Abstract
In Malaŵi, Malungo alibe odi is a saying that translates as: “Malaria does not ask permission before coming in”. The recent finding of a new severe malaria resistance locus next to a cluster of glycophorin genes involved in Plasmodium falciparum erythrocyte invasion seems to suggest otherwise: that evolutionary pressure is enabling erythrocytes to lock the door to keep malaria out.
Keywords: Plasmodium falciparum malaria, genome-wide association study
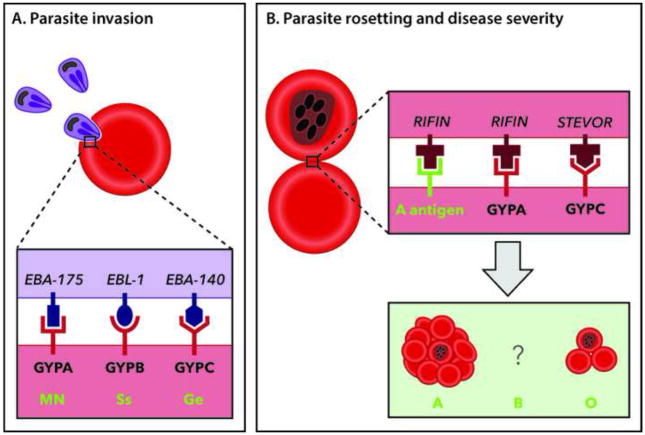
Human glycophorins and their effect on resistance to malaria has been a focus of attention for several years. Glycophorins are erythrocyte membrane proteins rich in sialic acids that carry blood group antigenic determinants, but also act as receptors for various intracellular pathogens. During the Plasmodium intra-erythrocytic cycle, the invasive merozoite stage of the parasite uses several glycophorins to penetrate erythrocytes. The glycophorin A (GYPA) and glycophorin B (GYPB) receptors are recognized by Plasmodium falciparum erythrocyte-binding antigen 175 (EBA-175) and erythrocyte-binding ligand 1 (EBL-1), respectively; amino acid variation in GYPA and GYPB determines the MN and Ss blood groups. On the other hand, glycophorin C (not homologous to GYPA and GYPB) codes for the Gerbich (Ge) blood group antigens, and also acts as a receptor for P. falciparum invasion, recognizing EBA-140 on the surface of merozoites (Figure 1A).
Figure 1. Polymorphisms in Human Glycophorin Receptors and Blood Groups Involved in Protection Against Severe Malaria.
(A) A purple merozoite is shown invading a red erythrocyte, with Plasmodium falciparum invasion ligands (EBA-175, EBL-1 and EBA-140), their receptors (GYPA, GYPB and GYPC) and the different blood group types they are responsible for (MN, Ss and Ge). Polymorphisms in the human glycophorin genes reduce the parasite’s invasion capacity, resulting in protection against malaria. (B) GYPA, GYPC and the A antigen contribute to the P. falciparum rosetting phenotype that is exacerbated in blood group A patients by GYPA binding to parasite encoded RIFINs. This suggests that the O blood group may be under selection pressure in humans in malaria-endemic areas as a means to avoid severe disease. The data concerning the role of blood group B remains conflicting and is represented with a question mark.
The role of glycophorins in malaria parasite invasion has once again been highlighted, this time by the Malaria Genomic Epidemiology Network (MalGEN), who identified a locus of resistance to severe malaria next to the glycophorin gene cluster on human chromosome 4 [1]. MalGEN, whose center is based at Oxford University and the Wellcome Trust Sanger Institute in England, is a consortium of researchers that are leading efforts to understand how genome variation in human, Plasmodium and Anopheles populations affects the biology and epidemiology of malaria. MalGEN has amassed an impressive set of genome sequence data for this triumvirate, and established policies for data sharing that facilitate large-scale projects across multiple populations, merging clinical and epidemiological data with genome sequencing, and developing population genetic tools for data analysis. In their paper, the authors describe the analysis of a staggering ~25 000 individuals from eight African countries, representing the largest genetic association study of malaria to date. Analysis of the data set revealed a haplotype at a locus either close or within the glycophorin gene cluster that reduces the risk of severe malaria by ~33%.
What is the significance of this finding? Malaria parasites have acted as a driving force in recent human and hominid evolution, leading to the emergence of several erythrocyte polymorphisms that confer resistance to both malaria parasite invasion and to severity of the disease. Because of their central role as receptors for merozoite invasion, glycophorins have been a target for this accelerated evolution, allowing the erythrocyte to change the lock against the parasite’s key. Indeed, other hominids such as chimpanzees and gorillas also present uniformly high substitution rates across glycophorin loci, indicating positive selection [2]. A key advantage of these glycophorin mutations is that even as homozygotes they do not encumber the host, unlike other genetic alterations associated with malaria protection such as sickle cell anemia or glucose-6-phosphate dehydrogenase deficiency.
