Abstract
Epigenetic alterations play a central role in the control of normal and malignant blood cell development. We demonstrate here that expression of a truncated DNA methyltransferase 3B isoform DNMT3B7, which has been shown to alter cellular epigenetic patterns, decreases the overall number of hematopoietic stem and progenitor cells (HSPCs), and markedly diminishes blood cell reconstitution within the female hormonal microenvironment. Gene expression profiling of HSPCs isolated from DNMT3B7 transgenic embryos identified Apolipoprotein E (Apoe) as overexpressed. The CpG island controlling Apoe expression had lower levels of modified cytosines in DNMT3B7 transgenic HSPCs, corresponding with the observed increase in gene expression. Furthermore, we observed that spleens and bone marrows of female mice transplanted with DNMT3B7 transgenic HSPCs express very high levels of Apoe. Finally, the introduction of Apoe-overexpressing HSPCs into male recipients decreased bone marrow engraftment, recapitulating our original observations in female recipients. Our work reveals a dynamic interplay between the intrinsic epigenetic changes in HSPCs and extrinsic endocrine factors acting on these cells to regulate the efficiency of hematopoietic stem and progenitor cell engraftment and reconstitution. We have identified a novel mechanism by which gender-specific hormones modulate HSPC function, which could serve as a target for augmenting hematopoiesis in cases with limited HSC functionality.
Keywords: Hematopoiesis, Epigenetics, DNA methyltransferase 3B, Gender-specific hormones, Apolipoprotein E
Introduction
Hematopoietic stem cell (HSC) self-renewal and differentiation are exquisitely controlled developmental processes that are regulated by several factors modifying signaling pathways and transcription factor binding, leading to gene expression changes. Signaling via gender-specific nuclear hormone receptors is known to regulate hematopoiesis. Androgens and estrogens regulate lineage-specific differentiation, although the results regarding the specific direction of this regulation are conflicting [1–5]. Despite extensive studies on the relationship of gender-specific hormones with HSC function, the molecular mechanism behind these interactions has not been defined.
Gene expression changes that regulate hematopoietic self-renewal and differentiation pathways are driven in part by epigenetic modifications. Deficiency of DNA methyltransferase 3a (Dnmt3a), one of the enzymes that converts cytosine to 5-methylcytosine (5-mC), has been associated with the expansion of the HSC pool and a progressive loss of their differentiation capacity [6]. Sequencing of genomes of patients with acute myeloid leukemia (AML) identified the presence of DNMT3A mutations in ~20% of cytogenetically normal patients [7–9]. Subsequent studies indicated that at least some of these mutations conferred a dominant-negative phenotype to the mutated DNMT3A allele [10]. In addition to the de novo DNA methyltransferases, the maintenance methyltransferase Dnmt1 has been demonstrated to be required for HSC self-renewal, niche retention, and lineage-specific differentiation [11, 12]. We showed recently that erythroid differentiation is associated with dynamic changes in the levels of a second modified cytosine base, 5-hydroxymethylcytosine (5-hmC), with altered erythroid and myeloid differentiation observed in the context of mutations in Ten-Eleven Translocase protein 2 (TET2), the enzyme that converts 5-mC to 5-hmC [13]. These observations illustrate the central role played by epigenetic modifiers in the self-renewal and differentiation of hematopoietic stem and progenitor cells (HSPCs).
We showed previously that the expression of a truncated DNA methyltransferase 3B (DNMT3B) isoform, DNMT3B7, which is expressed in cancer cell lines and tissues isolated from cancer patients, promotes developmental defects and accelerates tumorigenesis in Eμ-Myc mice, which are predisposed to developing B-cell lymphomas [14]. Subsequently, we demonstrated that DNMT3B7 likely acts as a dominant negative inhibitor of full-length Dnmt3b, since Dnmt3b heterozygosity within Eμ-Myc mice accelerated tumorigenesis similar to Dnmt3b heterozygosity [15]. Dnmt3b knockout mice demonstrate developmental defects and have characteristics similar to patients who have Immunodeficiency, Centromeric instability, Facial anomalies (ICF) syndrome [16]. Given that DNMT3B7 transgenic mice demonstrate defects in hematopoiesis and lineage-specific differentiation that are very similar to Dnmt3b deficiency [14], we tested the effect of DNMT3B7 expression on hematopoiesis. Our work reveals a mouse model in which recipient gender modulates the rate of engraftment and reconstitution of HSPCs. Using this novel experimental model, we sought to identify the molecular mechanisms that operate in the hormone-mediated control of hematopoiesis.
Materials and Methods
Mice and transplantations
DNMT3B7 transgenic mice were maintained as described previously [14, 15, 17]. Dnmt3b knockout mice were a kind gift from En Li [16, 18]. Apoe−/− mice on the C57Bl/6 background were obtained and maintained as described [19, 20]. Transplantations were performed using ten million E14.5 fetal liver cells derived from timed mating of DNMT3B7-transgenic mice. Competitive transplantations were performed using 2 million cells from E14.5 CD45.2-bearing WT or DNMT3B7-transgenic fetal livers or from E12.5 fetal liver cells derived from timed mating of Dnmt3b-heterozygous mice, mixed with 0.5 million E14.5 fetal liver CD45.1-bearing cells as the competitor cells. Bone marrow transplantations using WT or Apoe−/− mice were performed using 1.25 million experimental cells mixed with 1.25 million competitor cells derived from bone marrows of CD45.1 mice. Recipient C57Bl/6 mice carrying either CD45.1 or CD45.1/CD45.2 markers were conditioned for transplantation with lethal irradiation (960 rads) using a cesium source. Four hours after irradiation, cells for transplantation were injected intravenously into the retro-orbital sinus of recipient mice under Ketamine/Xylazine anesthesia. Irradiated mice were provided with antibiotics (Bactrim) for 2 weeks after transplantation. Blood counts were measured at 2 weeks, 4 weeks, and monthly thereafter on the Hemavet CBC analyzer (CDC technologies). All mice were maintained in pathogen-free conditions according to an IACUC-approved protocol at The University of Chicago.
Flow cytometry
For analysis of stem cell numbers, E14.5 fetal livers from WT or DNMT3B7-transgenic embryos were made into single cell suspensions, and stained for lineage markers using FITC-tagged antibodies. The lineage negative population was further analyzed for Sca-1 and c-Kit positive populations or for the SLAM markers CD48 and CD150 [21]. For competitive transplantation experiments, engraftment potential was analyzed in the blood of mice at 2, 4 and 8 weeks after transplantation using antibodies against CD45.1 and CD45.2.
RNA-Sequencing
LSK cells were isolated from wild-type, DNMT3B7 hemizygous and DNMT3B7 homozygous embryos at E14.5. We obtained between 10,000 and 100,000 cells per embryo and pooled the cells from four to five embryos of each genotype to perform RNA-Sequencing. Total RNA was isolated using the RNeasy Micro Kit (Qiagen), and 1–3 μg of total RNA used to isolate mRNA by polyA selection. Libraries were generated, and single end sequencing was performed on the Illumina HiSeq 2000 machine.
The RNA-Sequencing reads for pooled LSK cells derived from WT, DNMT3B7-hemizygous, or DNMT3B7-homozygous E14.5 fetal livers were mapped to the reference genome NCBI37/mm9 using Tophat [22, 23]. Gene expression analysis was performed using Cufflinks [24]. Detailed description of RNA-Sequencing analysis and quantification is included in Supplemental Methods.
