Abstract
A catalytic self-assembled DNA dendritic complex was herein reported and used for siRNA-based gene silencing. This kind of one-pot DNA dendrimer can be conveniently prepared as needed, and it was demonstrated to have better silencing efficiency and lower cytotoxicity than commercial cationic lipid transfection agents.
RNA interference (RNAi)-based gene silencing interferes with the expression of specific genes with complementary nucleotide sequences, causing mRNA to deteriorate after transcription.1 As such, it has powerful implications for biological studies and gene therapy.2 However, because of the negative backbone, siRNAs with 21–23 base pairs cannot traverse the cell membrane without additional instruments or carriers.3 To overcome this problem, numerous viral and nonviral approaches have been developed for efficient siRNA delivery.4 Although promising, viral vectors are hindered by some obvious drawbacks, such as low scalability, high immunogenicity, potential malignant transformation, and high manufacturing cost, thus constraining their applications in biological research.5 Previously reported nonviral carriers, such as cationic lipids,6 nanogels,7 cell-penetrating peptides (CPPs)8 and cationic polymers,9 often suffer from inhomogeneous size, complex components, high biotoxicity and sophisticated preparation, making their implementation difficult for researchers with a basic background in the biological sciences. The lack of specificity also limits further applications of these traditional nonviral carriers, such as liposomes.
DNAs are naturally water-soluble and biocompatible molecules,10 and they have shown potential in nanocarrier construction based on their programmable sequence design and regular Watson-Crick hydrogen-bonding interactions.11 Our group formerly reported a DNA dendrimer scaffold as an efficient nanocarrier to deliver functional nucleic acids (FNAs) for in situ monitoring of biological molecules.12 Such DNA dendrimers were demonstrated to possess obvious advantages like easy preparation, enhanced enzymatic stability, high stability under very low concentration, and good self-delivery capability. These properties, in our opinion, could also make DNA dendritic structures efficient siRNA delivery platforms and, hence, overcome the above-noted weaknesses of viral/nonviral approaches. However, the formerly reported DNA dendritic carriers were prepared by a multi-step strand annealing and hybridization process,13 which is somewhat time-consuming.
In order to develop an effective and convenient siRNA delivery method, we herein reported an auto-catalyzed DNA dendritic nanostructure for targeted gene silencing.14 As shown in Scheme 1a, a cascade hybridization reaction was triggered by an initial strand I, and nine kinds of hairpins were opened in order, leading to a 5-generation DNA dendritic structure. The outermost layer of dendrimer consists of two kinds of overhands and can be hybridized with aptamer15 and siRNA.16 The whole preparation process is very simple and convenient, and can, therefore, be performed as needed. Our experimental results showed that this kind of one-pot DNA dendrimer could selectively bind to target cells and act as a gene silencing carrier with an better efficiency and lower cytotoxicity than that of a commercial cationic lipid transfection agent, e.g., Lipofectamine.
Scheme 1.
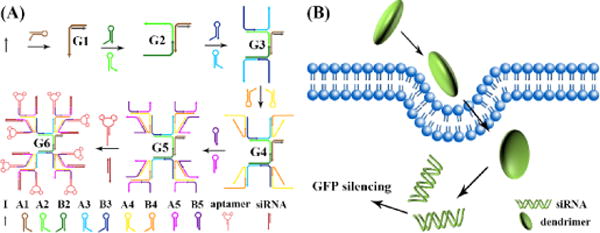
Catalytic self-assembly of DNA dendritic complex for gene silencing. A) Initial strand triggered catalytic self-assembly of DNA dendritic complex. In this system, nine kinds of hairpins were opened in order by a series of isothermal cascading hybridization reactions, thus generating a 5-layer DNA dendrimer G5. Aptamer and siRNA were then hybridized to the outermost layer of G5 to form a G5-aptamer-siRNA complex (G6) for efficient gene silencing. B) G5-aptamer-siRNA complex could act as an efficient gene silencing carrier and was used to silence the GFP expression in A549 cells.
