Abstract
Understanding the molecular landscape of cancer has facilitated the development of diagnostic, prognostic, and predictive biomarkers for clinical oncology. Developments in next generation DNA sequencing technologies have increased the speed and reduced the cost of sequencing the nucleic acids of cancer cells. This has unlocked opportunities to characterize the genomic and transcriptomic landscapes of cancer for basic science research through projects such as The Cancer Genome Atlas. The cancer genome includes DNA-based alterations such as point mutations or gene duplications. The cancer transcriptome involves RNA-based alterations including changes in messenger RNAs. Together the genome and transcriptome can provide a comprehensive view of an individual patient’s cancer and is beginning to impact real-time clinical decision-making. We discuss several opportunities for translating this basic science knowledge into clinical practice including a molecular classification of cancer, heritable risk of cancer, eligibility for targeted therapies, and the development of innovative genomic-based clinical trials. In this review, we outline key applications and new directions for translating the cancer genome and transcriptome into patient care in the clinic.
Keywords: Genomics, Transcriptome, Neoplasm, Gene fusion, Patient care (nlm.nih.gov/mesh)
Introduction
The molecular classification of cancer has informed novel approaches for clinical practice in oncology such as diagnosis, prognosis, and treatment decisions. In 2011, the National Research Council convened a committee to develop a framework for the precision taxonomy of human disease, with molecular classification as the foundation for advancing personalized or precision cancer medicine1, 2. Since 2003, new DNA sequencing technologies called next generation sequencing (NGS) have increased the speed and reduced the cost of sequencing cancer by nearly one million fold each3. These technologies have advanced our understanding of various subtypes of cancer through several national and international large-scale basic science cancer profiling efforts4, 5. Since 2011, clinicians have begun to translate genome and transcriptome sequencing approaches for patients with cancer in the clinic6. In this review, we will discuss the current and future impact of genome and transcriptome sequencing for patient care in clinical oncology.
Omics and defining the cancer genome and transcriptome
Through technology innovations that have miniaturized and parallelized laboratory tests to allow testing of thousands of molecules, we have entered the “Omics Era.” Omics refers to collection and analysis of large data sets of biologic variables or phenomenon, and is regularly applied to genes (genomics), proteins (proteomics), and their subunits including nucleotides, amino acids (metabolomics). Prior to the Omics Era, researchers have studied the traditional paradigm where genes encode a sequence of nucleotides that is transcribed into a messenger RNA (mRNA). These mRNA are processed by ribosomes where the sequence information is translated into a protein with the sequential addition of transfer RNAs (tRNA) and their associated amino acids. Collectively, proteins can affect biological processes such as metabolism by functioning as enzymes, structural proteins, or by regulating genes themselves through transcription or translation. Today, rather than studying a few genes, transcripts, or proteins, researchers can utilize new technologies to rapidly test or evaluate 1,000 to 10,000’s of data points.
The human genome includes two haploid sets of 23 chromosomes each comprised in total of six billion nucelotides7. One haploid set of chromosomes encodes approximately 20,000 genes that are transcribed into RNAs including the classical messenger RNAs (mRNA), ribosomal RNAs (rRNA), and transfer RNAs (tRNA). Collectively, these RNAs constitute the transcriptome. The mRNA sequence provides the recipe for a protein and is translated through ribosome machinery (including rRNA and ribosomal proteins) by consecutively adding amino acids carried by tRNAs. Additional RNAs have been discovered that do not encode protein, termed non-coding RNAs (ncRNA). These ncRNAs include microRNAs and long ncRNAs and have more recently been proven to have regulatory functions affecting gene expression and protein function8. Prior to 2001, the scientific community did not have a complete road map or dictionary of the entire human genome9, 10. The Human Genome Project was initiated by the National Institutes of Health and coordinated the sequencing of the Human Genome over 14 years at a cost greater than three billion U.S. dollars11. The Project provided the necessary reference for researchers to study the role of genetics in human diseases such as cancer. The Project also inspired the development of new technologies that have changed the landscape of genomics research by accelerating the speed and reducing the cost of DNA sequencing by one million fold12.
Historical impact of Omics on clinical medicine
The impact of Omics data on precision medicine can already be seen in clinical practice today. Clinical practices includes the application of data from genes, transcripts, and proteins towards diagnosis, disease monitoring, risk determination, counseling, and development of novel therapies13. In an early application for metabolomics, Koenig et al. described the value of assessing glycated hemoglobin (hgb A1c) as an every day metabolic measure of long term glucose levels for patients with glucose intolerance or diabetes14. Factor V Leiden is a genetic risk factor that occurs in 5% of North American Caucasians as a heterozygote mutation and is routinely applied towards risk assessment for thrombosis, and has led to the avoidance of prothrombotic drugs or prophylaxis recommendations in high-risk situations for patients who are at increased risk. Genetics has also made an impact on the most common form of dementia, Alzheimer’s dementia, with the discovery of several genetic factors involved in this disease15. Mutations in the gene for Apolipoprotein E have been identified as a risk factor for late-onset Alzheimer’s dementia, and research efforts are underway to study other genetic factors16. The genetic basis of cystic fibrosis was described in 1989 and 3–4% of Caucasians are carriers for this disease. Early on genetic testing has been important for counseling and diagnosis, more recently metabolic research has led to the development of novel therapies17. In cancer, Omics research led to the application of chromosome karyotyping of leukemias that guides diagnosis, risk stratification, and therapy selection. The discovery of the Philadelphia chromosome In chronic myeloid leukemia and subsequent characterization of the BCR-ABL1 gene translocation would pave a path for the development of imatinib, the first smart drug for cancer18. The majority of these Omics discoveries preceded the Human Genome Project (1989–2002) which has opened new doors for genomic medicine.
Next generation sequencing technologies and cancer
DNA sequencing has consistently relied on utilizing one strand of DNA as a template to synthesize the other strand by adding complementary nucleotides with the enzyme DNA Polymerase. The first method to determine which nucleotides are added was described in 1977 and involved termination of the sequencing reaction with nucleotides that cannot allow further DNA synthesis (so called Sanger or dideoxy termination)19. This strategy relied on radiographic detection of radioactively-labeled nucleotides and gel electrophoresis, and over time was supplanted for practical reasons by fluorescent-labeled nucleotides and electrophoresis in small capillary tubes. Thus, the Human Genome Project was completed with capillary sequencers. In 2005, new technologies known as next generation sequencing (NGS) miniaturized and parallelized the sequencing process to improve yield and reduce cost3, 12. All NGS methods start by fragmenting DNA into small segments, followed by the addition of adaptors that allow the fragment to be sequenced. The addition of specific nucleotides can be measured by light (Illumina, Pyrosequencing) or pH changes (Ion Torrent). The resulting sequence data is comprised of millions of pieces ranging in length from 50 to 250 base pairs and must be matched and assembled to a reference genome or map and this is akin to a jigsaw puzzle. Base pairs that are different from the expected reference may be mutations. This field is called bioinformatics data analysis and utilizes high performance computing to process this so-called big data set. In addition to DNA sequencing, RNA can be similarly sequenced by first converting RNA into complementary DNA (cDNA) using the reverse transcriptase enzyme, and then following the same procedure for DNA. Today, there are third generation technologies for NGS that have effectively decreased the cost of sequencing by one million fold since the Human Genome Project. Technologies for NGS are further detailed and reviewed elsewhere3.
