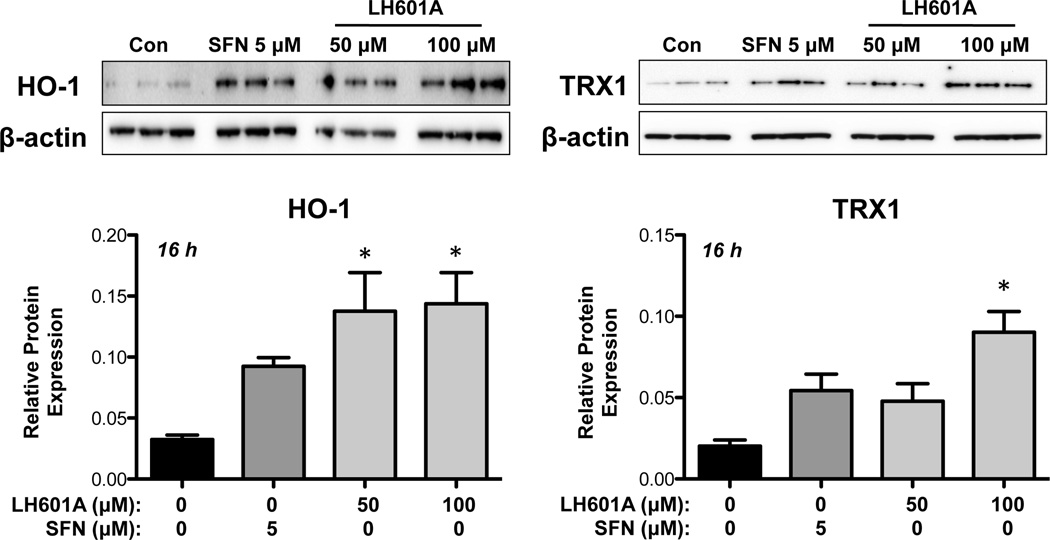
LH601A Induces HO-1 and TRX1 Proteins in HEK293 Cells
In addition to up-regulating mRNA expression, LH601A enhanced HO-1 and TRX1 protein expression at 16 h (Fig. 5). A similar HO-1 induction (3.5-fold) was observed with 50 and 100 µM LH601A, whereas SFN increased HO-1 protein by 2.5-fold. TRX1 protein levels were enhanced 2-fold by SFN and LH601A (50 µM) and 4-fold at the higher concentration of LH601A (100 µM). NQO1 protein was not detected in HEK293 cell lysates (data not shown).
Fig. 5. Regulation of HO-1 and TRX1 Proteins.
Protein expression of HO-1 and TRX1 protein was semi-quantified in HEK-293 cells at 16 h after exposure to vehicle, LH601A, or SFN. Representative western blot images and semi-quantified data are shown. Data are presented as mean ± SE (n=3). Asterisks (*) represent statistically significant differences (p < 0.05) compared to vehicle controls.
DISCUSSION
The present study investigated the ability of LH601A, a novel inhibitor of the KEAP1-NRF2 interaction, to activate NRF2 signaling in cultured human kidney cells. We have shown that LH601A-treated cells had increased NRF2 protein and nuclear localization compared to vehicle treatment. LH601A was shown to up-regulate a subset of NRF2 target genes including NQO1, HO-1, and TRX1. Despite lower HO-1 mRNA expression with LH601A compared to SFN, HO-1 protein changes were detected at both the 50 µM and 100 µM LH601A concentrations. However, LH601A had minimal effects on the expression of glutathione-related NRF2 targets, with only modest induction of GCLM and GSR at 16 h. The exact mechanism for the contrary glutathione expression profiles between SFN and LH601A is not known but may be related to the different mechanisms by which they alter the interaction of KEAP1 and NRF2, namely alterations in MAP kinase activity for SFN (17) and direct inhibition of the ETGE motif binding for LH601A (12). Additional NRF2 targets, including glutathione S-transferase a1, aldo ketoreductase 1c1, and the multidrug resistance-associated proteins 1–4 were assessed in the current study; however, baseline expression was low in HEK293 cells or not inducible by SFN (data not shown).