Analysis and visualization of RNA-Sequencing data
Using the FPKM values generated from the above analyses, an ‘adjusted’ gene list was generated for WT, DNMT3B7 hemizygous and DNMT3B7 homozygous LSK cells, consisting of genes that are uniquely expressed, or are 2-fold overexpressed in that genotype. To visualize these lists and compare the genes that are expressed in each genotype, a 3-way Venn diagram was generated using BioVenn [25]. More detailed descriptions are included in the Supplemental Methods.
From the Venn diagram, lists of genes that are uniquely expressed or overexpressed in a particular genotype were analyzed using gene ontology analysis pathways, specifically in the WebGestalt website [26]. Genes were subsequently ordered by the DNMT3B7 dose-dependent decrease or increase in FPKM values, and the ontology of these genes was analyzed similar to above.
Analysis of modified cytosine levels
Bisulfite sequencing was performed on fetal liver cell DNA isolated from WT, DNMT3B7 hemizygous, and DNMT3B7 homozygous embryos at E14.5 as described [13, 14] using primers listed in Supplemental Table 1.
Reverse transcription and PCR amplification
Total RNA was made using Trizol (Invitrogen). Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Invitrogen). Real time PCR was performed using the Applied Biosystems 7500 Fast PCR system using the Power SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences are listed in Supplemental Table S1. Genotyping of DNMT3B7 mouse embryos was performed by real-time PCR using the TaqMan mastermix, using primers and probes as listed in Supplemental Table S1. Genotyping of Dnmt3b mouse embryos was performed by end-point PCR using the primers listed in Supplemental Table S1.
GEO Accession number
The data corresponding to the RNA-Sequencing run are provided in GEO accession number GSE56170.
Results
Embryonic fetal livers with altered epigenetic patterns contain fewer HSPCs
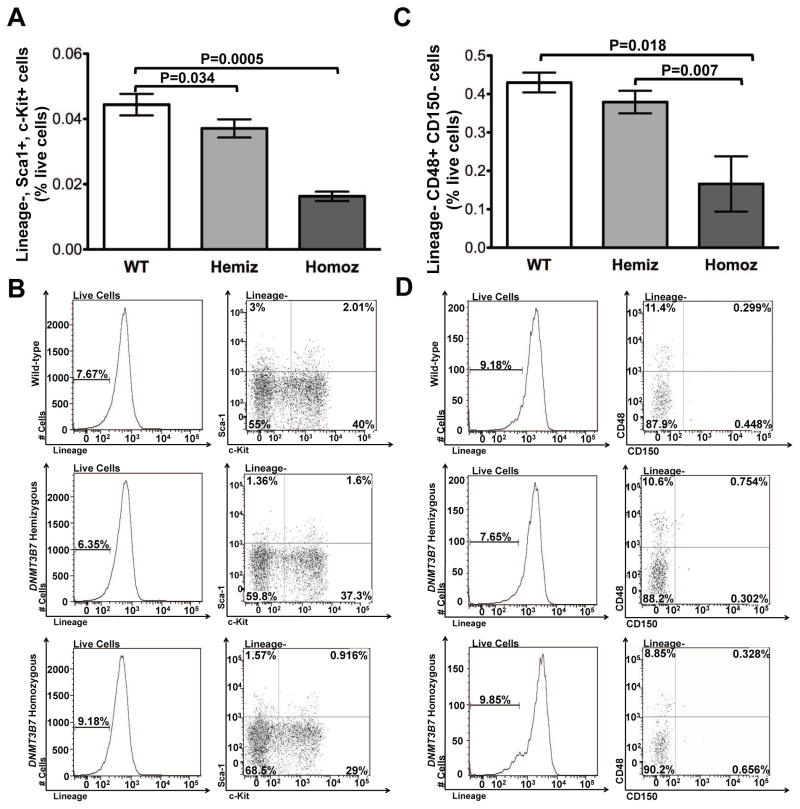
Given that alterations in DNA methylation patterns disrupt normal proliferation and differentiation of hematopoietic stem and progenitor cells [6, 11, 12], and that DNMT3B7 transgenic mice display defective B-lymphopoiesis [14, 15], we sought to assay the total number of stem and progenitor cells produced in DNMT3B7 transgenic mice. Flow cytometry was used to analyze the number of HSPCs in E14.5 fetal livers using Lineage negative (Lin-) Sca1+ c-Kit+ (hereafter referred to as LSK) cells as well as the SLAM markers, CD48 and CD150 [21]. Using the LSK population, we found 3-fold lower numbers of phenotypic stem and progenitor cells in embryos expressing DNMT3B7 (Fig. 1A). Representative flow plots are shown in Fig. 1B. The number of later, committed progenitor cells, marked by the SLAM marker CD48 within the Lin-population, is reduced by more than 3-fold in the DNMT3B7-expressing embryos (Fig. 1C). Representative flow plots are shown in Fig. 1D. We did not observe any significant difference in numbers of LT-HSC, marked by CD150+ CD48-SLAM markers (Supplemental Fig. S1A). This observation illustrates that the introduction of DNMT3B7 results in a fewer number of HSPC and committed progenitor cells within the fetal livers, and argues that the predominant effect of DNMT3B7 expression is within the committed progenitor cell compartment.
Figure 1. DNMT3B7 homozygous E14.5 embryos have fewer numbers of HSPCs.
(A) Lineage (FITC)-, Sca-1 (PE)+, c-Kit (APC)+ cells were stained in fetal livers isolated from E14.5 embryos that were Wild-type (WT: white), hemizygous DNMT3B7 transgenic (Hemiz: light grey) and homozygous DNMT3B7 transgenic (Homoz: dark grey). Average percentages from n ≥ 5 per group ± SEM are plotted. Two-tailed Student’s t-test was used to determine statistical significance. (B) Representative plots analyzed using FlowJo to quantify the relative levels of the analyzed populations. (C) Lineage (FITC)-, CD48 (PE)+, CD150 (APC)+ cells were stained in fetal livers isolated from E14.5 embryos that were Wild-type (WT: white), hemizygous DNMT3B7 transgenic (Hemiz.: light grey) and homozygous DNMT3B7 transgenic (Homoz.: dark grey). Average percentages from n ≥ 4 per group ± SEM are plotted. Two-tailed Student’s t-test was used to determine statistical significance. (D) Representative plots analyzed using FlowJo to quantify the relative levels of the analyzed populations.