Cancer genome (DNA) sequencing
Two major collaborative efforts, The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC), have utilized NGS to profile the landscape of the 30 most common cancer types. Each of these efforts has collected primary tumors from surgical resections for tumor sequencing. In addition to these collaborative multi-center projects, there are numerous independent research groups that have contributed to cancer genomic profiling for cancers subtypes with lower prevalence. In general, rather than sequencing the whole genome, these profiling projects have focused on 1% of the genome containing the approximate 20,000 known genes, also known as the whole exome. Exome sequencing uses probes or baits to allow NGS platforms to focus on DNA fragments containing the 20,000 known genes, thereby reducing the cost of sequencing by 100-fold. Despite the improvements in cost and throughput for NGS, whole genome sequencing remains expensive for both basic and clinical research applications. Today, the majority of basic and clinical applications use a targeted or whole exome approach20.
These large-scale cancer-profiling projects have revealed a landscape of the cancer genome that includes a diverse variety of genomic alterations including point mutations, copy number variation, and translocations. These genomic alterations can affect a variety of cellular processes from cell signaling and metabolism to gene expression21. Point mutations are single base pair substitutions that can change the function of a gene’s protein product. One example is the clinically relevant BRAF oncogene mutation V600E (amino acid change from valine to glutamic acid) that occurs commonly in melanoma and leads to constitutive activation compared to the wild type gene22–25. Copy number variation refers to either extra or missing copies of a gene. Tumors with copy number amplification have extra copies beyond the expected two genes, such as 50–100 copies of HER2 (ERBB2) as seen in approximately 20% of breast cancer26–28. Alternatively, loss of gene copies, also known as deletion, often occurs in tumor suppressor genes such as PTEN in prostate and other cancers29. Translocations or gene rearrangements involve two genes that are brought together and have a new function as a chimera. These events can create new functions for the gene fusion or inactivate functions. Gene fusions in cancer commonly involve kinases (enzymes that have phosphorylating activity) and transcription factors that are deregulated by the fusion event. With the advent of chromosomal karyotyping and banding in the 1960s, the first appearances of chromosomal rearrangements were uncovered in hematological malignancies and sarcomas due to the ability to easily obtain tumor tissue and metaphase chromosomes in these cancers. Visible chromosomal rearrangements in lymphomas, leukemias, and sarcomas facilitated the identification of novel oncogenes and tumor suppressors in cancer and characterization of their role in cancer biology and clinical applications30. As an example, chronic myeloid leukemia, characterized by the Philadelphia chromosome and BCR-ABL1 gene rearrangement31, 32, would later become a model for understanding kinases and the application of targeted therapies for treatment of cancer33. Meanwhile, outside of sarcomas and select solid tumors, there initially was a paucity of oncogenic gene fusions or translocations recognized in solid tumors. This was largely due to limited tissue access and technological restrictions, however it was predicted that gene fusions would be recurrent genomic alterations in solid tumors34. Since 2005, cancer genome and transcriptome sequencing has revealed additional clinically relevant novel gene fusions in solid tumors35.
Cancer transcriptome profiling
The transcriptome is comprised of “classical” RNAs (mRNA, rRNA, and tRNA) as well as there are multiple subtypes of noncoding RNA (microRNAs and long ncRNAs) that have been discovered to have novel regulatory functions in cell biology8, 36. Gene expression can be characterized using earlier microarray technology or the more recent transcriptome sequencing (RNAseq) methods. Transcriptome sequencing has significant advantages including precise detail about base pairs and ability to detect novel RNAs that cannot be detected on microarrays. For clinical applications of the cancer transcriptome, efforts have focused on using gene expression to classify cancer subtypes (that differ with regard to prognosis and response to specific treatments) and detection of gene fusions or rearrangements. In routine clinical practice, fluorescence in situ hybridization (FISH) and RT-PCR is used to detect gene rearrangements but is limited by only testing for one gene at a time. Hence, the advantage of sequencing approaches is the ability to detect multiple gene rearrangements as well as novel ones. Although genome sequencing can detect fusions, whole genome sequencing of cancer remains costly, and RNAseq is a fraction of the cost of whole genome sequencing, and has been applied with new bioinformatics approaches to detect fusions. Using a paired-end sequencing approach of RNA, gene fusions that are expressed at the transcript level can be detected37, 38. More recently, Stransky et al., performed a comprehensive analysis of publically available tumor RNA sequence (RNAseq) data for nearly 7000 cancers in The Cancer Genome Atlas to catalog a diverse landscape of known and novel candidate kinase gene fusions39. Furthermore, Klijn et al., performed RNAseq on 675 cancer cell lines and similarly cataloged kinase fusions40. The application of RNAseq to detect novel clinically relevant gene fusions in cancer is still in its infancy, and we anticipate additional fusions will be discovered as the number of cancers profiled increases, especially in rare or previously uncharacterized cancer subtypes. Importantly, detection of novel gene fusions involving kinases has also lead to novel treatment opportunities and therapeutic benefit with kinase inhibitors in patients with advanced cancer35. In pediatric B cell acute lymphoblastic leukemia, Roberts et al., recently identified kinase fusions involving genes such as ABL1, JAK2, CSFR1, and NTRK3 that have corresponding targeted therapies, which opens up new treatment hypotheses for patients with this type of leukemia to be tested in clinical trials41, 42.
Gene expression signatures can be utilized to classify cancer types into molecular subsets that have clinical relevance. For example, early studies applied transcriptome profiling of B cell lymphoma using microarray technologies to further classify this disease into clusters of activated B cell (ABC) and germinal center B cell (GCB) subtypes43. ABC lymphoma bears poorer prognosis compared to GCB44. In another pivotal study of breast cancer, Perou et al., used microarray-based transcriptome profiling on primary breast cancer samples and classified this disease into five molecular subsets with biological and clinical relevance45. Moving beyond microarray technologies, RNAseq has the potential to enable study of other diverse components of the cancer transcriptome.
For example, in addition to the classical elements of the transcriptome including messenger (mRNA), ribosomal (rRNA), and transfer (tRNA) RNAs, multiple subtypes of RNA have been discovered with novel regulatory functions in cell biology. In fact, the majority of the transcriptome are non-protein encoding RNAs (ncRNAs) including but not limited to microRNAs (miRNAs)46–48, small interfering RNAs (siRNAs)49, and long noncoding RNAs (lncRNAs)36. Beyond the classical function for mRNA that encode proteins, these novel RNAs can play multiple roles in cell biology ranging from regulation of transcription, post-transcriptional events, gene silencing, translation, and protein level function50. Much like prototypical genes encoded by DNA, miRNAs are subject to genomic alterations including mutation, deletion, amplification and epigenetic modifications51. Similarly, miRNAs can function as tumor suppressor or oncogenes52. siRNA are small RNAs that mediate a highly specific gene-silencing mechanism that is conserved from nematodes and plants to mammalian biology49 and have emerged as tools for biomedical research and potential strategies for gene-silencing therapies53. Newly described lncRNA are ubiquitous in cancer, have diverse regulatory functions, and are only recently being systemically characterized54–56.
Cancer genomes and transcriptomes: What have we learned?
