Abstract
The intestinal epithelium is very peculiar for its continuous cell renewal, fuelled by multipotent stem cells localized within the crypts of Lieberkühn. Several lines of evidence have established the evolutionary conserved RNA-binding protein Musashi1 as a marker of adult stem cells, including those of the intestinal epithelium, and revealed its roles in stem cell self-renewal and cell fate determination. Previous studies from our laboratories have shown that Musashi1 controls stem cell-like features in medulloblastoma, glioblastoma and breast cancer cells, and has pro-proliferative and pro-tumorigenic properties in intestinal epithelial progenitor cells in vitro. In order to undertake a detailed study of Musashi1’s function in the intestinal epithelium in vivo, we have generated a mouse model, referred to as v-Msi, overexpressing Musashi1 specifically in the entire intestinal epithelium.
Compared with wild type litters, v-Msi1 mice exhibited increased intestinal crypt size accompanied by enhanced proliferation. Comparative transcriptomics by RNA-seq revealed Musashi1’s association with gut stem cell signature, cell cycle, DNA replication and drug metabolism. Finally, we identified and validated three novel mRNA targets that are stabilized by Musashi1, Ccnd1 (Cyclin D1), Cdk6 and Sox4.
In conclusion, the targeted expression of Musashi1 in the intestinal epithelium in vivo increases the cell proliferation rate and strongly suggests its action on stem cells activity. This is due to the modulation of a complex network of gene functions and pathways including drug metabolism, cell cycle and DNA synthesis and repair.
Keywords: Intestinal epithelium, Musashi1, RNA binding protein, stem cells
Introduction
The intestinal epithelium is characterized by a rapid and continuous renewal throughout life. This process depends on multi-potent stem cells located in the intestinal crypts [1]. These adult stem cells self-renew and give rise to undifferentiated progenitors that proliferate, differentiate while migrating, die by apoptosis and finally shed into the lumen. In mice, this whole process lasts 3 to 4 days [1]. Given the recent characterization of a certain number of bona fide gut stem cell markers, a growing amount of data is now available concerning intestinal stem cell physiology. Several reports suggest that two pools of stem cells exist within the crypts. The first pool is located at the very bottom of the crypts and is constituted by the actively cycling crypt basal columnar (CBC) stem cells that express Lgr5 and Ascl2 markers [2, 3]. The second pool is located at the “+4” position from the crypt bottom, is considered quiescent and more resistant to irradiation [4–6], and is characterized by the expression of Bmi1, m-Tert and Lrig1 markers [5–8]. Despite the observation of distinct stem cell populations, other studies have shown that the best-characterized stem cell markers are expressed in a gradient throughout a “stem zone”, and not exclusively in a single stem cell pool [9, 10]. The RNA binding protein Musashi1 (MSI1) was proposed several years ago as an intestinal epithelial stem cell marker [4, 11] and confirmed in a more recent study [10]. We also recently corroborated this observation and demonstrated that Msi1-expressing cells of the intestinal crypts correspond to Lgr5 and m-Tert populations of stem cells [12].
MSI1 was initially characterized in neural precursor sensory cells of Drosophila where it regulates asymmetric cell division [13]. Other studies in this same organism have shown that MSI1 is implicated in the maintenance of stemnesss [14] and in cell fate control [15]. In mammals, in addition to intestinal epithelium, MSI1 has been described as a marker of adult stem cells and progenitors in the central nervous system [16], hair follicle [17] and mammary gland [18]. However, its function and repertoire of targets in these organs is not well known.
MSI1 exists at the intersection of stem cell function and tumor development; its participation in tumor initiation has been previously posited (reviewed in [19]). Supporting this notion, we previously determined that MSI1 is highly expressed in and required for the survival of tumor-spheroids which are enriched in “stem-like” cells; similar results have been obtained with medulloblastoma, breast and lung cancer cultures [20–22], where knockdown of MSI1 affected the expression of stem cell markers [20, 21]. High MSI1 expression is clearly required for tumor development and growth, as its knockdown affected the size of tumors generated by xenografted intestinal adenocarcinoma, medulloblastoma, glioblastoma and breast cancer cell lines [21, 23–25]. In agreement with this observation, high MSI1 expression has been reported in several solid tumors [25–37] and correlated with poor prognosis [25, 30, 36, 38]. Our recent studies in intestinal epithelial primary cultures and normal crypt cell lines revealed that cells with increased MSI1 expression acquire tumor-inducer potential. More specifically, we showed that MSI1 overexpression promoted progenitor cell proliferation via an action on Wnt and Notch pathways and induced tumoral growth of xenografted cells [39]. Despite all we have learned about MSI1 in recent years, we still lack detailed information concerning its function in the intestinal epithelium and the mechanisms governing its action. Towards this goal, here we have developed a murine model of targeted Msi1 expression in the intestinal epithelium, the v-Msi1 mice, and examined the impact of MSI1 ectopic expression on intestinal physiology. We observed an increased size of the progenitor compartment, enhanced proliferation and augmented ex vivo crypt growth, strongly suggesting an action of high MSI1 expression on stem cell activity. RNA sequencing (RNA-seq) analysis of v-Msi1 intestinal mucosa positioned MSI1 as a regulator of the stem cell signature and revealed numerous alterations in genes associated with cell cycle, DNA replication and drug metabolism. Finally, we determined that Msi1 regulates Ccnd1/CCND1 (Cyclin D1), Cdk6/CDK6 and Sox4/SOX4 expression through stabilization of their mRNAs, thus identifying them as novel MSI1 targets.