DNMT3B7 transgenic HSPCs are outcompeted in competitive transplantation experiments
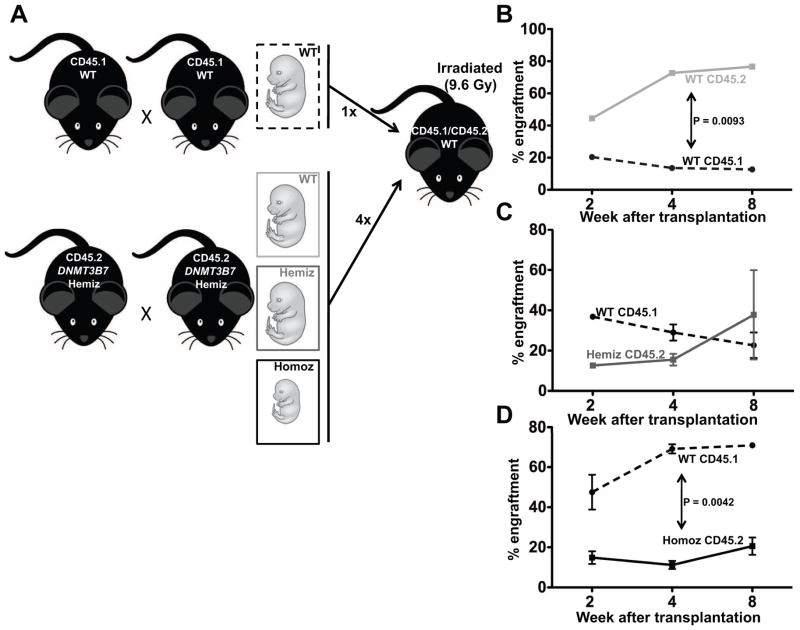
To measure the functionality of HSPCs isolated from CD45.2-expressing DNMT3B7 transgenic embryonic fetal livers, we performed competitive transplantation experiments in female CD45.1/CD45.2 recipient mice, using cells from wild-type CD45.1-expressing E14.5 embryonic fetal livers as competitor cells (Fig. 2A). The presence of both CD45.1 and CD45.2 markers on recipient mice helps distinguish endogenous cells within the recipients from the transplanted competitor (CD45.1) and experimental (CD45.2) cells. To give a competitive advantage to the experimental cells, only a quarter of the total number of cells injected comprised of competitor cells. As expected, we observed that donor-repopulation of CD45.2-expressing wild-type (WT) cells obtained from the DNMT3B7 transgenic breeding was 4-fold better than the competitor CD45.1-expressing WT cells (Fig. 2B). The donor repopulation of CD45.2-expressing cells from DNMT3B7 hemizygous (Hemiz) embryos demonstrated an early defect, which was overcome by two months post-transplantation (Fig. 2C). The most striking observation came from the transplantation of CD45.2-expressing DNMT3B7 homozygous (Homoz) cells, which were outcompeted by the CD45.1-expressing WT cells that were provided at a quarter of the total number of cells (Fig. 2D).
Figure 2. DNMT3B7 homozygous cells are functionally incapable of repopulating the recipient bone marrow.
(A) Schematic of competitive transplantation experiments. Briefly, CD45.2-bearing DNMT3B7 hemizygous mice were timed-mated and embryos were isolated at E14.5. Simultaneously, CD45.1-bearing wild-type (WT) mice were timed-mated and embryos isolated at E14.5. Fetal liver cells were made into single-cell suspensions, and the CD45.1-bearing competitor WT cells were mixed with the CD45.2-bearing experimental cells at 1:4, and injected into lethally irradiated (at 960 rads) CD45.1/CD45.2-bearing WT female mice. Mice were monitored for two months and assessed for the engraftment of CD45.2+ cells. (B) Percent engraftment of CD45.1-bearing WT competitor cells (dashed line) and CD45.2-bearing WT experimental cells (light grey solid line). (C) Percent engraftment of CD45.1-bearing WT competitor cells (dashed line) and CD45.2-bearing DNMT3B7-hemizygous (Hemiz) experimental cells (dark grey solid line). (D) Percent engraftment of CD45.1-bearing WT competitor cells (dashed line) and CD45.2-bearing DNMT3B7-homozygous (Homoz) experimental cells (black solid line). In panels B–D, average percentages from n ≥ 4 per group ± SEM are plotted.
The diminished repopulation of donor cells in DNMT3B7 homozygous cells raised the possibility that the phenotype is due to the loss of expression of one or more genes due to the transgene insertion site, rather than the expression of DNMT3B7 itself. Therefore, we performed competitive transplantation experiments using fetal liver cells retrovirally transduced with DNMT3B7. Consistent with our observation that the expression of DNMT3B7 is responsible for the observed phenotype, the introduction of MigR1-DNMT3B7 into CD45.2-bearing wild-type fetal liver cells diminished the engraftment of CD45.2+ cells in irradiated female mice relative to those that received only MigR1 (vector only). Furthermore, cells expressing MigR1-DNMT3B7 were outcompeted by the CD45.1-expressing competitor cells at two weeks post-transplantation (Supplemental Fig. S1B–C). Thus, transgenic or retroviral expression of DNMT3B7 compromises the ability of HSPCs to engraft and reconstitute in recipients.
To determine whether the altered engraftment and reconstitution of DNMT3B7 transgenic cells was a result of a dominant negative effect on Dnmt3b activity [14–16], we performed competitive transplantation experiments using cells isolated from E12.5 fetal livers of WT, Dnmt3b+/−, or Dnmt3b−/− embryos (Supplemental Fig. S2A–C). We observed that regardless of Dnmt3b expression, the CD45.2-expressing experimental cells engrafted better than CD45.1-expressing WT competitor cells within female recipient mice (Supplemental Fig. S2A–C), consistent with previous work [6]. In the control of engraftment and reconstitution, DNMT3B7 transgenic cells act differently than Dnmt3b knockout cells, suggesting that the effects of DNMT3B7 on hematopoiesis is not due to a dominant negative phenotype.
DNMT3B7-expressing HSPCs exhibit delayed reconstitution of blood counts in female mice
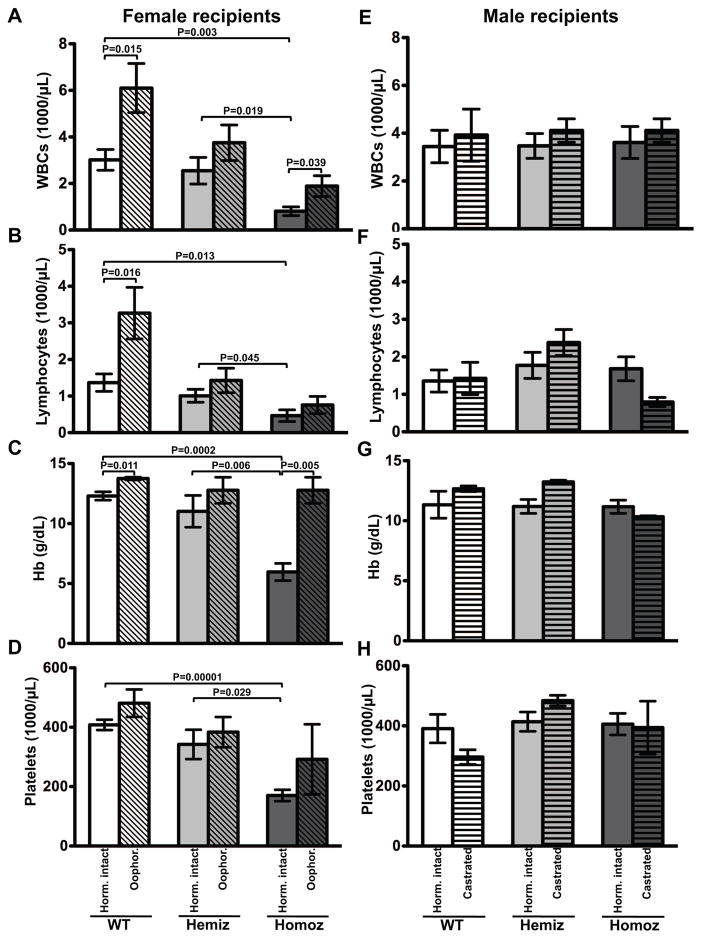
To assess the effects of DNMT3B7 expression on peripheral blood cell counts, we performed transplantation experiments using cells collected from fetal livers of WT, DNMT3B7 hemizygous or DNMT3B7 homozygous embryos at embryonic day E14.5. Since DNMT3B7 homozygous mice do not survive past the day of birth, transplantation is the most effective method to monitor hematopoietic effects of DNMT3B7. We observed that multilineage reconstitution was compromised in female recipients of DNMT3B7 homozygous cells, especially in the first two months after transplantation (Fig. 3, Supplemental Fig. S3). Specifically, female mice receiving DNMT3B7 homozygous cells were leukopenic, lymphopenic, anemic, and thrombocytopenic as early as two weeks after transplantation (Fig. 3A–D, solid bars), whereas male mice engrafted and reconstituted cells normally, irrespective of the genotype of the cells received (Fig. 3E–H, solid bars). While the effect was more dramatic in certain female mice that succumbed to low blood counts by three months post-transplant, generally, the overall blood counts of the female mice stabilized and were equivalent to male mice two months after transplantation (Supplemental Fig. S3). We measured apoptotic potential and cell cycle profiles in cells isolated from bone marrows of mice receiving WT or DNMT3B7 homozygous cells, and found that whereas there was no significant difference in the number of apoptotic cells in male versus female recipients, there were trends consistent with an increase in early-stage apoptotic cells in female recipients relative to males (Supplemental Fig. S4A). We also found a significant increase in cells in the S-phase in female mice receiving DNMT3B7 homozygous receiving females compared to males or female mice receiving WT cells (Supplemental Fig. S4B–C). DNMT3B7 was expressed both in fetal livers being transplanted into the mice (Supplemental Fig. S5A) and in spleens of mice that had been transplanted with DNMT3B7 transgenic cells at four months post-transplantation (Supplemental Fig. S5B). We found that donor cell gender had no effect on the engraftment phenotype within our model (Supplemental Fig. S6). Complete blood count data was supported by the counts generated from peripheral blood smears (data not shown).