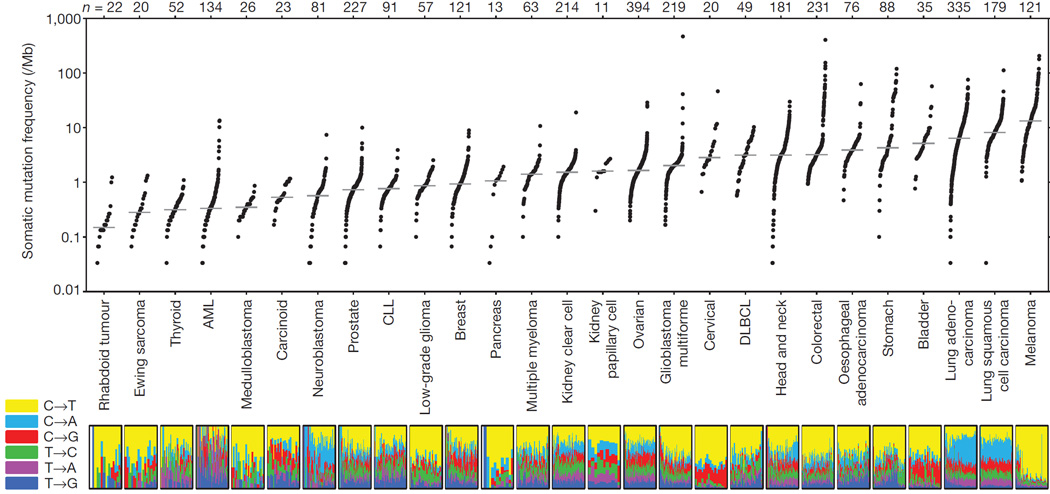
Over 10,000 cases of cancer have undergone DNA sequencing and published through collaborative projects and this has revealed diverse heterogeneity within and across cancer types classified by tissue-of-origin (e.g. breast, lung). We have also learned that signatures of mutational patterns for point mutations can aid classification through association with an underlying mechanism such as defects in DNA repair, radiation exposure, tobacco exposure57, 58. As an example, several cancer types such as lung and melanoma have abundant point mutations due to carcinogen exposures of tobacco smoke and ultraviolet radiation, respectively59. (Figure 1. With permission from Lawrence et al., reproducing Figure 1 from Nature 2012). In contrast, acute myeloid leukemia and prostate cancer generally have few point mutations, and rather have more copy number variation and gene fusions59. Ciriello et al., retrospectively assessed over 3000 cases from the TCGA and evaluated 12 types of cancer as having predominantly point mutations (M class), copy number variations (C class), while the majority are mixture of both M and C within a single disease type60. Along with the genome, the cancer transcriptome has informed classification of lymphoma and breast cancer into clinically relevant molecular subsets61. Lung cancer is an ideal example where genome and transcriptome profiling have affected measureable clinical outcomes, through moving from histology to a genomics-based classification based on point mutations (BRAF V600E), copy number alterations (MET amplification), and gene fusions(ALK fusion) that lead to treatment with matching targeted therapies62.
Figure 1. Mutational heterogeneity with and across cancer.
(Adapted from Lawrence, Nature 2013, Figure 1). Authors assessed over 3000 cases of cancer from The Cancer Genome Atlas and plotted the number of mutations identified, patterns of mutations, and grouped them by tumor tissue-of-origin. The plot illustrates that variation of point mutation burden across different cancer types and also within a given cancer type such head and neck cancers, and the importance of performing personalized genomic testing. Further, some cancers appear to be hyper-mutated with 100s of mutations such as lung and melanoma cancers, while others such as acute myeloid leukemia have few point mutations.
Moving forward to the clinic, there are several lessons to consider for translation. It has become clear that clinical decision making for cancer will require a personalized approach based on an individual’s cancer that is likely to be unique compared to other patients even with the same histologic type of cancer. Second, with slightly more than just 10,000 cases analyzed, researchers have only detected the clinically relevant mutations that represent more than 20% of common cancer types63. Therefore, based on limited sampling, we have not yet uncovered clinically relevant mutations with an estimated prevalence less than 20% in common cancers or in rare cancers that have simply not yet been sequenced yet. Therefore, more cancer sequencing data is necessary to advance a comprehensive catalog of cancer. Third, the majority of cancer genome data available are based on primary tumors rather than metastatic or advanced cancers, which may have acquired additional mutations, are cancers that behave more aggressively, and display more heterogeneity due to selective pressures from therapy. With this knowledge in hand, how are we proceeding to apply cancer genome and transcriptome biomarkers in the clinic?
Types of Molecular Biomarkers Applied in the Clinic: Diagnostic, Prognostic, and Predictive
Research discoveries derived through cancer genome and transcriptome studies have the potential for clinical impact as biomarkers64. There are three key types of biomarkers employed for clinical decision-making including diagnostic, prognostic, and predictive biomarkers. Diagnostic biomarkers facilitate identification of a cancer type or subtype. Prognostic biomarkers aid clinicians in determining the risk of relapse or disease progression after therapy, wherein patients with high risk are selected for aggressive screening or adjuvant therapy to prevent recurrence. Clinicians utilize predictive biomarkers to select one therapy over others, based on associations between biomarker results and likelihood of response to certain therapies. In practice, predictive biomarkers often identify the molecular targets of relevance to targeted anticancer drugs.
Each type of biomarker could be assayed to detect changes in a tumor’s genome (DNA), transcriptome (RNA), proteome (protein), or by phenotypic characteristics (such as histopathologic classification). As examples of methods to detect these biomarkers, BRAF gene mutation testing for melanoma is a DNA-based predictive biomarker that can guide therapy selection. For RNA-based biomarkers, FISH methods are standardly used for diagnostic subtyping of lymphoma to measure Epstein-Barr virus RNA expression. Immunohistochemistry is utilized to detect estrogen receptor protein (ER) in breast cancer and is an example of a biomarker that has both predictive and prognostic value. Oncotype Dx testing for breast cancer assesses the expression of 21 transcripts in women with node-negative estrogen-positive breast cancer is another example of a biomarker that is both predictive and prognostic, facilitating identification of patients after surgery who need further therapy (prognosis) and are most likely due to benefit from adjuvant chemotherapy (predictive)65.
However before any biomarker can be translated to the clinic for use in standard practice, the clinical utility of the biomarker must be tested through clinical trials to establish its impact and association with clinical outcomes66. The presence of ALK gene fusions in patients with metastatic lung cancer is an example of predictive biomarker for clinical response to ALK inhibitors67, 68. Preceding the use of NGS, the clinical utility of ALK gene fusions was completed in pivotal clinical trials using standard FISH methods to detect the gene fusion. However, it costly to develop FISH and Sanger sequencing tests for single genes and their relevant mutation, and NGS has been translated to the clinic to cost-effectively broaden the number of genes and type of mutations tested.
Clinical examples: whole genome (DNA) sequencing
Early efforts to employ whole genome sequencing for patients with cancer began as case-by-case research endeavors. In 2011, Welch et al., applied whole genome sequencing for a patient with acute promyelocytic leukemia (APL) which is characterized by having a gene fusion involving the retinoic acid receptor or RARA69. Patients with APL and RARA fusions are typically very sensitive to therapy (predictive biomarker) with oral all-trans retinoic acid with a significantly improved long-term survival70. However, this patient’s standard of care testing with cytogenetics and FISH did not detect the expected chromosome 15 and 17 translocation or the RARA fusion. Welch and team hypothesized that the RARA gene might be involved in a cryptic fusion that is not visible by standard cytogenetics or FISH methods and therefore evaluated the leukemia via NGS. They chose whole genome sequencing over exome sequencing since they were trying to detect potential chromosomal breakpoints that might not involve the exons tested by whole exome methods. After seven weeks of sequencing and analysis, they were able to identify a PML-RARA gene fusion, and this subsequently changed the course of treatment for the patient who received all-trans retinoic acid therapy instead of allogeneic stem cell transplantation.