Materials and Methods
Transgenesis, animal breeding and sample preparation
Mouse Musashi1-V5 cDNA followed by SV40 polyadenylation signal was cloned into a pGEMT-easy vector (Promega) (Figure S1A). After verification by DNA sequencing, the fragment was inserted into the vector pVill-AatII [40], excised by Xho1, and purified with the QIAexII extraction kit (Qiagen). The microinjection of the vector was performed at the Plateau de Biologie Expérimentale de la Souris, Ecole Normale Supérieure de Lyon (Lyon, France) and nine founders were obtained. The transgenic lines were backcrossed into the C57BL6 genetic background. Genotyping and expression analysis were done by PCR or RT-PCR; primers are described in Table S1A.
Animals received standard mouse chow and water ad libitum. They were euthanized at the indicated ages and the intestine was quickly removed, flushed with cold PBS and fixed in 4% paraformaldehyde for paraffin embedding, or immediately frozen in liquid nitrogen for DNA, RNA and protein extraction.
The transgenesis protocol, mouse housing and experimentation were approved from the animal experimental committee of the Ecole Normale Supérieure de Lyon (Lyon, France), the Comités d’Ethique en Experimentation Animale de l’Université de Lyon (C2EA55 and C2EA15; registration number DR2013-55) and in accordance with European legislation on animal care and experimentation.
Studies on animals
Histological analysis, Immunolabeling and Western blot
Fragments of the intestinal mucosa were fixed in 4% PFA for histology and immunohistochemical studies. For the different studies at least six animals per genotype have been analyzed. Paraffin sections (5 μm thickness) were used for indirect immunostaining; whole-protein extracts were obtained by homogenizing intestinal samples in RIPA buffer. Whole protein extracts (50 μg/lane) from WT or v-Msi1 animals were analyzed. We used the following antibodies for immunofluorescence (IF) and western blots (WB): anti-MSI1 (Abcam, WB; Chemicon, IF); anti-Ki67 (Labvision, IF); anti-CCND1 (Labvision, IF and WB); anti-activated β-catenin (Upstate, WB), anti-c-MYC (Santa Cruz, WB); anti-SOX4 (Santa Cruz, IF); anti-CDK6 (Santa Cruz, IF); anti-Actin (Sigma, WB). For immunolabeling experiments, we used appropriated fluorescent secondary antibodies (Jackson Laboratories and Life Technology). All nuclei were counter-stained with Hoechst (Sigma). For western blot analysis, we used secondary IgG-horseradish peroxidase conjugated antibodies (Promega). The signal was then analyzed using the enzymatic chemiluminescence detection kit (LumiLight, Roche).
RNA extraction, RT-PCR, RT-qPCR and RIP-RT-qPCR
Tissue fragments were lysed using a Precellys 24 homogenizer (Bertin); cultured 2D epithelium and 3D organoids were lysed by several passages through micropipette tips. Total RNA was extracted with the QIAGEN RNeasy kit (Qiagen). To avoid the presence of contaminating DNA, DNase digestion (Qiagen) was performed in all preparations. Reverse transcription was performed using MMLV reverse transcriptase (Promega) on 1 μg of total RNA according to the manufacturer’s instructions, using random hexanucleotide primers (Invitrogen). PCR and qPCR analysis were performed to amplify specific cDNAs, by using Euroblue Taq (Eurobio) or SYBR Green (Takara) and a BioRad thermocycler or a BioRad CFX Connect system, respectively. The RIP-RT-qPCR protocol used for detection of MSI1 targets is the same as described for 293T cells (see below). Primers and probes information are reported in Table S1B, C.
Intestinal epithelium 2D primary cultures
Intestinal epithelial primary cultures were derived from 4–6 day old neonatal mice [41] and the isolated epithelial fragments were prepared and plated on culture surfaces coated with Matrigel™ Basement Membrane Matrix (BD Biosciences) as previously described [39]. For immunolabeling experiments, cover-slips were inserted in the wells before coating and the following primary antibodies were used: anti-Msi1 (Chemicon); anti-GFP (Millipore); anti-Ki67 (Labvision, IF).
Crypt isolation and 3D cultures
Crypt isolation was performed according to Sato et al. [42]. Briefly, mouse small intestine (<10 cm) was excised, opened longitudinally, and washed with ice-cold PBS. The intestine was cut into small pieces (<4- to 5-mm diameter) and incubated in ice-cold PBS containing 2 mM EDTA for 30 minutes. After one wash with ice-cold PBS, the intestinal fragments were suspended four successive times in ice-cold PBS (10 ml) by repeated and vigorous pipetting. After each re-suspension the tissue fragments were recovered by sedimentation on ice, the supernatant from the first two rounds was discarded while that from the last two re-suspensions was collected and filtered through a 40 μm cell strainer (BD Bioscience) to remove large tissue fragments. After filtration, the crypts were separated from single cells by centrifugation (200 g, 5 minutes at 4°C), suspended with cold DMEM/Ham’s F12 (1:1 vol/vol) and then mixed 1:1 vol/vol with Matrigel (BD Bioscience) for plating (100 μl in 35mm dish or 40 μl in 10mm well). After polymerization of the Matrigel, culture medium composed of DMEM/Ham’s F12 containing 2% penicilline/streptomycine (Life Technology), 1% glutamax (Life Technology), 1% B27 (Life Technology), 50 ng/ml Noggin (PreProTech), 250 ng/ml R-spondin1 (R&D Systems), 50 ng/ml EGF (Sigma) and 10 ng/ml FGF (Sigma) was added and changed every 2 days. Note that the concentration of R-spondin1 and Noggin are 50% of those used in the method by Sato et al. [41] to slow the organoid development. After 7 days in culture, the dishes were washed twice with ice-cold PBS before addition of the cell recovery solution (BD Bioscience) for 10 minutes on ice. The organoids within the cell recovery solution were then collected from the dishes and transferred to 1.5 ml tubes for a further 30-min incubation on ice. They were finally centrifuged at 200 g (5 min at 4°C), and the crypt-containing pellet washed once with ice-cold PBS (<1 ml), centrifuged and frozen for RNA extraction.