The MalGEN paper complements previous studies showing how the evolutionary pressure exerted by malaria parasites has affected glycophorins across the globe. For example, Ko et al., [3] identified an excess of genetic variation in the region of GYPA that plays a critical role in parasite erythrocyte entry, and this signature of selection was found to be strongest in African populations with the highest malaria exposure. On the other hand, GYPB polymorphisms are especially high in Africans where multiple mutations exist, including the S-s-U-blood group (GYPB-null) that has a gene frequency of 59% in the Efe pygmies found in the Ituri Forest of the Democratic Republic of Congo [4]. In Melanesia, the deletion of exon 3 in the GYPC gene results in Ge negativity and, concomitantly, provides protection to malaria [5]. The geographical distribution of this specific allele closely matches the distribution of malaria transmission, and its frequency reaches ~50% in regions where malaria is hyperendemic, such as coastal areas of Papua New Guinea.
By lowering the invasion rate of the malaria parasite, glycophorin gene polymorphisms may help the host’s immune system to keep the infection under control. But they may also act together with the ABO blood group system to protect the host against malaria severity. Several reports have described a favorable clinical outcome in P. falciparum-infected individuals that are blood group O, while individuals with blood groups A and AB are associated with a higher risk of severe disease (reviewed in [6]). One such severe disease manifestation is rosetting, a phenotype whereby clusters of cells composed of uninfected erythrocytes bound to a central infected erythrocyte are generated (Figure 1B). The ability to form rosettes is a P. falciparum virulence factor, since it likely aggravates microvessel obstruction and shields infected erythrocytes from the immune system. Research by Rowe and colleagues has shown that the O blood group confers protection against severe malaria through reduced rosetting [7].
In this context, it is interesting to note the recent findings by Goel et al. [8], who showed that GYPA on erythrocytes of blood group A also binds to parasite-expressed RIFINs (repetitive interspersed families of polypetides, the largest P. falciparum antigen family), forming large rosettes of ten or more infected erythrocytes. Although RIFINs are also able to bind GYPA on the surface of infected O blood group erythrocytes, this interaction seems to form smaller rosettes (Figure 1B). Therefore, it is plausible that maintaining high polymorphism in GYPA would be beneficial to the host by avoiding the formation of large rosettes, especially in patients with blood group A. Overall, the findings of Goel et al. [8] support the hypothesis that type O alleles might have been evolutionarily selected in malarious areas to cause less severe disease.
Although the MalGEN study moves us one step forward in our understanding of the evolution of resistance to malaria, there are several key questions that remain. For example, why do so few P. falciparum-infected patients develop severe malaria? And what are the host and/or parasite-related factors associated with its development? Several field studies that aim to identify both host [9] as well as parasite [10] factors associated with the development of severe malaria are currently underway. Let us hope that these too will shed more light on the mechanisms of Plasmodium pathogenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Malaria Genomic Epidemiology. A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature. 2015;526:253–257. doi: 10.1038/nature15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang HY, et al. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003;20:1795–1804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 3.Ko WY, et al. Effects of natural selection and gene conversion on the evolution of human glycophorins coding for MNS blood polymorphisms in malaria-endemic African populations. Am J Hum Genet. 2011;88:741–754. doi: 10.1016/j.ajhg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer DC, et al. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A. 2009;106:5348–5352. doi: 10.1073/pnas.0900878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maier AG, et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med. 2003;9:87–92. doi: 10.1038/nm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cserti CM, Dzik WH. The ABO blood group system and Plasmodium falciparum malaria. Blood. 2007;110:2250–8. doi: 10.1182/blood-2007-03-077602. [DOI] [PubMed] [Google Scholar]
- 7.Rowe JA, et al. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci U S A. 2007;104:17471–17476. doi: 10.1073/pnas.0705390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goel S, et al. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med. 2015;21:314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 9.Wassmer SC, et al. Investigating the Pathogenesis of Severe Malaria: A Multidisciplinary and Cross-Geographical Approach. Am J Trop Med Hyg. 2015;93:42–56. doi: 10.4269/ajtmh.14-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlton JM, et al. Population Genetics, Evolutionary Genomics, and Genome-Wide Studies of Malaria: A View Across the International Centers of Excellence for Malaria Research. Am J Trop Med Hyg. 2015;93:87–98. doi: 10.4269/ajtmh.15-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]