In another clinical application, whole genome sequencing was able to identify a germline or heritable risk of cancer in a 37-year old woman who had personal history of ovarian cancer, breast cancer, and secondary therapy-related acute myeloid leukemia. While the patient did not have a significant family history of cancer, her clinicians were suspicious of her multiple primary cancers. Based on the personal history that suggested hereditary breast and ovarian cancer syndrome, gene testing for BRCA1 and BRCA2 was completed but no heritable genetic cause was identified. Link et al. performed whole genome sequencing of the patient’s skin biopsy and bone marrow leukemia sample, and identified a deletion in the TP53 gene which can confer a heritable risk of cancer as part of the Li-Fraumeni syndrome71. This case is illustrative of the advantages of NGS approaches to detect multiple genomic alteration types when the etiology is not apparent based on clinical presentation or standard testing. As a comparison, conventional comprehensive sequencing of BRCA1 and BRCA2 in 2011 would have cost $4,000 alone. This information has substantial clinical impact for the patient’s family where early screening measures to detect cancer are standard of care for relatives carrying the same mutation.
More recently, Demeure et al., evaluated a patient with papillary thyroid cancer whose disease was progressing despite thyroidectomy, radical neck dissections and radioiodine treatment. They performed whole genome sequencing of the patient’s thyroid tumor and identified a gene fusion involving EML4-ALK, which is a targetable fusion seen in 3–5% of lung cancer but it not commonly found in thyroid cancer72. This led to treatment with crizotinib, an oral ALK inhibitor approved for lung cancer, and stabilization of the patient’s tumor growth. This case example illustrates the advantage of NGS approaches in finding uncommon genetic changes in cases tests for the common genetic changes were negative.
These represent a sample of case reports demonstrating how whole genome sequencing technologies can affect the care of patients by providing an individualized treatment or screening plan that could affect the patient and even the family.
Clinical examples: integrating whole exome (DNA) and transcriptome (RNA) sequencing
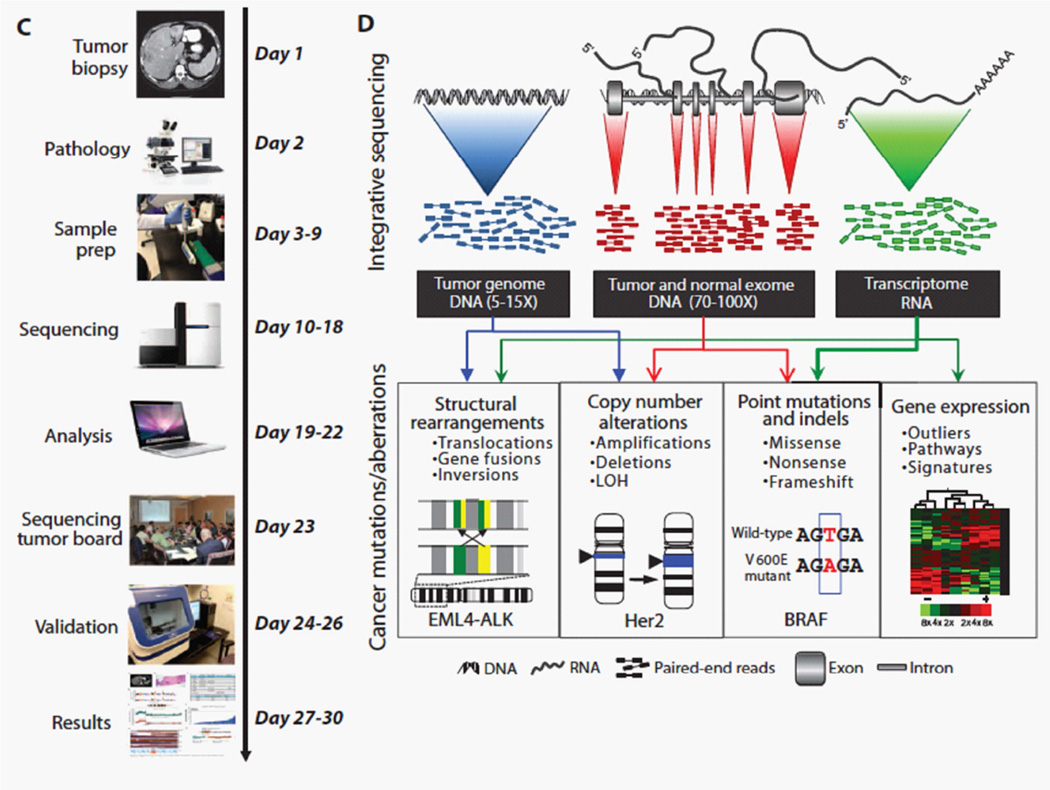
Several groups have developed trials for clinical tumor sequencing to offer patients with advanced cancer new molecular diagnostic tests to classify their tumors, gain molecular eligibility for investigational therapies in trials, and track clinical outcomes6, 73–75. In 2011, we completed a pilot study offering a combination of whole genome, whole exome, and transcriptome sequencing for patients with advanced cancer and returned clinically significant results within a clinically relevant time frame6 (Figure 2: Figure 1C+1D adapted from Roychowdhury et al., Science Translational Medicine 2011). This study demonstrated the feasibility of offering cancer genomic testing and addressed some of the early logistical challenges related to informed consent, incidental findings, and interpretation. The study also demonstrated the need for multi-disciplinary team effort required by oncologists, genomics scientists, bioinformaticians, pathologists, and genetic counselors. Currently, the majority of NGS assays are focusing on targeted DNA sequencing for 25–300 gene panels (discussed below74, 76–79), but several academic cancer centers continue to study the merits of whole exome (~20,000 genes) and transcriptome sequencing. Nevertheless, there are several advantages to be gained by incorporating RNA sequencing concurrently with DNA sequencing, including data on gene expression, enhanced variant calling, splice variants, novel RNAs, noncoding RNAs, and gene fusions6, 80 81. For example, whole transcriptome sequencing lead to the discovery of novel gene fusions involving fibroblast growth factor receptors (FGFR) occurring in an estimated 5–7% of solid tumor cancers82, 83. FGFR signaling is an important pathway for cancer biology and there are multiple inhibitors of FGFR in clinical development. The discovery of FGFR fusions subsequently led to the development of clinical trials of tyrosine kinase inhibtors ponatinib and BGJ398 to treat patients whose tumors have FGFR fusions with FGFR inhibitors (NCT02272998, NCT02160041).
Figure 2. Strategies for integrative clinical tumor sequencing.
(Adapted from Roychowdhury, Science Translational Medicine 2011, Figure 1C, 1D). This pilot study for clinical tumor sequencing demonstrated the feasibility and the need for multi-disciplinary collaboration. A, Shows a clinical relevant timeline that is dependent collaboration with oncologists, radiologists, pathologists, genetics labs, and bioinformaticians. B, Several strategies for sequencing tumor DNA and RNA can contribute to characterizing the landscape of alterations in an individual’s cancer. Whole genome, whole exome, and transcriptome sequencing can be integrated to evaluate for point mutations, copy number alterations, gene fusions, and gene expression.