The cascade hybridization reaction was triggered by an initial strand I, as shown in Scheme 1a. Strand I hybridized with hairpin A1 and generated complex G1. G1 had an overhang and could open hairpin A2 and B2 to form complex G2. Overhangs on G2 could further open hairpin A3 and B3, forming complex G3. Following the same strategy, hairpin A4 and B4 were opened by G3 to form G4, and hairpin A5 and B5 were subsequently opened by G4 to form G5. Complex G5 has two kinds of overhangs at the outermost layer and, hence, could hybridize with aptamer and siRNA individually. The generation of DNA dendrimers with 5 layers was first characterized by agarose electrophoresis (Figure 1a) and polyacrylamide gel electrophoresis (Figure S1). A reducing band shift was observed from G1 (Figure 1a, lane 2) to G5 (lane 6) because of the increasing molecular weight. In the presence of strand I and all nine kinds of hairpins, this result indicated that the catalytic self-assembly of DNA dendrimer was successful (lane 6, G5). Aptamer sgc8c, which can specifically recognize PTK7 overexpressed on many cancer cells,17 was first hybridized to the dendrimer and then used here as a model to construct aptamer-G5 DNA dendritic complex. Sgc8c was demonstrated to selectively recognize target CEM cells, but not nontarget Ramos cells. A shorter migration was observed after the integration of sgc8c (lane 7), indicating hybridization of sgc8c to the outermost layer of G5 DNA dendrimer. AFM imaging of Sgc8c-G5 complex, as shown in Figure 1b, also illustrated the formation of DNA dendrimer with a size of ~50 nm, which corresponds to former reports.14 The monodispersity and stability of DNA dendrimer was shown in Figure S2.
Figure 1.
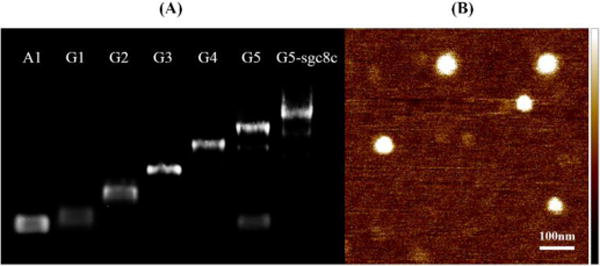
Agarose gel electrophoresis and atomic force microscopy (AFM) demonstrating triggered self-assembly. A) 1% agarose gel electrophoresis was used to characterize the formation of G1 (line 2), G2 (line 3), G3 (line 4), G4 (line 5), G5 (line 6) and sgc8c-G5 complex (line 7). Line 1 indicates the monomer hairpin A1. A reducing band shift was observed from lane 1 to lane 7 because of the increasing molecular weight. B) AFM characterization of G5-sgc8c dendritic complex.
The selective cell recognition of G5-sgc8c complex was studied by confocal microscopy and flow cytometry. In order to give a visible signal, FITC fluorophore was modified at the 3′-terminal of initial strand I. The fluorescent imaging results were shown in Figure S3. After incubating with G5-sgc8c dendritic complex for 2 h, target CEM cells, but not nontarget Ramos cells, gave an obvious fluorescence at 520 nm, indicating the selective endocytosis of FITC-labelled G5-sgc8c dendritic complex by CEM cells. The selective recognition was further evidenced by flow cytometry results shown in Figure 2a (2h incubation) and Figure S4 (8 hour incubation) because a signal shift was observed after incubating CEM cells with FITC-labelled G5-sgc8c complex. Again, no obvious signal change was seen in the Ramos cell lines. Endocytosis efficiency was then estimated to be about 2% by lysing the cells and quantifying fluorescence signal (Figure S5).
Figure 2.
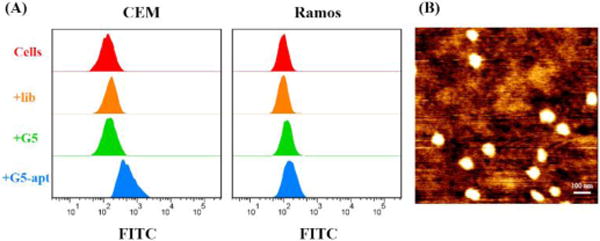
A) Flow cytometric analysis indicating the selective binding of sgc8c-modified DNA dendrimers toward target CEM cells. Cells were incubated with 50 ul of 150 nM FITC-labeled DNA dendrimers for 2 h. B) AFM image of G5-s11e-siRNA dendritic complex with high monodispersity.