Lentiviral infections
For ShRNA experiments, primary 2D cultures or crypt 3D cultures were infected 24h after seeding or from the beginning of the culture respectively, with Sh viral supernatant at a 3:2 ratio with culture medium for 72h. ShRNA lentiviral vectors were Mission-shRNA (derived from pLKO.1-puro, Sigma). Lentiviral particles expressing Sh-Scr (scrambled control), Sh1-1 and Sh1-4 (targeting Msi1 mRNA) were obtained from the Vectorology Facility at IFR128 Lyon-Biosciences. At the end of the culture, dishes were washed twice with phosphate-buffered saline and frozen at −80°C for RNA extraction, or alternately, fixed in 2% paraformaldehyde for immunostaining experiments.
Studies on 293T cell line
RIP-RT-qPCR
293T were maintained at 37°C, 5% CO2 and grown in DMEM supplement with 10% fetal bovine serum, penicillin and streptomycin. RIP experiments were performed according to established protocols using rabbit anti-MSI1 from Millipore and normal rabbit IgG from R&D as a negative control. Briefly, cell extracts were prepared from 293T cells in polysomal lysis buffer, diluted in NT2 buffer and incubated for 3 hours with antibody coated protein A-sepharose 4Fast flow from GE. After immuno-precipitation, the RNA was extracted using acid phenol:ChCl3 from Ambion. cDNA was prepared from experimental and control samples with high capacity cDNA reverse transcription kit; RT-qPCR with specific probes (Table S1 C) was done with a Taqman assay kit (Applied Biosystems). To normalize the data, we spiked in identical amounts of cDNA from DH5a in each sample and conducted amplification with specific bacterial probe. Experiments were conducted in triplicate. Primers and probes information for the different approaches are reported in Table S1B, C.
Transfection
293T cells were transfected using GeneJammer (Agilent Technologies) with either a pcDNA3.1 plasmid (Life Technologies) containing Msi1 coding sequence or GST coding sequence (negative control). Cells were harvested for western analysis, using protocols described bellow, 72 hours post-transfection.
RT-qPCR
RNA was extracted with Trizol (Invitrogen). The cDNA synthesis and RT-qPCR were performed using Taqman small RNA assays kit (Applied Biosystems).
Musashi1 siRNA knockdown
siMSI1 and siControl were transfected into 293T using Lipofectamine RNAiMax reagent (Invitrogen) by reverse transfection according to the manufacturer’s instructions. Transfection complexes were prepared and added directly to the cells at a final concentration of 25nM; cells were then incubated for 72 hours. Finally, cells were collected, resuspended and sonicated in SDS Laemmli sample buffer.
Western Blot
12% SDS-PAGE gels with a 4% stocking gel were run in Tris-glycine-SDS buffer. A wet transfer procedure was carried out on to PVDF membrane. After transfer, membrane were blocked in TBS with Tween-20 and 5% milk or 5% BSA. The membranes were probed with mouse monoclonal anti-CDK6 (Cell Signaling), mouse monoclonal anti-SOX4 (Santa Cruz), rabbit monoclonal anti-MSI1 (Millipore), rabbit monoclonal anti-CCND1 (Millipore), or mouse monoclonal anti-α-TUBULIN (Sigma). HRP-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology) was used as a secondary antibody for MSI1 and CCND1 and HRP-conjugated goat anti-mouse antibody (Zymed Laboratories) was used as a secondary antibody for CDK6, SOX4 and α-TUBULIN. Proteins were detected by using Immobilon Western chemiluminescent HRP substrate (Millipore).
RNA-seq study
RNA preparation
RNA samples from wild type and v-Msi1 intestinal mucosae were prepared with Trizol (Life Technologies) followed by further purification with RNAeasy (Qiagen). RNA quality and concentration were checked with Nanodrop and Bioanalyzer.
Library construction and sequencing
RNA-seq libraries were prepared according to Illumina protocols and sequenced in a HiSeq 2000 (50bp, single reads) at the Greehey CCRI Genome Sequencing Facility.
Read mapping and gene expression analysis
The libraries were mapped to the mouse reference genome (mm10, downloaded from UCSC Genome Browser) using Tophat [43] version 2.0.8b and Ensembl (release #71) to the transcriptome annotations. Only uniquely mapping reads with mapping quality (Q) > 20 were used. The set of expressed genes from each library was defined counting reads falling within each transcript using Cufflinks [44] pipeline.
The identification of differentially expressed genes from RNA-seq was performed using edgeR [45], DESeq [46] and Cuffdiff2 [47]. In order to select only high-confidence differentially expressed genes, we selected only those up- or down-regulated genes indicated by at least two out of three methods. Only differentially expressed genes presenting a q-value < 0.05 were selected. Detailed sequencing information can be obtained from ENA (https://www.ebi.ac.uk/ena) number: ERP006988.
Cellular pathway annotations
All expressed genes (23,387) in WT sample were used as background of expression. The set of up- and down-regulated genes in Msi1-transgenic samples were analyzed for enrichment in KEGG pathways with DAVID (http://david.abcc.ncifcrf.gov/). Only pathways enriched (adjusted p-value < 0.05, Benjamini correction) were selected. Drug metabolism and Cell cycle pathways analysis was performed with Cytoscape (http://www.cytoscape.org/). Adult stem cell-enriched genes were obtained from [10].