To expand on this pilot study of integrative sequencing approach for clinical tumor sequencing, we collaborated with others on efforts to expand clinical cancer genomics in pediatric oncology and also overcome logistical barriers to launch a multi-center clinical study in adults. For pediatric oncology, we addressed issues for provided informed consent and assent to minors and their guardians, obtaining permission for research biopsy and tumor sequencing. This study enrolled 102 patients with refractory cancer, performed exome and transcriptome sequencing of tumor, demonstrating feasibility for pediatric oncology, and was able to identify clinically relevant alterations in 46% of patients84. In one case example, an infant who was diagnosed with spindle cell sarcoma was found to have a novel translocation involving NTRK1, which lead to treatment with crizotinib resulting in a partial response, followed by stable disease. To demonstrate feasibility for multi-center trials, our collaborative team designed a study for advanced prostate cancer deployed across multiple clinical sites and evaluated 150 men with metastatic castration-resistance prostate cancer85. These patients had progressed after receiving standard anti-androgen hormonal therapies for prostate cancer. We consented patients to tumor biopsy, tumor and germline testing with whole exome and transcriptome sequencing approaches. The study has identified pathways with known mutations, but also pathways that were previously not observed in prostate cancer including WNT pathway signaling and somatic defects in DNA repair. This international study has demonstrated multi-center feasibility and brought attention to additional considerations for scaling the volume of patients in the study related to testing (turnaround time, use centralized testing, and quality control) and availability of therapies in clinical trials.
EXPANDING CANCER GENOMIC MEDICINE
Clinical Sequencing Exploratory Research (CSER) Program: Systematically advancing genomic medicine
To help address the need to systematically apply genomics to the practice of medicine beyond a case-by-case basis, the National Human Genome Research Institute established and funded the CSER program to study and provide guidelines for bringing genomics to clinical practice (https://cser-consortium.org). The consortium includes sites that study adult and pediatric cancer, cardiovascular disease, and hereditary diseases. The projects include expertise in clinical specialties, laboratory scientists, bioinformaticians, clinical genetics, legal experts, bioethicists, and patient advocates. CSER has implemented working groups to evaluate specialized issues related to return of results, the electronic medical record, genetic counseling, informed consent, outcomes, pediatrics, phenotype measures, standards for sequencing, and cancer.
Physician and patients attitudes on genomic testing
As academic cancer centers begin to deploy clinical trials to evaluate how to deliver genomic medicine, it is vital to assess how both physicians and patients view genomic testing approaches. Gray et al. surveyed 160 physicians at an academic cancer center about use of somatic testing, and their genomic confidence. Interestingly 22% of physicians reported “low confidence in their genomic knowledge” and the authors suggested a need for guidelines and education to support understanding of genomic tests for physicians86. Miller et al. completed structured interviews with 17 physicians about genomic testing in practice and they similarly observed a need for decision tools and education to aid physicians87. Gray and Blanchette et al., completed studies by interview and questionnaire respectively of patients with cancer about genomic testing and found that patients are interested and motivated to undergo genomic testing and potentially improving their cancer care, but also learned that some patients had concerns about incidental findings, discrimination, and a need for more information or genetic counseling88, 89. Studies such as these and through the CSER Program are critical to address barriers for translating cancer genomic medicine into the larger clinical oncology and patient community.
Clinical interpretation of tumor sequencing results
Interpretation of mutations is critical for translation of genomic testing results into the clinic. There are many software tools to predict or model the potential impact of mutations in basic science research90, but there is a need for expert clinical annotation of specific mutations that provides the exact level of clinical or pre-clinical evidence that a physician will need for decision-making. When physicians receive genomic test results, they are faced with mutations in a large number of genes, across multiple pathways, and this is a significant obstacle for busy practicing oncologists who cannot keep up with the vast volumes of data that are emerging. Not all mutations are driver mutations that confer a selective advantage (to the cancer) for its survival, growth, or spread, and the majorities are so called passenger mutations or variants of unknown significance. Several databases and websites have been developed as clinical decision support tools to aid in the interpretation of mutations including MyCancerGenome (mycancergenome.com), Knowledge base for precision oncology (pct.mdanderson.org), and Cancer Driver Log (candl.osu.edu). Moving forward, CSER’s Tumor Working Group and ClinGen’s Somatic Working Group are working together to provide overarching infrastructure and leadership to support a more comprehensive database and framework for clinical interpretation of somatic mutations.
Genomic tests that inform clinical decision-making
Bringing NGS-based cancer genomic testing up to clinical grade standards to support clinical decision-making equates to understanding and following standards for molecular diagnostics. While NGS is relatively new to the molecular pathology and diagnostics community, several groups have already offered guidelines to address quality for NGS-based cancer genomic testing91–93. Assays must undergo analytic validation that includes determination of the assay’s sensitivity and specificity for detecting mutations using standards. Clinical validation of the assay refers to the broader application of the assay on clinical samples such as formalin-fixed, paraffin-embedded or frozen tumors, and association of the test results with real world clinical diagnoses66. This often includes confirmation of the test result by another assay such as Sanger sequencing, PCR, or FISH. All of this is performed in a clinical grade laboratory that has been inspected by a certifying body such as a State Department of Health or the College of American Pathologists to ensure that the labs have standard operating procedures, training, and quality assurance programs in place to deliver quality tests93. Demonstrating clinical utility of the assay is separate from building the tests with analytic validity and is subsequently completed through clinical trials that may look at clinical outcomes retrospectively or prospectively66.
Cancer gene panels and case reports
Developing NGS-based genomic tests in clinical grade laboratories has considerable constraints that include ensuring a rapid turnaround time, keeping costs of the assay down, and limiting the complexity (size) of the assay for analysis. As a consequence, many commercial and academic laboratories have developed targeted gene panels focused on 25–400 genes that are known to be important for cancer biology or disease management (Table 1). Because of the ease of testing for gene panels, thousands of patients are undergoing genomic testing with cancer gene panels in the U.S. since 2012. Ou et al., reported a patient with lung cancer who was found to have a novel ROS1 gene fusion based on a NGS test, that was missed by standard FISH or PCR approaches because it involved a novel fusion partner with TMEM106B94. Unfortunately, testing was not initiated until the patient had progressed on standard chemotherapy, and they passed from disease before receiving a ROS1 inhibitor. Chalmer et al., found a patient with a myeloid neoplasm with eosinophilia to have a novel gene fusion involving PDGFRa, who subsequently benefited from therapy with imatinib95. Once again, standard FISH testing missed this particular gene fusion since it is designed to detect a specific fusion only. Ali et al., observed a patient with metastatic kidney cancer with a TSC1 mutation, which is predicted to result in activation of MTOR signaling, and who clinically responded and benefited from MTOR inhibitors96, 97.
Table 1.
Commercial Targeted DNA Pan-Cancer NGS Assays
| Vendor | Assay Name | Number of genes |
Results | Turnaround Time |
|---|---|---|---|---|
| Foundation Medicine |
Foundation One | 315 | SNVs, CNVs, Fusions |
12–14 days |
| University of Washington |
UW-Oncoplex | 234 | SNVs, CNVs, Fusions |
6 weeks |
| ParadigmDx | PCDx | 114 | SNVs, CNVs, Fusions |
4–5 days |
| Washington University GPS |
Solid Tumor Gene Set |
48 | Hot spot mutations, 6 fusions |
3 weeks |
| ARUP Labs | Solid Tumor Mutation Panel |
48 | Hot spot mutations |
14 days |
| Caris Life Sciences |
MI Profile | 46 | Hot spot Mutations |
14 days |
| Knight Diagnostic Labs |
GeneTrails Solid Tumor Panel |
37 | Hot spots mutations |
10–14 days |
Abbreviations: SNV, single nucleotide variation or point mutation; CNV, copy number variation; NGS, next generation sequencing.