The findings in this manuscript hold promise for the development of specific NRF2 activators that work by directly blocking the interaction between NRF2 and KEAP1 to prevent degradation of the complex by the ubiquitin-proteasome system. While initial interest led to the investigation of renal NRF2 as a target to treat chronic kidney disease in diabetic patients, there has recently been a generalized lack of enthusiasm for this approach secondary to the cardiac adverse effects reported in a recent Phase III study (18). However, there is the potential for drugs such as LH601A to exert kidney protection, while avoiding off-target toxicities associated with repression of NFκB signaling. Additionally, the potential global therapeutic role for NRF2 specific drugs is great given the role of NRF2 signaling in other organ systems. For example, mice lacking Nrf2 are more susceptible to ultraviolet B-induced inflammation of the skin, doxorubicin-induced cardiotoxicity, iron-induced hepatotoxicity, and corneal epithelial wounds as compared to their wild-type counterparts (19–22).
The results of this study showed that relatively high concentrations of LH601A (50 or 100 µM) were needed to up-regulate NRF2 target genes, demonstrating the low potency of LH601A. Additional research will be necessary to identify the structure-activity relationships within LH601A and its analogs that are critical for KEAP1 inhibition. The effects of structural modifications on potency can then be fully evaluated. Nonetheless, this proof-of-concept study demonstrates the utility of a specific KEAP1 inhibitor as a targeted approach to activate NRF2 signaling in human kidney cells. Future studies will employ primary human cells including tubule cells, mesangial cells and podocytes, in addition to studies in preclinical animal models that represent a variety of kidney pathologies to characterize the ability of LH601A and related compounds to activate NRF2 signaling.
Acknowledgments
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK080774, DK093903] and the National Institutes of Environmental Health Sciences [Grants ES020721, ES005022], components of the National Institutes of Health. Funds were also provided to Gabriell Thorne from the Society of Toxicology Intern Program.
Non-Standard Abbreviations
- HEK293
human embryonic kidney 293 cells
- HO-1
heme oxygenase-1
- KEAP1
Kelch-like ECH-associated protein 1
- NRF2
Nuclear factor (erythroid-derived 2)-like 2
- NQO1
NAD(P)H quinone oxidoreductase 1
- SFN
sulforaphane
- TRX1
thioredoxin 1
REFERENCES
- 1.Huang HC, Nguyen T, Pickett CB. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc Natl Acad Sci U S A. 2000;97(23):12475–12480. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275(21):16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz S, Pergola PE, Zager RA, Vaziri ND. Targeting the transcription factor Nrf2 to ameliorate oxidative stress and inflammation in chronic kidney disease. Kidney Int. 2013;83(6):1029–1041. doi: 10.1038/ki.2012.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka Y, Aleksunes LM, Goedken MJ, Chen C, Reisman SA, Manautou JE, Klaassen CD. Coordinated induction of Nrf2 target genes protects against iron nitrilotriacetate (FeNTA)-induced nephrotoxicity. Toxicol Appl Pharmacol. 2008;231(3):364–373. doi: 10.1016/j.taap.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aleksunes LM, Goedken MJ, Rockwell CE, Thomale J, Manautou JE, Klaassen CD. Transcriptional regulation of renal cytoprotective genes by Nrf2 and its potential use as a therapeutic target to mitigate cisplatin-induced nephrotoxicity. J Pharmacol Exp Ther. 2010;335(1):2–12. doi: 10.1124/jpet.110.170084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu QQ, Wang Y, Senitko M, Meyer C, Wigley WC, Ferguson DA, Grossman E, Chen J, Zhou XJ, Hartono J, et al. Bardoxolone methyl (BARD) ameliorates ischemic AKI and increases expression of protective genes Nrf2, PPARgamma, and HO-1. Am J Physiol Renal Physiol. 2011;300(5):F1180–F1192. doi: 10.1152/ajprenal.00353.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan SM, Sharma A, Stefanovic N, Yuen DY, Karagiannis TC, Meyer C, Ward KW, Cooper ME, de Haan JB. Derivative of Bardoxolone Methyl, dh404, in an Inverse Dose-Dependent Manner Lessens Diabetes-Associated Atherosclerosis and Improves Diabetic Kidney Disease. Diabetes. 2014;63(9):3091–3103. doi: 10.2337/db13-1743. [DOI] [PubMed] [Google Scholar]