Figure 3. DNMT3B7 transgenic HSPCs reconstitute normal blood counts in male, but not in female recipients.
(A–D) Female and (E–H) Male recipients of wild-type (WT: white), hemizygous DNMT3B7 transgenic (Hemiz: light grey) and homozygous DNMT3B7 transgenic (Homoz: dark grey) cells at two weeks post-transplantation. Ten million cells isolated from fetal livers of E14.5 embryos of the three genotypes were transplanted into hormonally intact (solid bars) or hormonally compromised (hatched bars) females (A–D) or males (E–H) that had been lethally irradiated at 960 rads. (A), (E) Average WBC number (x1000/μL); (B), (F) lymphocyte number (x1000/μL); (C), (G) Hb concentration (g/dL); and (D), (H) platelet number (x1000/μL) from n ≥ 4 mice per genotype per gender are plotted ± SEM. Two-tailed Student’s t-test was used to determine statistical significance.
These observations suggested that hormonal microenvironment within the recipient mice influences gender-related differences. We hypothesized that within our mouse model, one or both of the following occurred: (i) female hormones repressed normal hematopoiesis and/or (ii) male hormones enhanced hematopoietic function. We used hormonally compromised females (oophorectomized females) and males (castrated males) to uncover the role played by the major female and male hormones on hematopoietic stem cell engraftment and repopulation. Oophorectomy reduces plasma estradiol levels to <5% of normal levels (34 pg/mL to 2 pg/mL) [27], and castration reduces circulating testosterone levels to <0.1 ng/nmL [28], allowing us to test the specific roles of female and male hormones on hematopoiesis. Oophorectomized mice receiving DNMT3B7 transgenic cells had partially recovered white blood counts (WBC) (1,900±800/μL vs. 810±420/μL in hormonally intact females) and lymphocyte numbers (760±410/μL vs. 470±360/μL in hormonally intact females) (Fig. 3A–B, hatched bars). Interestingly, oophorectomized mice that received wild-type cells reconstituted their WBC and lymphocyte number to a better extent than hormonally intact females that received wild-type cells, suggesting that the leukocyte phenotype is at least partially independent of DNMT3B7 expression. There was complete recovery of hemoglobin levels in oophorectomized female mice receiving DNMT3B7 homozygous cells (11.13±1.59 g/dL vs. 5.96±1.61 g/dL in hormonally intact females) (Fig. 3C, hatched bars). Platelet numbers in oophorectomized female mice were improved compared to hormonally intact females, although the variance in platelet number within this group of mice was high (Fig. 3D, hatched bars). Castrated male mice did not demonstrate any significant differences in engraftment and reconstitution of cells compared to the hormonally intact male mice, regardless of the genotype of cells they received (Fig. 3E–H, hatched bars), suggesting that male hormones did not influence hematopoiesis. These data suggest that the female hormones repress hematopoiesis, especially within the context of cells with aberrant DNA methylation. We also found that the genotype of the donor fetal liver stem cells, but not their gender, predicted total stem cell numbers (Fig. 1, Supplemental Table S2).
DNMT3B7 expression in HSPCs upregulates genes involved in hormone signaling
We hypothesize that expression changes in DNMT3B7 transgenic HSPCs relative to WT cells drive the observed reduction in engraftment and repopulation within female recipients. To determine whether the introduction of DNMT3B7 regulated the expression of specific genes that may alter hematopoietic function, we performed RNA-Sequencing of the LSK population isolated from wild-type, DNMT3B7 hemizygous or DNMT3B7 homozygous embryos at E14.5.
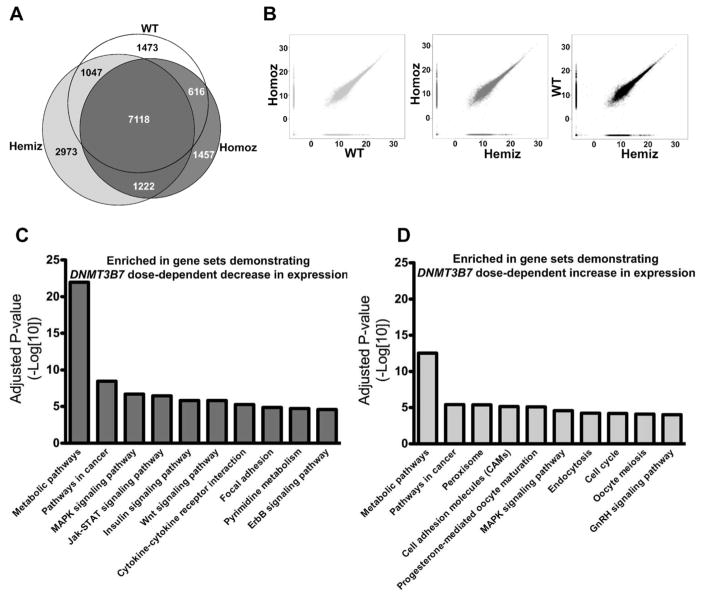
To distinguish genes that demonstrated altered expression with the introduction of DNMT3B7, we first identified genes that were uniquely expressed within LSK cells obtained from WT, DNMT3B7 hemizygous, and DNMT3B7 homozygous embryos (Fig. 4A). Further, to detect the overlap of the genes upregulated in WT, DNMT3B7 hemizygous, and DNMT3B7 homozygous LSK cells, we charted MA (M: log intensity ratios; A: log intensity averages) plots in a pair-wise fashion, and found that >60% of genes were virtually indistinguishable across all three genotypes (Fig. 4B). Using the gene ontology pathway analysis software on Webgestalt [26], we determined that homing-associated pathways were enriched in WT LSK cells (Supplemental Fig. S7A), whereas many receptor-ligand signaling pathways were enriched in the DNMT3B7 hemizygous and DNMT3B7 homozygous cells (Supplemental Fig. S7B–C). Furthermore, we performed gene ontology analysis on the list of genes that were up- and down-regulated in a dose-dependent manner with DNMT3B7 expression (Fig. 4, C and D). Mostly, genes involved in metabolic pathways were enriched with both DNMT3B7 up-and down-regulation. Genes that were downregulated with DNMT3B7 expression were enriched in signaling pathways including the Jak-STAT, Wnt, and ErbB signaling pathways, all of which are known to be involved in the maintenance of normal hematopoietic function (Fig. 4C). Interestingly, genes that were upregulated with DNMT3B7 expression were enriched in hormone-specific pathways, such as progesterone mediated oocyte maturation and GnRH signaling (Fig. 4D), suggesting that gender-specific hormones may play a role in the observed hematopoietic defects.