After confirming the selective binding of G5-aptamer dendritic complex, siRNA was then integrated into DNA dendrimers to construct a G5-aptamer-siRNA complex (Figure 2b). A549 cells with stable and heritable GFP expression were used as model cells in these experiments. Aptamer s11e,18 which can specifically bind to A549 cells, and siRNA for GFP silencing16 were hybridized to the outermost layer of G5 dendrimer for selective gene silencing. The gene silencing efficiencies of DNA dendrimers were studied using confocal microscopy, flow cytometry and Western blot. Confocal imaging results in Figure 3a and Figure S6 showed that dendrimer-mediated gene silencing obviously reduced the fluorescent intensity of A549 cells, indicating effective RNA interference. Flow cytometry results, as shown in Figure 3b, demonstrated the same results. In order to quantitatively estimate the silencing efficiency, Western blot was also carried out (Figure 3c). As a comparison, the commercial cationic lipid transfection agent lipofectamine was also used for siRNA delivery and gave a slightly lower silencing result. Confocal imaging, flow cytometry and Western blot results of lipofectamine-mediated GFP silencing were shown in Figure S6, Figure S7 and Figure 3c.
Figure 3.
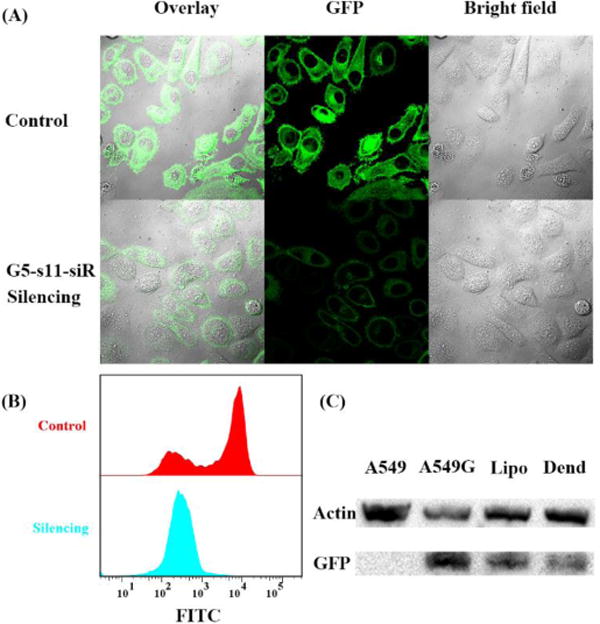
Confocal laser-scanning microscopy imaging, flow-cytometric analysis and Western blot results of DNA dendritic complex-mediated gene silencing. (A) Fluorescent imaging of stable GFP-expressing A549 cell lines with/without G5-s11e-siRNA dendritic complex treatment. (B) Flow cytometric analysis of DNA dendrimer-mediated gene silencing. (C) Western blot result of DNA dendrimer/lipofectamine-mediated gene silencing. Line 1: no GFP-expressing A549 cells without DNA dendrimer treatment. Line 2: stable GFP-expressing A549 cells without DNA dendrimer treatment. Line 3: stable GFP-expressing A549 cells with lipofectamine treatment. Line 4: stable GFP-expressing A549 cells with DNA dendrimer treatment. The average silencing efficiency was calculated to be 70.88% for siRNA-aptamer-G5 dendritic complex and 57.08% for lipofectamine-siRNA complex.
Another advantage of DNA dendritic complex as a siRNA carrier is its outstanding biocompatibility when compared with other artificial carriers, as further validated by MTS assay. Data shown in Figure S8 demonstrated negligible inhibition of proliferation in cancer cells when treated with G5-aptamer-siRNA complex. In contrast, cationic lipofectamine encapsulating the same amount of siRNA showed relative higher cytotoxicity.
Conclusions
In summary, we reported catalytic self-assembly of DNA dendritic complex for efficient gene silencing. In this paper, we demonstrated that DNA dendritic complexes generated by a series of cascading hybridization reactions could be uptaken after selective binding to target cells. Also, for siRNA-based gene silencing, this kind of dendritic complex demonstrated better silencing efficiency and lower cytotoxicity than the commercial siRNA delivery vehicle lipofectamine. Given the convenient preparation, selective targeting, high silencing efficiency and low cytotoxicity, this type of DNA dendrimer could find wide application in biological research as a substitute for other nonviral siRNA carriers.
Supplementary Material
Acknowledgments
This work is supported by NIH grants (GM079359, CA133086), the National Key Scientific Program of China (2011CB911000), the Foundation for Innovative Research Groups of the NSFC (21221003), the China National Instrumentation Program (2011YQ03012412).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here].