Other methods used in the data analysis
Circos was used for visualization of KEGG data [48]. Custom Perl scripts (http://perl.org), Linux shell scripts, BioPerl (http://www.bioperl.org), samtools (http://samtools.sourceforge.net), bedtools (http://bedtools.readthedocs.org), and R (http://www.r-project.org/) were also used to perform data analyses.
Results
Generation and intestinal phenotype of v-Msi1 animals
Musashi1-V5 cDNA was cloned downstream of the Villin promoter (scheme Figure S1A), leading to the expression of Msi1 specifically in gut endoderm/intestinal epithelium starting from E11 [40]. The presence of the transgene in the genome and the corresponding mRNA expression were verified by PCR and RT-PCR respectively in F5, F7 and F9 v-Msi1 lines (Figure S1B, C). Figure S1C shows in particular the presence of an amplified cDNA only in the intestine of v-Msi1 animals compared to wild type littermates, while Figure S1D demonstrates that the transgene was absent from the other organs analyzed of both v-Msi1 and WT litters. It has been reported that the Villin promoter in transgenic mice can be active also in kidney (rate of one out five transgenic lines) [40]. However, none of the v-Msi1 lines (Figure S1D and not shown) showed expression of Msi1 in this organ. Given that all transgenic lines presented similar features (not shown), further in depth analyses were performed using the F7 line.
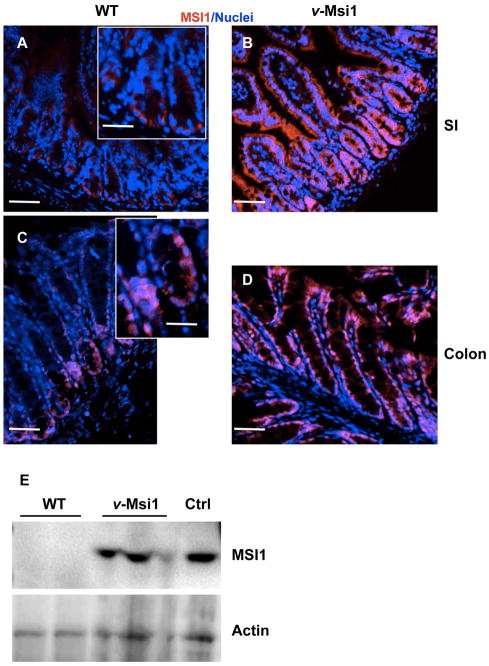
Next, we performed MSI1 immunostaining experiments on small intestine and colon sections. As expected only few cells were positive for MSI1 at the bottom of the WT crypts (Figure 1A, C), while specific over- and ectopic MSI1 expression all along the intestinal epithelium in both intestines was observed in v-Msi1 mice (Figure 1B, D). Western blots and RT-qPCR of whole intestinal lysates further confirmed the increased MSI1 protein and mRNA expression respectively in v-Msi1 intestines (Figures 1E, S1E).
Figure 1. Increased MSI1 expression in v-Msi1 intestine.
Anti-MSI1 antibodies were used to analyze the expression pattern of MSI1 along the epithelial axis of the small intestine (A, B) and colon (C, D) of v-Msi1 (B, D) and WT (A, C) animals. Pictures show the merging of nuclear (blue) and MSI1 (red) staining. Bar=15μm. (E) Analysis of MSI1 protein levels by WB in v-Msi1 and WT animals. Actin was used as the loading control. The Ctrl lane (positive control) corresponds to lysate from Cos7 cells transfected with a Msi1-expressing vector.
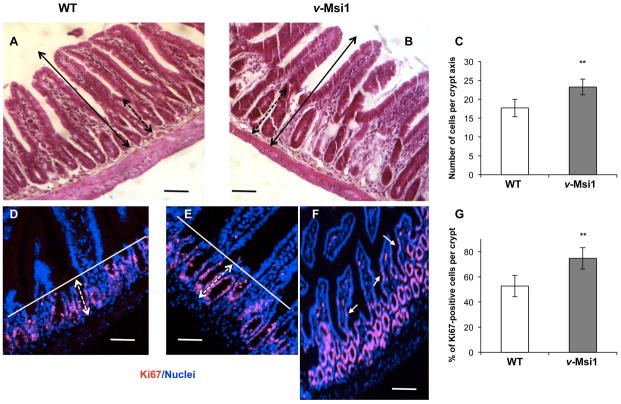
Analysis of the intestinal features of transgenic adult animals showed a slight but significant increase of crypt depth, due to an enhanced number of cells per crypt axis in both proximal and distal small intestine compared to WT (Figures 2A–C, S2A), while the whole crypt-villus height looked unchanged in size. In the colon however, we could not observe any clear difference in morphology (Figure S2C). As Msi1 overexpression has been shown to enhance proliferation, we performed immunostaining for the proliferation marker Ki67 on sections of small and large intestine in both v-Msi1 and WT mice (Figures 2D–G, S2B, D). Interestingly, the percentage of proliferative Ki67-positive cells was significantly increased in both proximal and distal small intestine of mutant compared to WT animals. Furthermore, Ki67 positive nuclei were present in the villi of some v-Msi1 mice (arrows in Figure 2F). In the colon no clear hyperproliferative phenotype was observed, according to the generally unaltered morphology (Figure S2D). We also conducted Msi1-knockdown experiments by transducing Sh-interfering viral particles in intestinal epithelial primary cultures from the small intestine as previously described [39]. Interestingly, we observed a significant reduction of proliferating Ki67-positive cells in Sh-Msi1 (Sh1-1 and Sh1-4) condition compared with controls (GFP-infected or Sh-Scrambled infected cells). The decrease paralleled the expression levels of Msi1 mRNA in the cells maintained in the different culture conditions (Figure S3A–C). Finally, given the role described for MSI1 in cell fate determination (reviewed in [49]) we also analyzed the major epithelial differentiation markers, but found no major defects (data not shown).