Unmet needs: Cancers of unknown primary
Cancers of unknown primary (CUP) represents 2–3% of adult cancers, up to 80,000 cases per year in the U.S., and are defined by the inability to identify the anatomic organ or tissue of origin for these patients using traditional radiologic imaging and immunohistochemical (IHC) assessment of the tumor98. Without knowledge of the tissue of origin (TOO), it is challenging for oncologists to select the appropriate treatment for these patients, and overall survival outcomes for these patients are poor99. In clinical practice, the focus has been to identify the most probable TOO based on clinical presentation and available pathologic data, recognize favorable subsets of cancer when possible, and choose therapies that match the suspected disease. Up to 20% of CUP may be favorable subsets including prostate cancer, ovarian cancer, breast cancer, germ cell tumors, or neuroendocrine cancers, all of which have established effective therapies. However, most CUP patients have tumors with poorly differentiated histology, limited markers, and no clear evidence of primary tumor origin. Consequently, empiric chemotherapy has been the standard of care with generally poor survival outcomes100. Several assays have been developed to classify the TOO on the basis of mRNA or miRNA expression signatures and may aid in choosing chemotherapy101, 102. More recently, NGS-based genomic testing assays have revealed potentially actionable genomic alterations in patients with CUP that could more directly guide selection of a targeted therapy103.
DESIGN OF CLINICAL TRIALS
Genomics-based classification of cancer has changed the outlook on how clinical trials are designed. Traditionally, an investigational agent is developed for a specific cancer type such as lung or breast cancer. Since these cancers can now be characterized and split into different molecular subsets, there is a rationale for enrolling patients for treatment based on molecular eligibility. As an example, for early trials of BRAF inhibitors in melanoma, the initial Phase 1 trial included patients with any solid tumor to determine dosing and assess toxicity and they observed that patients with BRAF V600E activating mutations were more likely to respond. In the subsequent expansion phase, only patients with BRAF mutations were enrolled, and 80% of patients had a response based on overall response rate24. Similarly, molecular eligibility with ALK gene fusions was a requirement for entry into early trials of ALK inhibitors in lung cancer104.
A special challenge for conducting such clinical trials is that the number of eligible patients with molecular eligibility is dramatically reduced, and the traditional approaches for statistical trial design may not be feasible. For example, a clinical trial for a mutation with prevalence of 1% in a common cancer type will be difficult to accrue and complete. In contrast, this may in fact represent an advantage, as a molecularly enriched trial may be more likely to have patients who respond to therapy and may display a greater magnitude of response. Traditionally, patients are randomized to receive the targeted therapy and either previous best standard therapy or placebo, but there may be insufficient numbers of patients to complete these trials. Alternative endpoints such response rate and magnitude of response105 may be necessary for rare mutations or rare cancers such as the non-randomized trial for imatinib, as a CKIT inhibitor, in patients with gastrointestinal stromal cell tumors106. Meeting the demands of trial accrual for patients with rare mutations may be partially accomplished via screening across many clinical sites and multi-center trials through networks such as the National Cancer Institute and its cooperative groups. One limitation for multi-center trials is the substantially increased regulatory cost, and reduced funding, for institutions to open trials for potentially enrolling only 0–2 patients per year, and difficulty in implementing complex correlative studies within those trials. Nevertheless, there are several examples in this regard for lung cancer. The Lung Cancer Mutation Consortium developed over a dozen pathway-based trials for lung cancer across 16 clinical sites (www.golcmc.com). Selected trials may ultimately be more feasible than others towards meeting accrual goals based on gene or mutation prevalence. More recently, the LungMap trial for squamous cell carcinoma similarly includes pathway-based trials for FGFR, PI3-Kinase, cyclin-dependent kinase pathways, and an immunotherapy arm for patients who lack an actionable driver mutation (www.lung-map.org).
Another emerging approach for trials is exclusively mutation-based and pathway-based eligibility and thus truly tumor site agnostic. One example is a so-called “basket trial” for patients with any solid tumor that has alterations in FGFRs including point mutations, amplifications, or fusions. In this Phase 2 study, patients receive an oral pan-FGFR inhibitor (ponatinib) and the endpoints are to identify clinical responders in disease or mutation subsets to guide future trials and drug development (NCT02272998). The National Cancer Institute has recently laid out a strategic plan for precision medicine trials including the MATCH program (Molecular Analysis for Therapy Choice)107. The NCI-MATCH Trials can be opened at any NCI-designated cancer center and entail centralized tumor testing for each patient at one of four genomic testing labs, and multiple trials each with eligibility for a targeted therapy that is mutation-based. While a majority of trials are tumor site agnostic, some trials could be focused on a mutation pathway in a disease group such as MTOR signaling in genitourinary cancers.
Learning from exceptional responders in trials
In addition to prospective trials to match patients to targeted therapies based on the mutations in their cancer, other efforts from clinical trials are learning from rare patients who experienced an exceptional response to a therapy, but the mechanism for that response is unknown107, 108. In this approach, clinicians make phenotypic observations in rare patients who have complete responses to a therapy for their metastatic disease, and retrospective genomic and transcriptome sequencing of the patients archival tumor can potentially reveal the underlying biology. For example, Iyer et al., evaluated a patient with metastatic bladder cancer who had a sustained complete response to an MTOR inhibitor clinical trial, while the majority of patients in that trial did not109. They performed whole genome sequencing on the patient’s archival tumor and identified a point mutation in TSC1, a tumor suppressor gene that negatively regulates MTOR signaling. Subsequently, they identified other patients who had TSC1 mutations who were more likely to have response to MTOR inhibition. In another example, Wagle et al., observed an exceptional response in a patient with metastatic urothelial cancer receiving an MTOR inhibitor and identified alterations in another gene, specifically activating point mutations in MTOR110. Each of these exceptional responder evaluations has now identified mutations that represent new treatment hypotheses for the development of MTOR inhibitors in patients with mutations in MTOR and TSC1 genes. On a national level the NCI has established an Exceptional Responders Initiative to utilize genomic sequencing to facilitate drug development for advanced cancer107. The mission of the NCI Exceptional Responders Initiative is to identify and confirm patients who have had remarkable responses to systemic therapy, and use genomic technologies to characterize their tumor to study the molecular mechanisms underlying why these patients benefit from systemic therapy, particularly chemotherapies111.
Challenges and opportunities
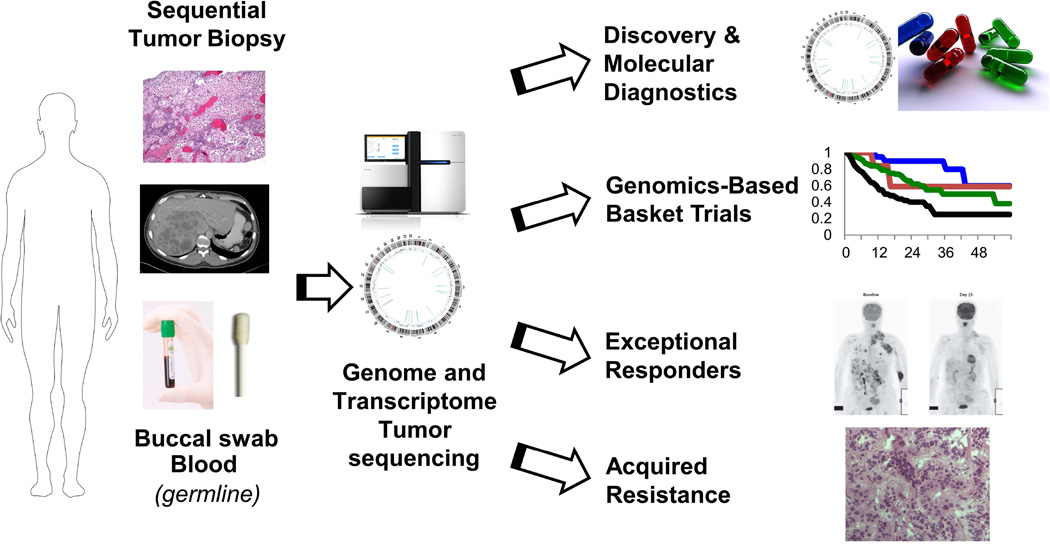
While the new framework for precision medicine in cancer has great promise, the full realization of this approach has several challenges as well as opportunities. Beyond the molecular characterization of cancer through whole and transcriptome sequencing, there are additional complexities for cancer biology to consider. First, basic science research on epigenetic alterations are continuing to reveal how methylation regulates gene expression, and can contribute to our view of individual cancers112. Second, new omics approaches for the proteome and the metabolome can reveal additional layers to cancer biology. As new “Omics” approaches became more practical, we should consider new standards for integrating data analysis and transparency in clinical trials as recommended by the Institute of Medicine’s recent assessment of translational omics113. Third, additional aspects of cancer biology including tumor heterogeneity114, mechanisms of drug resistance115, 116, the tumor microenvironment117, and stem cell properties118 of cancer can influence how patients respond to therapy. Together, these challenges can be met with new opportunities created by investment in science. Beyond genomics-guided therapy, we envision combination therapies with other modalities including immunotherapy, oncolytic viruses, and stem cell/metabolism targeting inhibitors. For immunotherapy, genomic sequencing approaches can be applied to identify the burden of neo-tumor antigen or molecular defects in DNA repair such as mismatch repair genes that may predict response to novel therapies that inhibit immune regulatory checkpoints to boost the immune response against cancer119–121. These challenges can be met through a collaborative network of innovative clinical trials with systematic collection of tumor tissue, clinical data, and transparency (Figure 3).