Notes and references
- 1.Agrawal N, Dasaradhi PVN, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. Microbiol Mol Biol R. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, Benedicenti F, Vallanti G, Biasco L, Leo S, Kabbara N, Zanetti G, Rizzo WB, Mehta NAL, Cicalese MP, Casiraghi M, Boelens JJ, Del Carro U, Dow DJ, Schmidt M, Assanelli A, Neduva V, Di Serio C, Stupka E, Gardner J, von Kalle C, Bordignon C, Ciceri F, Rovelli A, Roncarolo MG, Aiuti A, Sessa M, Naldini L. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]; (b) Alabi CA, Love KT, Sahay G, Yin H, Luly KM, Langer R, Anderson DG. Proc Natl Acad Sci USA. 2013;110:12881–12886. doi: 10.1073/pnas.1306529110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mok H, Lee SH, Park JW, Park TG. Nature Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 3.(a) Dufès C, Uchegbu IF, Schätzlein AG. Adv Drug Delivery Rev. 2005;57:2177–2202. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]; (b) Wagner E. Acc Chem Res. 2012;45:1005–1013. doi: 10.1021/ar2002232. [DOI] [PubMed] [Google Scholar]; (c) Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]; (d) Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]; (e) Mastrobattista E, Hennink WE. Nature Mater. 2012;11:10–12. doi: 10.1038/nmat3209. [DOI] [PubMed] [Google Scholar]; (f) Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Wang M, Liu H, Li L, Cheng Y. Nat Commun. 2014;5:3053. doi: 10.1038/ncomms4053. [DOI] [PubMed] [Google Scholar]; (b) Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]; (c) Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Proc Natl Acad Sci USA. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wang M, Cheng Y. Biomaterials. 2014;35:6603–6613. doi: 10.1016/j.biomaterials.2014.04.065. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni A, DeFrees K, Hyun SH, Thompson DH. J Am Chem Soc. 2012;134:7596–7599. doi: 10.1021/ja300690j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas S, Deshpande PP, Navarro G, Dodwadkar NS, Torchilin VP. Biomaterials. 2013;34:1289–1301. doi: 10.1016/j.biomaterials.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn SS, Tian S, Blake S, Wang J, Galloway AL, Murphy A, Pohlhaus PD, Rolland JP, Napier ME, DeSimone JM. J Am Chem Soc. 2012;134:7423–7430. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suh JS, Lee JY, Choi YS, Chong PC, Park YJ. Biomaterials. 2013;34:4347–4359. doi: 10.1016/j.biomaterials.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 9.(a) Zhao T, Zhang H, Newland B, Aied A, Zhou D, Wang W. Angew Chem. 2014;126:6209–6214. doi: 10.1002/anie.201402341. [DOI] [PubMed] [Google Scholar]; (b) Ma D, Lin QM, Zhang LM, Liang YY, Xue W. Biomaterials. 2014;35:4357–4367. doi: 10.1016/j.biomaterials.2014.01.070. [DOI] [PubMed] [Google Scholar]
- 10.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 11.(a) Bath J, Turberfield AJ. Nature Nanotech. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]; (b) Jiang Q, Song C, Nangreave J, Liu X, Lin L, Qiu D, Wang ZG, Zou G, Liang X, Yan H, Ding B. J Am Chem Soc. 2012;134:13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 12.Meng HM, Zhang X, Lv Y, Zhao Z, Wang NN, Fu T, Fan H, Liang H, Qiu L, Zhu G, Tan W. ACS Nano. 2014;8:6171–6181. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Zhang H, Ma Y, Xie Y, An Y, Huang Y, Zhu Z, Yang CJ. Sci Rep. 2015;5:10099. doi: 10.1038/srep10099. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lee JB, Roh YH, Um SH, Funabashi H, Cheng W, Cha JJ, Kiatwuthinon P, Muller DA, Luo D. Nature Nanotech. 2009;4:430–436. doi: 10.1038/nnano.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Li Y, Tseng YD, Kwon SY, D’Espaux L, Bunch JS, McEuen PL, Luo D. Nature Mater. 2004;3:38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]; (d) Zhou T, Chen P, Niu L, Jin J, Liang D, Li Z, Yang Z, Liu D. Angew Chem Int Ed. 2012;51:11271–11274. doi: 10.1002/anie.201205862. [DOI] [PubMed] [Google Scholar]
- 14.Yin P, Choi HM, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 15.(a) Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; (b) Xiong X, Lv Y, Chen T, Zhang X, Wang K, Tan W. Annu Rev Anal Chem. 2014;7:405–426. doi: 10.1146/annurev-anchem-071213-015944. [DOI] [PubMed] [Google Scholar]; (c) Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W. Nat Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 16.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nature Nanotech. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