Figure 4. Enrichment of genes associated with hormone signaling pathways in LSK cells isolated from DNMT3B7 transgenic fetal livers from E14.5 embryos.
(A) Venn diagram depicting the number of unique genes expressed in WT (white), in DNMT3B7 hemizygous (light grey), and in DNMT3B7 homozygous (dark grey) LSK cells. Number of genes that overlap in all the three genotypes is in the center, and overlap of each pair of genotypes is shown at the intersection of the respective colors. (B) Pairwise M-A plots to visualize the extent of overlap between each pair of samples (WT vs. DNMT3B7 Homoz; DNMT3B7 Hemiz vs. DNMT3B7 Homoz; DNMT3B7 Hemiz vs. WT). Genes ordered by DNMT3B7 dosage effects were input into gene ontology pathway analyses, and enriched pathways are shown in (C) genes showing DNMT3B7 dose-dependent decrease and (D) genes showing DNMT3B7 dose-dependent increase in expression.
Lower levels of DNA cytosine modifications within the CpG island of Apolipoprotein E in DNMT3B7 transgenic fetal liver cells
Previously, we had observed that the expression of DNMT3B7 in several types of cells was associated with global changes in DNA cytosine modifications [14, 15, 17]. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS) we found no differences in the global levels of 5-mC and 5-hmC in WT and DNMT3B7 transgenic fetal liver cells (Supplemental Fig. S8A–B). We tested the top twenty genes demonstrating dose-dependent alterations in gene expression with DNMT3B7 introduction for regional cytosine modifications. We focused on genes that had CpG islands (in bold) and were known to be associated with hematopoiesis or were regulated hormonally (Table 1).
Table 1.
Top genes demonstrating DNMT3B7 dose-dependent (i) decrease and (ii) increase in expression.
| (i) Gene Name | WT FPKM > | HEMIZ FPKM > | HOMOZ FPKM | CpG Island |
|---|---|---|---|---|
| Mir669m-1 | 1,290,000.00 | 611,390.00 | 0.01 | No |
| Mir3471-2 | 538,646.00 | 509,252.00 | 0.01 | No |
| n-R5s89 | 493,694.00 | 466,753.00 | 0.01 | No |
| Mir1949 | 416,091.00 | 262,257.00 | 0.01 | No |
| n-R5s24 | 296,423.00 | 280,248.00 | 0.01 | No |
| Lpo | 35,307.00 | 15,335.10 | 0.01 | No |
| Rps25-ps1 | 20,768.30 | 11,389.00 | 0.01 | No |
| Trav6d-5 | 19,503.20 | 18,438.90 | 0.01 | No |
| Fam111a | 17,283.50 | 0.01 | 0.01 | No |
| Fbxo4 | 14,219.60 | 8,131.64 | 0.01 | Yes |
| Hist1h2aI | 12,046.40 | 5,694.52 | 0.01 | Yes |
| Marcksl1-ps4 | 11,428.50 | 10804.9 | 0.01 | No |
| Dnajc5b | 10,455.30 | 4,942.37 | 0.01 | Yes |
| Rbfox3 | 10,423.30 | 9,854.53 | 0.01 | No |
| Lsm10 | 10,217.00 | 6,568.01 | 0.01 | No |
| Trav15d-1- | 9,852.60 | 9,314.95 | 0.01 | Yes |
| AC158135.1 | 9,582.96 | 1,670.09 | 0.01 | Yes |
| Mnf1 | 8,979.29 | 4,468.84 | 0.01 | Yes |
| Ppp1r3g | 8,545.86 | 8,079.52 | 0.01 | Yes |
| Dek | 7,949.11 | 1,475.83 | 0.01 | Yes |
| (ii) Gene Name | WT FPKM < | HEMIZ FPKM | HOMOZ FPKM | CpG Island |
|---|---|---|---|---|
| Traj31 | 0.01 | 2,980,000.00 | 3,300,000.00 | No |
| n-R5s58 | 0.01 | 260,995.00 | 577,825.00 | No |
| n-R5s169 | 0.01 | 252,029.00 | 278,987.00 | No |
| Galp | 0.01 | 44,542.80 | 117,116.00 | No |
| Snora74a | 0.01 | 38,130.10 | 42,208.70 | No |
| Pcca | 0.01 | 6,097.24 | 36,947.60 | No |
| Blcap | 0.01 | 520.83 | 28,806.00 | Yes |
| Apoc2 | 0.01 | 17,140.30 | 19,495.80 | No |
| Rpl18-ps2 | 0.01 | 2,367.30 | 13,606.70 | No |
| Smt3h2-ps | 0.01 | 11,451.90 | 12,676.80 | No |
| Ptprv | 0.01 | 167.96 | 11,960.60 | No |
| Sirt4 | 0.01 | 8,527.06 | 10,355.40 | No |
| Lepre1 | 0.01 | 778.97 | 778.97 | Yes |
| Ldlrad2 | 0.01 | 8,818.54 | 9,761.81 | No |
| Tnp2 | 0.01 | 2,875.13 | 9,547.98 | No |
| Adarb1 | 0.01 | 4,016.42 | 8,892.06 | Yes |
| Apoe | 0.01 | 3,936.69 | 8,715.54 | Yes |
| Clstn3 | 0.01 | 1,765.51 | 8679.54 | No |
| Wrap53 | 0.01 | 5073.23 | 8433.84 | No |
| Chn2 | 0.01 | 4238.58 | 8175.30 | Yes |
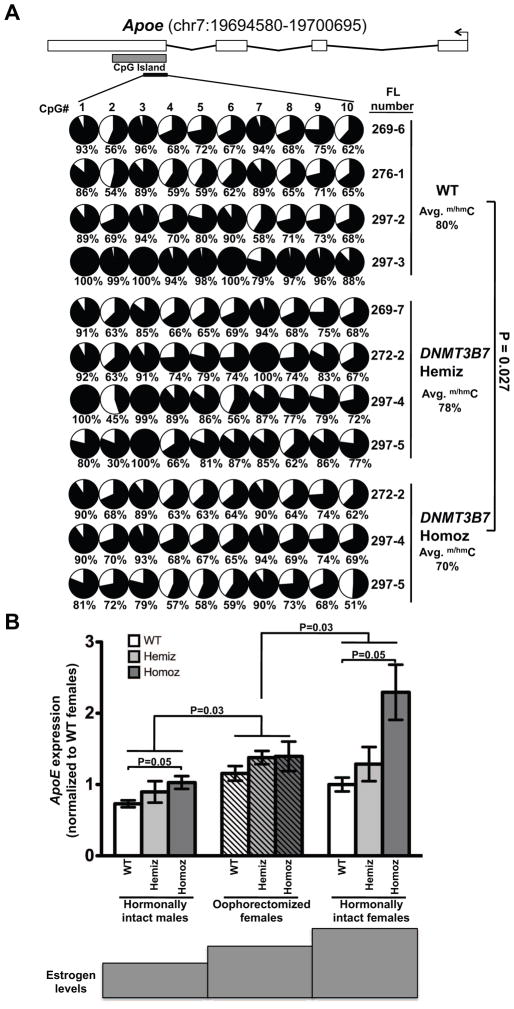
We first measured cytosine modifications in the CpG island of the selected genes in fetal liver gDNA from WT, DNMT3B7 hemizygous, and DNMT3B7 homozygous embryos at E14.5, using bisulfite sequencing. Hematopoietic genes, including Runx2, Hist1h2al, Dek, and Atrx did not demonstrate differences in cytosine modification between WT and DNMT3B7 transgenic fetal livers (data not shown). Apolipoprotein E (Apoe) encodes a protein associated with plasma lipoproteins that is a ligand for cell surface receptors and promotes cellular cholesterol efflux and thus functions in removing cholesterol from the blood and cells and transporting it to the liver for processing. In addition, it has recently been found to repress proliferation and differentiation of hematopoietic stem cells in mouse models [16, 18]. Apoe had significantly lower levels of DNA cytosine modification in the DNMT3B7 homozygous embryos compared to WT embryos (Fig. 5A). The Apoe CpG island is uniquely placed in exon 4 of Apoe, downstream of the transcriptional start site. Methylation at this island has been shown to regulate the transcription of APOE [19].