Figure 2. Morphological and proliferative properties of the v-Msi1 intestinal mucosa.
A, B) Morphological analysis of the distal small intestine from WT (A) and v-Msi1 (B) mice. C) Quantification of the number of cells per crypt axis, as indicated by the black dotted double arrows in A and B. Approximately 40 axes were scored under the microscope from at least four mice per genotype; histograms represent the mean ± SD. **: P<0.01, in comparison with WT, by Student’s t-test. (D–F) Ki67 immunolabeling of proliferating cells in intestinal sections from WT (D) or v-Msi1 mice (E, F). Images show merged Ki67 immunolabeling (red) and nuclear staining (blue). G) Quantification of the percentage of Ki67 positive cells per crypt. Approximately 40 crypts were scored under the microscope from at least four mice per genotype; histograms represent the mean ± SD. **: P<0.01, in comparison with WT, by Student’s t-test. Bar=15μm. Black double arrows in A and B define the length of the vertical crypt-villus axis. Dotted double black arrows in A and B show the size of the crypts. White bars in D and E define the limit of the proliferative Ki67-positive cells and dotted double white arrows show the size of the proliferating zone. White arrows in F indicate some Ki67-positive cells in villi.
In summary, MSI1 ectopic expression in the intestinal epithelium is responsible for increased intestinal crypt size and increased cell proliferation, consistent with its function highlighted in other systems (reviewed in [19]) and in our previous in vitro studies [25, 39].
Molecular characterization of the v-Msi1 intestinal mucosa
To determine the signals regulated by MSI1 increased expression in the whole intestinal epithelium, we compared the transcriptomic profile of v-Msi1 vs. WT animals using RNA-seq. After stringent analyses (see Material and Methods), we identified a total of 1,365 genes up-regulated in v-Msi1 animals relative to wild-type, and 1,018 genes down-regulated (Figure S4, Tables S2, S3). Using a KEGG pathway enrichment analysis tool, we identified drug metabolism as the top enriched pathway among down regulated genes (Figure S5, Tables S4, S5). Specifically, genes of the Cytocrome P450 super family of mono-oxygenases (CYPs) and Glutathione-S-transferase families were down-regulated in v-Msi1 mucosa (Figures S5, S6). Surprisingly, few genes of the canonical Wnt pathway were differentially regulated (Figure S7A), and statistical analysis revealed no significant association between MSI1-dependent differentially expressed genes and those in the canonical Wnt pathway (14/148 genes corresponding to 9.4%; p-value, adjusted by Benjamini correction = 0.98). However, we did observe increased activated β-catenin, CCND1 and c-MYC protein levels in v-Msi1 intestinal mucosa compared with the WT counterpart (Figure S7B), indicating an increased Wnt activity dependent on high MSI1 expression as previously shown [39, 50].
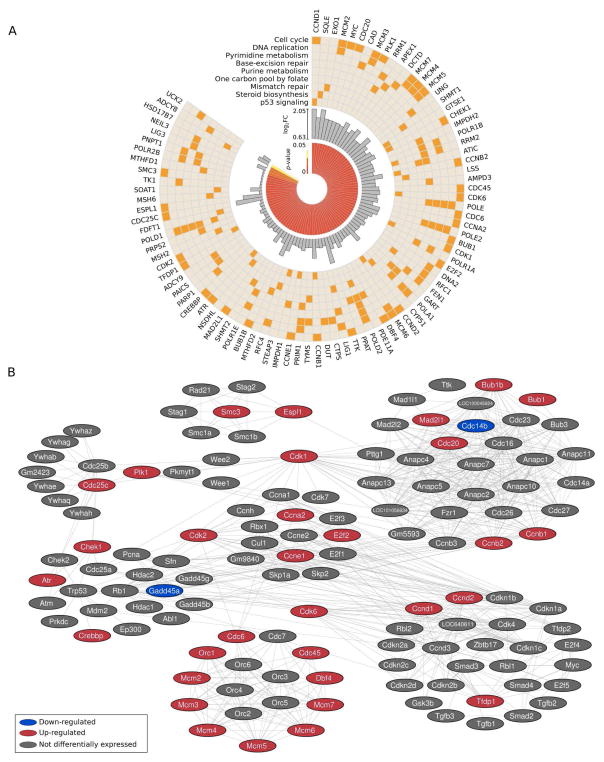
Among up-regulated genes, the top cellular pathways affected by MSI1 include cell cycle, confirming the observed increased proliferation in v-Msi1 crypts (Figures 1 and S2), as well as DNA replication and repair (Figure 3A). Network analysis established correlations among affected genes in these different pathways (Figure 3B). Some of these functional interactions represented the focus of more in depth analyses.
Figure 3. Pathways and functions affected by Msi1 altered expression.
A) Enriched KEGG pathways for genes up-regulated in v-Msi1 samples. Only pathways presenting adjusted P-value < 0.05 were selected. B) Increased expression of genes implicated in cell cycle, DNA replication and repair in v-Msi1 compared with WT intestine. Gene pathway data was obtained from DAVID (http://david.abcc.ncifcrf.gov/). Cytoscape (http://www.cytoscape.org/) was used for pathway visualization.