Figure 3. Research and clinical opportunities for precision cancer medicine through genomic and transcriptome sequencing.
A systematic framework for implementing precision cancer medicine through genome and transcriptome sequencing can support multiple clinical and research efforts. Genomics can support the development of molecular diagnostics, drug target discovery, innovative genomics-based trials, evaluation of exceptional responders, and study of mechanisms of acquired resistance.
Future directions
Integrative profiling through DNA and RNA sequencing opens new doors for both basic and clinical cancer research. Molecular classification of cancer based on genomic and transcriptome alterations may reveal novel biomarkers for diagnosis, prognosis, and predicting response to therapies. Translating the cancer genome and transcriptome for patients will require continued multi-disciplinary collaboration between oncologists, pathologists, basic scientists, and computational biologists (Table 2). Additional resources and funding are necessary to support the ongoing profiling efforts for basic genomics research, tumor sequencing in the clinic, and data sharing networks to enable precision cancer medicine.
Table 2.
Summary Points (Box)
|
Acknowledgements
Thanks to Karen Giles and Jenny Badillo for administrative support. Thanks to Julie Reeser for helpful review of the manuscript.
Funding sources
S.R. is supported by the American Cancer Society (MRSG-12-194-01-TBG), the Prostate Cancer Foundation, the Clinical Sequencing Exploratory Research (CSER) Consortium grant (UM1HG006508-01A1), Fore Cancer Research, American Lung Association, and Pelotonia. A.M.C. is supported in part by the American Cancer Society Clinical Research Professorship Grant (CRP-14-112-06-COUN), the Prostate Cancer Foundation (PCF351883), the Clinical Sequencing Exploratory Research (CSER) Consortium grant (1UM1HG006508), the National Institutes of Health S.P.O.R.E. (P50CA69568), and the Early Detection Research Network (UO1 CA113913).
Financial Disclosures
S.R. receives funding from Novartis and Ariad Pharmaceuticals for conduct of clinical trials.
S.R.’s immediate family member owns stock in Johnson and Johnson.
A.M.C. serves as a consultant for Hologic, Paradigm and Ventana-Roche and is Co-Founder of Oncofusion Therapeutics.
References
- 1.Toward precision medicine : building a knowlege network for biomedical research and a new taxonomy of disease. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 2.Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1803–1805. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 3.Mardis ER. Next-generation sequencing platforms. Annual review of analytical chemistry. 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ, Barrette T, Siddiqui J, Kunju LP, Navone N, Araujo JC, Troncoso P, Logothetis CJ, Innis JW, Smith DC, Lao CD, Kim Roberts JS, Gruber SB, Pienta KJ, Talpaz M, Chinnaiyan AM. Personalized Oncology Through Integrative High-Throughput Sequencing: A Pilot Study. Science Translational Medicine. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. N Engl J Med. 2010;362:2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 8.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 9.McPherson JD, Marra M, Hillier L, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, Adams MD, Myers EW, et al. The Sequence of the Human Genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 12.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 13.Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 14.Koenig RJ, Peterson CM, Jones RL, Saudek C, Lehrman M, Cerami A. Correlation of glucose regulation and hemoglobin AIc in diabetes mellitus. N Engl J Med. 1976;295:417–420. doi: 10.1056/NEJM197608192950804. [DOI] [PubMed] [Google Scholar]
- 15.Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 16.Green RC, Roberts JS, Cupples LA, et al. Disclosure of APOE genotype for risk of Alzheimer's disease. N Engl J Med. 2009;361:245–254. doi: 10.1056/NEJMoa0809578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 18.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark MJ, Chen R, Lam HY, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 23.Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nature reviews Clinical oncology. 2011;8:426–433. doi: 10.1038/nrclinonc.2011.69. [DOI] [PubMed] [Google Scholar]
- 24.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. New England Journal of Medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slamon DJ, Clark GM. Amplification of c-erbB-2 and aggressive human breast tumors? Science. 1988;240:1795–1798. doi: 10.1126/science.3289120. [DOI] [PubMed] [Google Scholar]
- 27.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 28.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasso CS, Wu Y-M, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 doi: 10.1038/nature11125. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haluska FG, Tsujimoto Y, Croce CM. Oncogene activation by chromosome translocation in human malignancy. Annu Rev Genet. 1987;21:321–345. doi: 10.1146/annurev.ge.21.120187.001541. [DOI] [PubMed] [Google Scholar]
- 31.Nowell PC, Hungerford D. A minute chromosome in human chronic granulocytic leukemia (abstract) Science. 1960;132:1497. [Google Scholar]
- 32.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 33.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 34.Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–253. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- 35.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 37.Maher CA, Kumar-Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maher CA, Palanisamy N, Brenner JC, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2014 doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 41.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 44.Barton S, Hawkes EA, Wotherspoon A, Cunningham D. Are we ready to stratify treatment for diffuse large B-cell lymphoma using molecular hallmarks? Oncologist. 2012;17:1562–1573. doi: 10.1634/theoncologist.2012-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 46.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 47.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 48.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu Rev Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 50.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manjunath N, Dykxhoorn DM. Advances in synthetic siRNA delivery. Discov Med. 2010;9:418–430. [PubMed] [Google Scholar]
- 54.Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 55.Prensner JR, Iyer MK, Sahu A, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015 doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discovery. 2013;3:27–34. doi: 10.1158/2159-8290.CD-12-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clinical Lung Cancer Genome P, Network Genomic M. A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 65.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 66.Parkinson DR, McCormack RT, Keating SM, et al. Evidence of clinical utility: an unmet need in molecular diagnostics for patients with cancer. Clin Cancer Res. 2014;20:1428–1444. doi: 10.1158/1078-0432.CCR-13-2961. [DOI] [PubMed] [Google Scholar]
- 67.Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 69.Welch JS, Westervelt P, Ding L, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanz MA, Grimwade D, Tallman MS, et al. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113:1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- 71.Link DC, Schuettpelz LG, Shen D, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Demeure MJ, Aziz M, Rosenberg R, Gurley SD, Bussey KJ, Carpten JD. Whole-genome sequencing of an aggressive BRAF wild-type papillary thyroid cancer identified EML4-ALK translocation as a therapeutic target. World J Surg. 2014;38:1296–1305. doi: 10.1007/s00268-014-2485-3. [DOI] [PubMed] [Google Scholar]