Figure 5. Reduction in DNA cytosine modifications in the CpG island of Apoe leads to increased Apoe expression in female mice transplanted with DNMT3B7 transgenic cells.
(A) DNA cytosine modifications in WT, DNMT3B7 hemizygous, or DNMT3B7 homozygous fetal liver cells measured by bisulfite sequencing around the CpG island of mouse Apoe (located within exon 4). Schematic diagram for Apoe is shown with exons represented by vertical rectangles, and the location of the CpG island shown with a horizontal shaded rectangle. The black arrow indicates the transcriptional start site (TSS). The smaller, black horizontal rectangle indicates the location of CpGs analyzed for changes in DNA cytosine modifications. Each row represents DNA methylation in a single E14.5 fetal liver, identified by the number on the right. Numbers across the top indicate specific CpG dinucleotides in a region of the CpG island. Changes in DNA cytosine modification are indicated by shaded circles, with the shading indicating average amount of DNA cytosine modification at each CpG, and numbers below represent percent modified cytosine. Average percent modification is indicated to the right, and P-values calculated using the two-tailed Student’s t-test. (B) Apoe expression in spleens isolated from hormonally intact males (left set of bars), oophorectomized females (middle set of bars), and hormonally intact females (right set of bars) receiving WT (white), DNMT3B7 hemizygous (light grey) or DNMT3B7 homozygous (dark grey) fetal liver cells. Relative expression was measured by real-time PCR and with β-Actin serving as the internal control and plotted normalized to WT hormonally intact females. Two-way ANOVA was used for comparison of the three groups, using genotype and gender as the two factors.
Since fetal liver gDNA was used for the cytosine modification analyses, the change in expression of Apoe observed in the RNA-Seq was validated in total fetal liver RNA, using real-time PCR (Fig. S9A, GEO submission GSE56170). As expected, WT fetal liver RNA had the lowest levels of Apoe mRNA and DNMT3B7 homozygous fetal livers had very high levels of Apoe (Fig. S8A).
Apoe expression is regulated by levels of female hormones
We analyzed Apoe mRNA expression in the hematopoietic organs (spleen and bone marrow) of mice that had been transplanted with WT or DNMT3B7 transgenic cells. By RT-PCR, we found that male mice receiving WT cells had the lowest levels of Apoe, and as DNMT3B7 expression increased, there was a dose-dependent increase in Apoe levels (Fig. 5B, left solid bars). This dose-dependent increase in Apoe expression held true within each gender sub-group (Fig. 5B). Overall, female mice had significantly higher expression of Apoe than male mice (Fig. 5B, right solid bars). Strikingly, oophorectomized female mice had lower levels of Apoe than did hormonally intact females and exhibited trends very similar to the male mice (Fig. 5B, hatched bars), supporting the theory that Apoe levels are positively associated with the presence of female hormones. As indicated below the graph, female hormone levels increase from left to right, and the increase in Apoe expression levels followed this dynamic trend. ApoE protein levels were also up-regulated in a similar genotype and gender-dependent manner: female mice expressing the DNMT3B7 transgene had the highest levels of Apoe mRNA within the spleen, whereas male WT mice had the lowest levels (Supplemental Fig. S9B). We also observed a similar trend in Apoe mRNA expression within bone marrows isolated from transplanted male and female mice. Female mice consistently had higher levels of Apoe than male mice, and the expression of DNMT3B7 led to Apoe up-regulation (Supplemental Fig. S9C), suggesting that the regulation of ApoE by female hormones influences HSPC function in all hematopoietic organs.
Apoe overexpression decreases the engraftment of HSPCs into male recipients
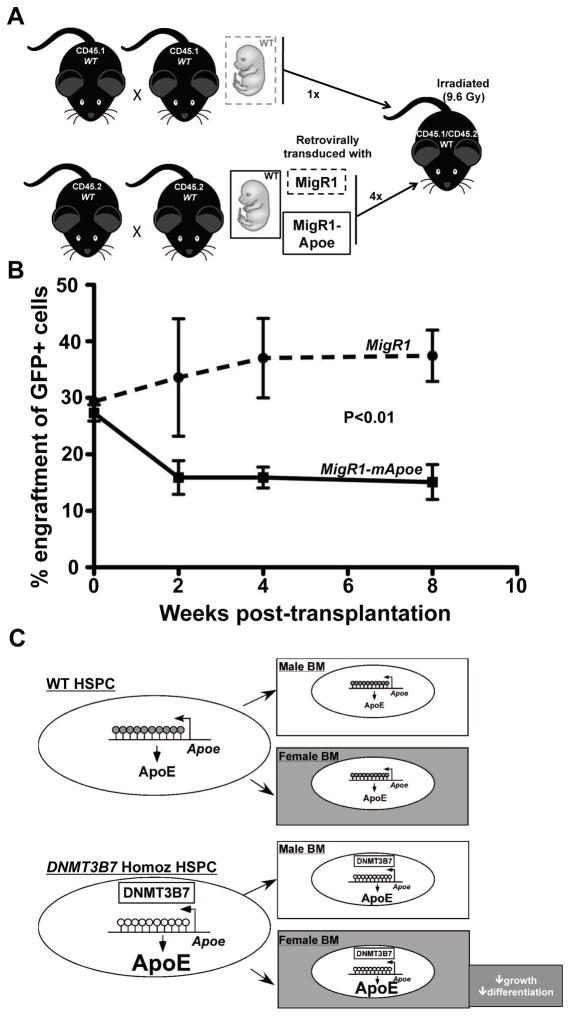
To test directly whether ApoE expression controls the engraftment and reconstitution of HSPCs within the bone marrow, competitive transplantation experiments were performed. Wild-type CD45.2+ E14.5 fetal liver cells were retrovirally transduced with MigR1 or MigR1-mApoe and were mixed at a proportion of 4:1 with competitor CD45.1+ cells. Since transduction efficiency for both MigR1 and MigR1-mApoe was ~30%, the final proportion of experimental GFP+ cells to competitor CD45.1+ cells was 1.2:1. The mixture of cells was transplanted into lethally irradiated CD45.1+/CD45.2+ male recipients. We found that the MigR1-tranduced cells engrafted normally, and even appeared to repopulate the hematopoietic system at a higher efficiency at the later time points of four and eight weeks. However, we found a significant deficit in the engraftment of MigR1-mApoe transduced cells in recipient male mice (Fig. 6A), with a parallel increase in the engraftment of competitor CD45.1 cells (Supplemental Fig. S10A). There was a 2-fold induction of ApoE expression in MigR1-mApoe-transduced cells relative to MigR1-transduced cells (Supplemental Fig. S10B), which was similar to the increase observed in mice transplanted with DNMT3B7 homozygous cells (Fig. 5B). We also tested whether Apoe deficiency would affect engraftment in female mice. Bone marrow cells isolated from CD45.2+ WT or Apoe−/− males or females were mixed in equal quantities with competitor bone marrow cells from WT CD45.1+ female mice, and transplanted into irradiated CD45.1+/CD45.2+ WT female recipients (Supplemental Fig. S11A). Since equal quantities of CD45.1+ and CD45.2+ cells were transplanted into recipients, we would expect 1:1 engraftment if there were no differences in engraftment potential. We found that cells isolated from WT CD45.2+ mice engrafted at the expected 1:1 ratio at 2, 4, and 8 weeks after transplantation. However, Apoe deficiency enhanced donor repopulation of CD45.2+ cells by ~60–80% (Supplemental Fig. S11B). This was significantly different from the 1:1 ratio obtained from WT CD45.2+ cells (Supplemental Fig. S11B). Of note, donor gender had no effect on the engraftment of HSPCs (data not shown).