Cdk6/CDK6 and Ccnd1/CCND1 mRNAs are direct targets of MSI1
Changes in MSI1 level of expression have been previously reported to influence the cell cycle of mammary progenitor cells [51]. Among cell cycle genes up-regulated in v-Msi1 mice, Cdk6 and Ccnd1 were determined to contain binding sites for MSI1 according to iCLIP data obtained from U251 glioblastoma cells [52] (Figure 4A). To obtain the iCLIP map, we conducted experiments in triplicate using the protocol described in [53]. More details are provided in GEO — accession number GSE68800. We hypothesized that the direct effect of MSI1 on the expression of these two genes functions as the initial driver to boost cell cycle and cell proliferation. We conducted experiments to validate Cdk6 and Ccnd1 as targets of MSI1 and to evaluate its direct impact on their expression.
Figure 4. CCND1 and CDK6 are direct targets of MSI1.
A) UCSC genome browser plots showing the position (x-axis) and count (y-axis) of iCLIP reads overlapping the 3′ UTR of CCND1 (top) and CDK6 (bottom). Highlighted are the sequences of two regions in each UTR exhibiting particularly high density of iCLIP reads and concordance between replicates. Each region shows the presence of several UAG and GUAG oligomers, the known core of the Msi1 binding site. B) CDK6 and CCND1 immunolabeling in intestinal sections from WT or v-Msi1 mice as indicated. Images show merged specific labeling (red) and nuclear staining (blue). Bar=15μm; insets in CDK6 panels=7μm. C) RIP-PCR performed with anti-MSI1 and control antibodies in 293T cells showing that CCND1 and CDK6 mRNAs are highly associated with MSI1 protein. D) RT-qPCR showing the impact of MSI1 knockdown on CCND1 and CDK6 mRNA levels. E) Representative western blot showing the impact of MSI1 knockdown on CCND1 and CDK6 protein levels. F) MSI1 knockdown affects CCND1 and CDK6 mRNA decay.
First, RIP-RT-qPCR analysis performed on intestinal mucosa of v-Msi1 showed that MSI1 preferentially associates with Ccnd1 and Cdk6 transcripts (Figure S8). Second, by RT-qPCR we confirmed that Ccnd1 and Cdk6 mRNA expression were significantly increased in v-Msi1 animals compared with WT (Figure S9A), and significantly down regulated in Msi1-KD primary 2D and 3D cultures (Figure S9B). Then, we performed immunostaining to evaluate CCND1 and CDK6 expression in v-Msi1 vs. WT animals. For both proteins, we observed a clear increase in their expression and in the number of positive crypt cells (Figure 4B). For an in depth analysis of MSI1 potential binding on CCND1 and CDK6 mRNAs we employed the human embryonic cell line 293T. First, we observed an increase in both protein levels after MSI1 transient transfection (Figure S10). Binding of MSI1 to CDK6 and CCND1 mRNAs was confirmed by RIP-RT-qPCR. RNPs were immuno-precipitated with either anti-MSI1 antibodies or control IgG and presence of CDK6 and CCND1 mRNAs were evaluated by RT-qPCR (Figure 4C). MSI1 knockdown experiments showed a decrease in mRNA and protein levels of CDK6 and CCND1 by RT-qPCR (Figure 4D) and western blot (Figure 4E). The results of an mRNA-decay assay (Figure 4F) confirmed that higher CDK6 and CCND1 expression upon increased MSI1 levels was due to decreased transcript decay rates, suggesting that MSI1 binding acts to stabilize these mRNAs.
Taken together these results demonstrate that Ccnd1/CCND1 and Cdk6/CDK6 are MSI1 target mRNAs and that there is a direct correlation between the expression levels of MSI1 and these two transcripts and proteins in both intestinal mucosa or epithelial primary cultures from v-Msi1 mice and in 293T cells.
Increased Musashi1 expression has an impact on crypt 3D cultures
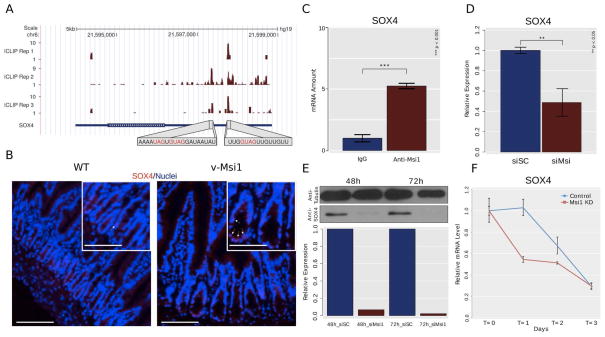
MSI1 has been proposed as a marker of gut stem cells [4, 10] and our previous study indicated an association between Msi1 expression and the stem cell zone of the intestinal crypts [12]. Furthermore, as increased MSI1 expression in v-Msi1 mice provoked a clear expansion of the crypt compartment, we specifically checked for the impact of its increased expression on the intestinal stem cell signature genes defined by Muñoz et al. [10] (Table S6). Interestingly, out of the 510 signature genes, 142 showed differential expression in v-Msi1 compared with WT intestines, with 14 genes being down- and 128 up- regulated (Figure 5). It is worth noting that the best-characterized markers of the crypt stem cells such as Lgr5, Ascl2, Olfm4, Lrig1, Smoc2, are up-regulated in v-Msi1 mice. Among these stem cells genes we focused in particular on Sox4 and investigated its potential direct regulation by MSI1, also according to our CLIP data [52] (Figure 6A). Binding of MSI1 to the Sox4 transcript in v-Msi1 intestinal mucosa was corroborated by RIP-RT-qPCR (Figure S8). SOX4 has well-described roles in stem cell biology [54, 55] and we speculated it could be the main vector of MSI1’s role in intestinal stem cells. In fact, we looked for described targets of SOX4 [56] and determined that 36 differentially expressed genes from our dataset were characterized as SOX4 targets (Table S7), a result that is statistically relevant (p = 0.0072; χ2 test 1-d.f.). By RT-qPCR we confirmed that Sox4 mRNA expression was significantly increased in v-Msi1 animals compared with WT (Figure S9A), and significantly down regulated in Msi1-KD primary 2D and 3D cultures (Figure S9B). Immunostainings for SOX4 protein in intestinal sections showed a clear increase in the number and intensity of SOX4-expressing cells in v-Msi1 vs. WT crypts (Figure 6B). Furthermore, using 293T cells we validated SOX4 as a direct target of MSI1 and determined that MSI1 knockdown decreases SOX4 expression levels and influences its mRNA decay ratios (Figure 6C–F). We also observed an increase in SOX4 protein levels after MSI1 transient transfection (Figure S10).