- 8.Zheng H, Whitman SA, Wu W, Wondrak GT, Wong PK, Fang D, Zhang DD. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes. 2011;60(11):3055–3066. doi: 10.2337/db11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang T, Tian F, Zheng H, Whitman SA, Lin Y, Zhang Z, Zhang N, Zhang DD. Nrf2 suppresses lupus nephritis through inhibition of oxidative injury and the NF-kappaB-mediated inflammatory response. Kidney Int. 2014;85(2):333–343. doi: 10.1038/ki.2013.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai PY, Ka SM, Chang JM, Chen HC, Shui HA, Li CY, Hua KF, Chang WL, Huang JJ, Yang SS, et al. Epigallocatechin-3-gallate prevents lupus nephritis development in mice via enhancing the Nrf2 antioxidant pathway and inhibiting NLRP3 inflammasome activation. Free Radic Biol Med. 2011;51(3):744–754. doi: 10.1016/j.freeradbiomed.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu L, Magesh S, Chen L, Wang L, Lewis TA, Chen Y, Khodier C, Inoyama D, Beamer LJ, Emge TJ, et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1-Nrf2 protein-protein interaction. Bioorg Med Chem Lett. 2013;23(10):3039–3043. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoyama D, Chen Y, Huang X, Beamer LJ, Kong AN, Hu L. Optimization of fluorescently labeled Nrf2 peptide probes and the development of a fluorescence polarization assay for the discovery of inhibitors of Keap1-Nrf2 interaction. J Biomol Screen. 2012;17(4):435–447. doi: 10.1177/1087057111430124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev. 2012;32(4):687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin MP, Reisman SA, Bakris GL, O'Grady M, Linde PG, McCullough PA, Packham D, Vaziri ND, Ward KW, Warnock DG, et al. Mechanisms contributing to adverse cardiovascular events in patients with type 2 diabetes mellitus and stage 4 chronic kidney disease treated with bardoxolone methyl. Am J Nephrol. 2014;39(6):499–508. doi: 10.1159/000362906. [DOI] [PubMed] [Google Scholar]
- 16.de Zeeuw D, Akizawa T, Audhya P, Bakris GL, Chin M, Christ-Schmidt H, Goldsberry A, Houser M, Krauth M, Lambers Heerspink HJ, et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N Engl J Med. 2013;369(26):2492–2503. doi: 10.1056/NEJMoa1306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, Han J, Agarwal A, Kong AN. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66(17):8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 18.Chin MP, Wrolstad D, Bakris GL, Chertow GM, de Zeeuw D, Goldsberry A, Linde PG, McCullough PA, McMurray JJ, Wittes J, et al. Risk factors for heart failure in patients with type 2 diabetes mellitus and stage 4 chronic kidney disease treated with bardoxolone methyl. J Card Fail. 2014;20(12):953–958. doi: 10.1016/j.cardfail.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Wang W, Niu T, Wang H, Li B, Shao L, Lai Y, Li H, Janicki JS, Wang XL, et al. Nrf2 deficiency exaggerates doxorubicin-induced cardiotoxicity and cardiac dysfunction. Oxid Med Cell Longev. 2014;2014:748524. doi: 10.1155/2014/748524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saw CL, Yang AY, Huang MT, Liu Y, Lee JH, Khor TO, Su ZY, Shu L, Lu Y, Conney AH, et al. Nrf2 null enhances UVB-induced skin inflammation and extracellular matrix damages. Cell Biosci. 2014;4:39. doi: 10.1186/2045-3701-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva-Gomes S, Santos AG, Caldas C, Silva CM, Neves JV, Lopes J, Carneiro F, Rodrigues PN, Duarte TL. Transcription factor NRF2 protects mice against dietary iron-induced liver injury by preventing hepatocytic cell death. J Hepatol. 2014;60(2):354–361. doi: 10.1016/j.jhep.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi R, Himori N, Taguchi K, Ishikawa Y, Uesugi K, Ito M, Duncan T, Tsujikawa M, Nakazawa T, Yamamoto M, et al. The role of the Nrf2-mediated defense system in corneal epithelial wound healing. Free Radic Biol Med. 2013;61C:333–342. doi: 10.1016/j.freeradbiomed.2013.04.008. [DOI] [PubMed] [Google Scholar]