Figure 6. Apoe overexpression decreases HSPC engraftment potential in male mice.
(A) Schematic of HSPC engraftment experiment. Retroviral vectors MigR1 or MigR1-mApoe were transduced into CD45.2 wild-type E14.5 fetal liver cells. These cells were mixed with CD45.1-bearing competitor WT fetal liver cells at 4:1, and injected into lethally irradiated CD45.1/CD45.2 bearing WT female mice. Mice were monitored for two months and assessed for engraftment of GFP+ CD45.2-bearing cells. (B) Percent engraftment of cells transduced with MigR1 (dashed line) or MigR1-mApoe (solid line) at two, four, and eight weeks post-transplantation. N= 5 for MigR1 and N= 6 for MigR1-mApoe recipient mice; 2-way ANOVA was used to calculate biological significance (C) Proposed model for the genotype and hormone-dependent differences observed in female mice. Expression of DNMT3B7 alters DNA cytosine modifications within the Apoe gene and enhances ApoE protein expression. Whereas there is no further significant increase in ApoE in the male bone marrow microenviroment, within the female hormonal environment, with exposure to estrogen/progesterone, there is an increase in ApoE that is associated with DNMT3B7 expression, which leads to downstream signaling of the IL-3 pathway and reduced growth and differentiation of HSPCs. Male bone marrow is depicted in white rectangular boxes and female bone marrow in light grey boxes.
Discussion
Our study demonstrates for the first time that expression of Apoe contributes to gender-specific differences in blood counts. We find that the epigenetic changes intrinsic to the stem cells cooperate with the effect of female hormones that work in an extrinsic manner to regulate the efficiency of HSPC engraftment and reconstitution. Specifically, we demonstrate that DNMT3B7-expressing HSPCs have fewer phenotypic stem and progenitor cells, and have a lower engraftment and repopulation potential compared to wild-type cells. While illustrating the molecular mechanism of this defect, we find that DNMT3B7 expression decreases overall cytosine modification within the CpG island of Apoe, associated with an increase in Apoe mRNA expression. The increase in Apoe mRNA expression mediates defective engraftment in female recipients of DNMT3B7 transgenic cells, given that overexpression of Apoe represses bone marrow engraftment of HSPCs in male mice.
The gender-mediated difference in engraftment is consistent with a recent study showing that oophorectomy, which diminishes the levels of female hormones, expanded the pool of short-term HSPCs and the eventual recovery after bone marrow transplantation via the CD40 ligand, a T-cell co-stimulatory molecule [29]. We also observed a short-term defect in stem cell reconstitution and engraftment, since female mice receiving DNMT3B7 homozygous cells had stabilized their blood counts after two months post-transplantation (Fig. 3 and Fig. S3). Recent data has shown that proliferative multipotent progenitors are more prone to apoptosis, unlike LT-HSCs [30]. Likewise, it was shown previously that the proliferation of early B-lineage precursors is negatively regulated by estrogen treatment [2, 3, 31]. Contrary to our observations, however, another recent study demonstrated that estrogens promoted HSC self-renewal and the expansion of splenic HSCs and erythropoiesis during pregnancy via estrogen receptor signaling pathways [4]. Differences in experimental conditions likely led to these discrepancies. First, the majority of our experiments used fetal liver stem and progenitor cells, which contain a larger proportion of non-quiescent, cycling cells than do bone marrow stem cells. HSPCs derived from fetal livers function differently than do the HSPCs from bone marrow. For example, IL-7 signaling has been shown to be important in B-cell development in bone marrow, but not in fetal livers [32]. Furthermore, the DNMT3B7 transgenic mice had disrupted epigenetic patterns resulting in the overexpression of Apoe and leading to the observed differences in engraftment. Nevertheless, together with our study, these observations substantiate the argument that gender-specific hormones play a key role in hematopoietic control by modulating hormone-mediated signaling pathways.
That Apoe mRNA levels are highest in female mice receiving DNMT3B7 transgenic cells in our model is reminiscent of historic studies that showed that adult females have higher APOE levels than age-matched males [33]. Estrogen has been shown to upregulate Apoe gene expression in C57Bl/6 mice by altering post-transcriptional mechanisms via the recruitment of the Apoe mRNA to the translating pool of polysomes [34], within most mouse strains tested, including C57Bl/6 [35]. Within an astrocytoma cell line, progesterone but not estrogen, was shown to induce APOE secretion [36], suggesting that cell-type specific differences in response to hormones may dictate the APOE-mediated control of HSPC function. Gender specific differences in APOE expression have been revealed within the context of Alzheimer’s disease, in which the female gender and the ε4 genotype of APOE increases risk for Alzheimer’s disease up to four-fold [37].
Based on our observations, a potential molecular model involves the interplay between epigenetic modifications and female hormones to regulate hematopoietic stem cell function. Apoe expression is upregulated in DNMT3B7 transgenic fetal liver cells, likely as a result of cytosine modifications within its CpG island. This increase in Apoe makes the cells more sensitive to exposure to female hormones like estrogen, which further increases the Apoe levels in DNMT3B7-expressing HSPCs within the female microenvironment. In contrast to the physiological response in Apoe knockout mice [18, 38], the increase in Apoe levels in female mice transplanted with DNMT3B7 transgenic cells leads to reduced proliferation of HSPCs and disrupted lineage-specific differentiation as a result of diminished growth and signaling pathways (Fig. 6C).
Our work emphasizes the importance of experimental design that includes both male and female investigative cohorts, an idea recommended by the NIH recently to make preclinical research more rigorous [39]. We could observe the striking difference in Apoe levels within the HSPC population in male and female bone marrows only by analyzing our transplantation data in gender-specific cohorts. This work also provides a basis for modifying the hormonal environment in patients experiencing bone marrow failure, or who have limited HSC populations, so rapid recovery of peripheral blood counts can be induced. Importantly, our work establishes a novel mechanism that includes hormonal action, epigenetic modifications, and cholesterol metabolism by which stem cell function is maintained, and provides several targets via which early stem cell function may be augmented.
Supplementary Material
Significance Statement.
Alteration of epigenetic patterns, specifically DNA methylation patterns, within blood forming stem cells led to diminished function and reduced blood counts in female transplantation recipients. Stem cells with epigenetically altered patterns expressed high levels of a protein involved in cholesterol metabolism, Apolipoprotein e (Apoe). Apoe was also highly expressed in bone marrow isolated from female mice, regardless of genotype. Introduction of Apoe-deficient stem cells into female mice rescued the defective function and recapitulated the effects seen in male mice. Our work has identified a novel mechanism whereby gender-specific hormones can modulate blood cell development.
Acknowledgments
This work was supported by F32-DK092030 (AV) and NIH CA129831 (LAG).