Figure 5. Stem cell marker expression is affected in v-Msi1 mice intestine.
Stem cell gene markers defined by Muñoz et al. [10] show altered expression in v-Msi1 mice according to RNA-seq analysis.
Figure 6. SOX4 is a direct target of Msi1.
A) UCSC genome browser plot showing the position (x-axis) and count (y-axis) of iCLIP reads overlapping the 3′ UTR of SOX4 mRNA. Highlighted are the sequences of two regions exhibiting particularly high density of iCLIP reads and concordance between replicates. Each region shows the presence of UAG and GUAG oligomers, the known core of the Msi1 binding site. B) SOX4 immunolabeling in intestinal sections from WT or v-Msi1 mice as indicated. Images show merged specific labeling (red) and nuclear staining (blue). Bar=15μm; insets=7μm. C) RIP-qPCR performed with anti-MSI1 and control antibodies in 293T cells shows that SOX4 mRNAs are highly associated with MSI1 protein. D) RT-qPCR showing the impact of MSI1 knockdown on SOX4 mRNA levels. E) Representative western blot showing the impact of MSI1 knockdown on SOX4 protein levels. F) MSI1 knockdown affects SOX4 mRNA decay.
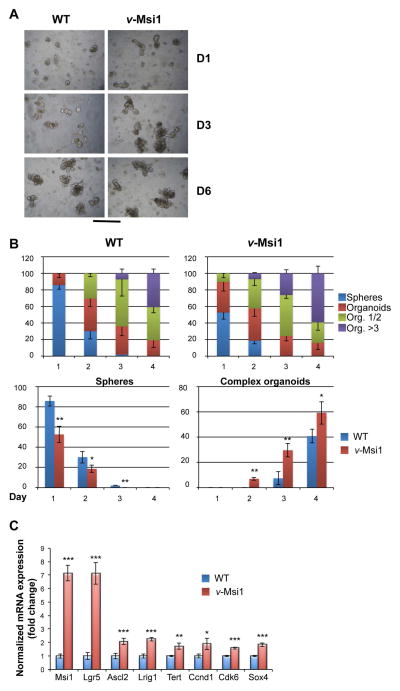
To further confirm that the increased expression of MSI1 in v-Msi1 animals has an impact on crypt cell physiology and on their potentiality ex vivo, we grew 3D crypt cultures [42]. We speculated that an increase in the expression of MSI1, as well as the associated increased expression of several stem cell signature genes, would impact the kinetics of organoids formation. As illustrated in Figure 7A, the crypt cultures established from v-Msi1 animals presented increased and faster budding, clearly visible after one or three days in culture. At six days, when the cultures are fully established, the difference between WT and v-Msi1 is less evident. We quantified the differences in the structures of the organoids over the time in culture and counted the number of simple (sphere) or complex (simple organoids or organoids bearing several buddings) conformations. As illustrated in Figure 7B, in WT cultures there is a significantly higher number of spheres after one day in culture and a slower appearance of organoids bearing more than three budded structures compared with the v-Msi1 crypt cultures. In fact, the v-Msi1 cultures display a higher number of organoids with several buddings at each time point and the differences are statistically significant. We completed our study by analyzing the expression of several stem cell markers and that of the three direct MSI1 mRNA targets Ccnd1, Cdk6 and Sox4. We observed a significant positive regulation of all the markers analyzed in v-Msi1 compared with WT organoid cultures (Figure 7C), and that were down regulated in Msi1-KD primary 2D and 3D cultures (Figure S9C).
Figure 7. Increased growth potentialities of v-Msi1 cultured crypts.
A) Crypts were prepared from WT or v-Msi1 intestine as indicated and maintained in culture for several days, allowing complex organoid development and structuration. Pictures in (A) have been taken under inverted microscope at the indicated days after the start of the culture, and are representative of two independent experiments, each conducted on six replicates. Bar=50μm. B) The number of simple structure (spheres) or organoids of increasing complexity (1 or 2 buds, more than 3 buds) were scored under the inverted microscope during the first four days of culture. Multilayered histograms in the upper panels represent the mean ± SD, n=6, of each counted structure in the cultured crypts of indicated genotype. Histograms in the lower panels show the direct comparison of the number of spheres and that of complex organoids depending on the genotype. *: P<0.05, **: P<0.01, in comparison with WT, by Student’s t-test. C) RT-qPCR analysis of indicated stem cell markers and MSI1 targets in organoids of different genotype. Values represent fold change ± SD, n=4, after normalization to WT organoids. *: P<0.05, **: P<0.01 and ***: P<0.001, in comparison with WT, by Student’s t-test.
In summary, high MSI1 levels in the intestinal crypts of v-Msi1 animals increase the potentialities of growth ex vivo, strongly suggesting an increased stem cell activity.