- 73.Mardis ER. The translation of cancer genomics: time for a revolution in clinical cancer care. Genome Med. 2014;6:22. doi: 10.1186/gm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cottrell CE, Al-Kateb H, Bredemeyer AJ, et al. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn. 2014;16:89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss GJ, Liang WS, Izatt T, et al. Paired tumor and normal whole genome sequencing of metastatic olfactory neuroblastoma. PLoS One. 2012;7:e37029. doi: 10.1371/journal.pone.0037029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nature biotechnology. 2013 doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. The Journal of molecular diagnostics : JMD. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 78.Beadling C, Neff TL, Heinrich MC, et al. Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. The Journal of molecular diagnostics : JMD. 2013;15:171–176. doi: 10.1016/j.jmoldx.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 79.Pritchard CC, Salipante SJ, Koehler K, et al. Validation and Implementation of Targeted Capture and Sequencing for the Detection of Actionable Mutation, Copy Number Variation, and Gene Rearrangement in Clinical Cancer Specimens. The Journal of molecular diagnostics : JMD. 2013 doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkerson MD, Cabanski CR, Sun W, et al. Integrated RNA and DNA sequencing improves mutation detection in low purity tumors. Nucleic Acids Res. 2014;42:e107. doi: 10.1093/nar/gku489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dhanasekaran SM, Alejandro Balbin O, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun. 2014;5:5893. doi: 10.1038/ncomms6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discovery. 2013 doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borad MJ, Champion MD, Egan JB, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. doi: 10.1371/journal.pgen.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mody RJ, Wu YM, Lonigro RJ, et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA. 2015;314:913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gray SW, Hicks-Courant K, Cronin A, Rollins BJ, Weeks JC. Physicians' attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32:1317–1323. doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller FA, Hayeems RZ, Bytautas JP, et al. Testing personalized medicine: patient and physician expectations of next-generation genomic sequencing in late-stage cancer care. Eur J Hum Genet. 2014;22:391–395. doi: 10.1038/ejhg.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gray SW, Hicks-Courant K, Lathan CS, Garraway L, Park ER, Weeks JC. Attitudes of patients with cancer about personalized medicine and somatic genetic testing. J Oncol Pract. 2012;8:329–335. doi: 10.1200/JOP.2012.000626. 322 p following 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blanchette PS, Spreafico A, Miller FA, et al. Genomic testing in cancer: patient knowledge, attitudes, and expectations. Cancer. 2014;120:3066–3073. doi: 10.1002/cncr.28807. [DOI] [PubMed] [Google Scholar]
- 90.Ding L, Wendl MC, McMichael JF, Raphael BJ. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15:556–570. doi: 10.1038/nrg3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nature biotechnology. 2012;30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aziz N, Zhao Q, Bry L, et al. College of American Pathologists' Laboratory Standards for Next-Generation Sequencing Clinical Tests. Arch Pathol Lab Med. 2014 doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
- 94.Ou SH, Chalmers ZR, Azada MC, et al. Identification of a novel TMEM106B–ROS1 fusion variant in lung adenocarcinoma by comprehensive genomic profiling. Lung Cancer. 2015;88:352–354. doi: 10.1016/j.lungcan.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 95.Chalmers ZR, Ali SM, Ohgami RS, et al. Comprehensive genomic profiling identifies a novel TNKS2-PDGFRA fusion that defines a myeloid neoplasm with eosinophilia that responded dramatically to imatinib therapy. Blood cancer journal. 2015;5:e278. doi: 10.1038/bcj.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ali SM, Miller VA, Ross JS, Pal SK. Exceptional Response on Addition of Everolimus to Taxane in Urothelial Carcinoma Bearing an NF2 Mutation. Eur Urol. 2015;67:1195–1196. doi: 10.1016/j.eururo.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 97.Ali SM, Stephens PJ, Miller VA, Ross JS, Pal SK. Selective Response to Mammalian Target of Rapamycin Inhibition in a Patient with Metastatic Renal Cell Carcinoma Bearing TSC1 Mutation. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 98.Varadhachary GR, Raber MN. Cancer of unknown primary site. N Engl J Med. 2014;371:757–765. doi: 10.1056/NEJMra1303917. [DOI] [PubMed] [Google Scholar]
- 99.Greco FA. Cancer of unknown primary site: improved patient management with molecular and immunohistochemical diagnosis. American Society of Clinical Oncology educational book / ASCO American Society of Clinical Oncology Meeting. 2013:175–181. doi: 10.14694/EdBook_AM.2013.33.175. [DOI] [PubMed] [Google Scholar]
- 100.National Comprehensive Cancer Network Guidelines (NCCN)
- 101.Hainsworth JD, Rubin MS, Spigel DR, et al. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J Clin Oncol. 2013;31:217–223. doi: 10.1200/JCO.2012.43.3755. [DOI] [PubMed] [Google Scholar]
- 102.Varadhachary GR, Spector Y, Abbruzzese JL, et al. Prospective gene signature study using microRNA to identify the tissue of origin in patients with carcinoma of unknown primary. Clin Cancer Res. 2011;17:4063–4070. doi: 10.1158/1078-0432.CCR-10-2599. [DOI] [PubMed] [Google Scholar]
- 103.Ross JSea. Comprehensive genomic profiling of carcinoma of unknown primary site: New routes to targeted therapies. JAMA Oncology. 2015;1:40–49. doi: 10.1001/jamaoncol.2014.216. [DOI] [PubMed] [Google Scholar]
- 104.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sharma MR, Schilsky RL. Role of randomized phase III trials in an era of effective targeted therapies. Nature reviews Clinical oncology. 2011 doi: 10.1038/nrclinonc.2011.190. [DOI] [PubMed] [Google Scholar]
- 106.Dagher R, Cohen M, Williams G, et al. Approval summary: imatinib mesylate in the treatment of metastatic and/or unresectable malignant gastrointestinal stromal tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:3034–3038. [PubMed] [Google Scholar]
- 107.Abrams J, Conley B, Mooney M, et al. National Cancer Institute's Precision Medicine Initiatives for the new National Clinical Trials Network. American Society of Clinical Oncology educational book / ASCO American Society of Clinical Oncology Meeting. 2014:71–76. doi: 10.14694/EdBook_AM.2014.34.71. [DOI] [PubMed] [Google Scholar]
- 108.Chang DK, Grimmond SM, Evans TR, Biankin AV. Mining the genomes of exceptional responders. Nat Rev Cancer. 2014;14:291–292. doi: 10.1038/nrc3723. [DOI] [PubMed] [Google Scholar]
- 109.Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagle N, Grabiner BC, Van Allen EM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takebe N, McShane L, Conley B. Biomarkers: exceptional responders-discovering predictive biomarkers. Nat Rev Clin Oncol. 2015;12:132–134. doi: 10.1038/nrclinonc.2015.19. [DOI] [PubMed] [Google Scholar]
- 112.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 113.Christine M, Micheel SJN, Gilbert S, Omenn Evolution of Translational Omics: Lessons Learned and the Path Forward. 2012 [PubMed] [Google Scholar]
- 114.Swanton C. Intratumor Heterogeneity: Evolution through Space and Time. Cancer Research. 2012 doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 116.Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3987–3996. doi: 10.1200/JCO.2012.45.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 118.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nature reviews Clinical oncology. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 119.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]