We gratefully acknowledge the constructive comments from Michelle Le Beau, Richard Larson, Jill deJong, Amittha Wickrema, and Angela Stoddart. This work was supported by F32-DK092030 (AV) and NIH CA129831 (LAG).
Footnotes
Author contributions
AV, CAR, and LAG designed the research; AV, HZ, JBL, KT, and NY performed research; AV, HZ, JA, and LAG analyzed data; AV and LAG wrote the paper. All authors edited and approved the final manuscript.
The authors have no conflicting financial interests.
References
- 1.Goldberg GL, Dudakov JA, Reiseger JJ, et al. Sex Steroid Ablation Enhances Immune Reconstitution Following Cytotoxic Antineoplastic Therapy in Young Mice. THE JOURNAL OF IMMUNOLOGY. 2010;184(11):6014–6024. doi: 10.4049/jimmunol.0802445. [DOI] [PubMed] [Google Scholar]
- 2.Medina KL, Kincade PW. Pregnancy-related steroids are potential negative regulators of B lymphopoiesis. PROC NATL ACAD SCI US A. 1994;91(12):5382–5386. doi: 10.1073/pnas.91.12.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina KL, Strasser A, Kincade PW. Estrogen influences the differentiation, proliferation, and survival of early B-lineage precursors. BLOOD. 2000;95:2059–2067. [PubMed] [Google Scholar]
- 4.Nakada D, Oguro H, Levi BP, et al. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. NATURE. 2014;505(7484):555–558. doi: 10.1038/nature12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer JW, Samuels AI, Adamson JW. Steroids and hematopoiesis. I. The effect of steroids on in vitro erythroid colony growth: structure/activity relationships. J CELL PHYSIOL. 1976;88(2):127–134. doi: 10.1002/jcp.1040880202. [DOI] [PubMed] [Google Scholar]
- 6.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. NAT GENET. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley TJ, Ding L, Walter MJ, et al. DNMT3AMutations in Acute Myeloid Leukemia. N ENGL J MED. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. LEUKEMIA. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita Y, Yuan J, Suetake I, et al. Array-based genomic resequencing of human leukemia. ONCOGENE. 2010;29(25):3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 10.Kim SJ, Zhao H, Hardikar S, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bröske A-M, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. NAT GENET. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 12.Trowbridge JJ, Snow JW, Kim J, et al. DNA Methyltransferase 1 Is Essential for and Uniquely Regulates Hematopoietic Stem and Progenitor Cells. CELL STEM CELL. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madzo J, Liu H, Rodriguez A, et al. Hydroxymethylation at Gene Regulatory Regions Directs Stem/Early Progenitor Cell Commitment during Erythropoiesis. CELL REP. 2014;6(1):231–244. doi: 10.1016/j.celrep.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah MY, Vasanthakumar A, Barnes NY, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. CANCER RES. 2010;70(14):5840–5850. doi: 10.1158/0008-5472.CAN-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasanthakumar A, Lepore JB, Zegarek MH, et al. Dnmt3b is a haploinsufficient tumor suppressor gene in Myc-induced lymphomagenesis. BLOOD. 2013;121(11):2059–2063. doi: 10.1182/blood-2012-04-421065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. CELL. 1999 doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 17.Ostler KR, Yang Q, Looney TJ, et al. Truncated DNMT3B isoform DNMT3B7 suppresses growth, induces differentiation, and alters DNA methylation in human neuroblastoma. CANCER RES. 2012;72(18):4714–4723. doi: 10.1158/0008-5472.CAN-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy AJ, Akhtari M, Tolani S, et al. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J CLIN INVEST. 2011;121(10):4138–4149. doi: 10.1172/JCI57559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu C-E, Cudaback E, Foraker J, et al. Epigenetic signature and enhancer activity of the human APOE gene. HUM MOL GENET. 2013;22(24):5036–5047. doi: 10.1093/hmg/ddt354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sontag TJ, Krishack PA, Lukens JR, et al. Apolipoprotein A-I protection against atherosclerosis is dependent on genetic background. ARTERIOSCLER THROMB VASC BIOL. 2014;34(2):262–269. doi: 10.1161/ATVBAHA.113.302831. [DOI] [PubMed] [Google Scholar]
- 21.Kiel MJ, Yilmaz ÖH, Iwashita T, et al. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. CELL. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. BIOINFORMATICS. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. NATURE BIOTECHNOLOGY. 2009 doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. NATURE BIOTECHNOLOGY. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC GENOMICS. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. NUCLEIC ACIDS RES. 2005;33(Web Server issue):W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altavilla D, Saitta A, Galeano M, et al. The Phytoestrogen |[agr]|-Zearalenol Reverses Endothelial Dysfunction Induced by Oophorectomy in Rats. LABORATORY INVESTIGATION. 2001;81(2):125–132. doi: 10.1038/labinvest.3780219. [DOI] [PubMed] [Google Scholar]
- 28.Christoffersen B, Raun K, Svendsen O, et al. Evalution of the castrated male Sprague–Dawley rat as a model of the metabolic syndrome and type 2 diabetes. INT J OBES RELAT METAB DISORD. 2006;30(8):1288–1297. doi: 10.1038/sj.ijo.0803261. [DOI] [PubMed] [Google Scholar]
- 29.Li JY, Adams J, Calvi LM, et al. Ovariectomy expands murine short-term hemopoietic stem cell function through T cell expressed CD40L and Wnt10B. BLOOD. 2013;122(14):2346–2357. doi: 10.1182/blood-2013-03-487801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez-Aguilera A, Arranz L, Martín-Pérez D, et al. Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. CELL STEM CELL. 2014;15(6):791–804. doi: 10.1016/j.stem.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Medina KL, Garrett KP, Thompson LF, et al. Identification of very early lymphoid precursors in bone marrow and their regulation by estrogen. NAT IMMUNOL. 2001;2(8):718–724. doi: 10.1038/90659. [DOI] [PubMed] [Google Scholar]
- 32.Kondo M, Wagers AJ, Manz MG, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. ANNU REV IMMUNOL. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 33.Phillips NR, Havel RJ, Kane JP. Sex-related differences in the concentrations of apolipoprotein E in human blood plasma and plasma lipoproteins. J LIPID RES. 1983;25:1525–1531. [PubMed] [Google Scholar]
- 34.Srivastava RA, Srivastava N, Averna M, et al. Estrogen up-regulates apolipoprotein E (ApoE) gene expression by increasing ApoE mRNA in the translating pool via the estrogen receptor alpha-mediated pathway. J BIOL CHEM. 1997;272(52):33360–33366. doi: 10.1074/jbc.272.52.33360. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava RA, Krul ES, Lin RC, et al. Regulation of lipoprotein metabolism by estrogen in inbred strains of mice occurs primarily by posttranscriptional mechanisms. MOL CELL BIOCHEM. 1997;173(1–2):161–168. doi: 10.1023/a:1006896131186. [DOI] [PubMed] [Google Scholar]
- 36.Fan J, Shimizu Y, Chan J, et al. Hormonal modulators of glial ABCA1 and apoE levels. J LIPID RES. 2013;54(11):3139–3150. doi: 10.1194/jlr.M042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altmann A, Tian L, Henderson VW, et al. Sex modifies the APOE-related risk of developing Alzheimer’s disease. ANN NEUROL. 2014 doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westerterp M, Gourion-Arsiquaud S, Murphy AJ, et al. Regulation of hematopoietic stem and progenitor cell mobilization by cholesterol efflux pathways. CELL STEM CELL. 2012;11(2):195–206. doi: 10.1016/j.stem.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clayton JA, Collins FS. Policy: NIH to balance sex in cell and animal studies. NATURE. 2014;509(7500):282–283. doi: 10.1038/509282a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.