Discussion
MSI1 has long been described as a potential central actor in intestinal physiology (reviewed in [57]). Although Msi1-KO mice have been developed [58], eventual intestinal-developmental or adult-physiological abnormalities in those mice have never been reported. Using the opposite approach, following our previous results in ex vivo models [39], we developed a transgenic mouse model in which constitutively we expressed MSI1 in the entire intestinal epithelium, representing a model of targeted ectopic- and over-expression. Our data on the phenotypic analysis of the v-Msi1 mice indicate an increased cell proliferation rate in the intestinal epithelium, confirming our previous in vitro data [39]. However, this defect was evident only in the small intestine despite the fact that the transgene targets both the small intestine and the colon similarly. Some still unknown mechanism might be responsible for protecting the colon epithelium from the consequences of high Msi1 expression. Along the same lines, v-Msi1 young adult animals did not develop intestinal cancers as expected from the literature (reviewed in [57]). Although this result initially seems contradictory to our previous observations in primary cultures [39], a direct comparison is misleading. In fact, the over- and ectopic expression of MSI1 in our model was induced in a physiological context. It is likely that complex, still undefined mechanisms are at-play in vivo that ameliorates the deleterious effects of MSI1 overexpression. These results however do not dismiss MSI1 as a driver of tumor initiation in the intestine, but rather suggest that an altered microenvironment and/or additional mutations might be needed to boost MSI1 tumor-inducer potential. Nevertheless, our molecular analyses have revealed that MSI1 overexpression affects several functions and pathways related to stem cell physiology, drug metabolism, cell cycle and DNA synthesis and repair. Indeed, the observed up-regulation of several actors involved in DNA repair in v-Msi1 intestine, strongly suggests that this might be one of the protective mechanisms acting against increased MSI1 expression in a physiological context.
Gene expression analyses, aimed at defining the impact of increased MSI1 expression on the intestinal epithelium in vivo, revealed a strong influence on drug resistance pathways. MSI1 has been previously implicated in drug resistance due to its association with stem cells and “cancer stem cells” [20, 69, 60]. In fact, these cells express high levels of the ABC transporters (ATP binding cassette), which transport drugs outside the cell [61]. Moreover, MSI1-expressing cells in the intestine increase dramatically in number after 5-FU treatment, a routinely used anti-colon cancer drug [59]. MSI1-positive cells also express high levels of the cytokine IL-4, the inhibition of which renders cells more sensitive to 5-FU, oxaliplatin, and death ligand TRAIL treatments [60]. Intriguingly, our data suggests other associations between MSI1 overexpression and drug resistance, offering potential new alternatives to target tumor cells in colon cancer and/or in other tumors with high MSI1 expression. In particular, genes of the Cytocrome P450 super family of mono-oxygenases (CYPs) were down-regulated in v-Msi1 mucosa. CYPs have a large range of action, impacting the metabolism of hormones, drugs and toxic chemicals [62]. Similarly several members of the Glutathione-S-transferase family were down-regulated by MSI1 over-expression. These proteins are detoxification enzymes which catalyze the conjugation of glutathione with broad substrates including chemotherapeutic agents, and are involved in cell protection against apoptotic signals by inhibiting the stress-signaling cascade mediated by ASK1 (Apoptosis signal-regulating kinase)-JNK (c-Jun N-terminal kinase) [63].
Another cellular function highly affected by increased MSI1 expression was cell cycle. The impact of MSI1 on the cell cycle was previously reported in the context of colon cancer cells [23]. Several cyclins (B1, B2, D1, D2 and E1) along with CDK-1, -2 and -6, and CDC20, CDC25c and CDC45 were affected by MSI1-overexpression, suggesting an impact in all three steps of the cell cycle. Interestingly, our data show that Ccnd1/CCND1 and Cdk6/CDK6 mRNAs are stabilized by MSI1, strongly suggesting one mechanism by which MSI1 is implicated in cell cycle control and cell proliferation. Increased cell cycle activity demands a boost in DNA replication. All members of the mini-chromosome maintenance (MCM) 2–7 helicase complex were shown to be up-regulated in v-Msi1 mice. This complex has a role in both the initiation and the elongation phases of eukaryotic DNA replication, specifically the formation and elongation of the replication fork [64]. Other major players in DNA and RNA synthesis like POLE, POLR1B, POLE2, POLA1, POLD2, POLR2B, POLD1, POLR1E were also up-regulated. Similarly, an increase in cell proliferation and DNA synthesis requires more robust DNA repair mechanisms. The most relevant players showing an increased expression in v-Msi1 intestine are two core checkpoint proteins (ATR and CHK1). The ATR-CHK1 pathway is the principal direct effector of the DNA damage and replication checkpoints and it is essential for the survival of many cell types [65]. Given that the v-Msi1 mice did not display any obvious intestinal lesions, we can conclude that these control mechanisms are efficiently working. We can also speculate that this generalized overexpression of players involved in DNA repair might be responsible for preventing intestinal cell transformation and tumor development.
One prominent result from our study is the strong up-regulation of the gut stem cell signature in v-Msi1 mice, and the impact of MSI1 increased expression on ex vivo 3D cultures, strongly suggesting an action on crypt stem cell activity. The association between MSI1 and gut stem cells has already been established [4, 10, 66], but we lacked information concerning its mechanism of action specifically in these cells. Interestingly, we showed that MSI1-overexpression boosts the expression of a large group of stem cell related genes. In particular, driven by MSI1 iCLIP data [52], we focused on the Sox4/SOX4 mRNA and demonstrated that MSI1 directly stabilizes it. Of note, the expression of Sox4 is restricted in mammals to embryonic structures and some adult tissues, such as lymphoid organs, pancreas, intestine, and skin [67–74]. Indeed, SOX4 has well-described roles in stem cell biology [54, 55] and we hypothesize that it could act as a main target for MSI1’s role in crypt stem cell biology. This assumption is supported by the fact that a significant number of SOX4 targets [56] are also up-regulated in v-Msi1 mice.
In conclusion, we have reported here the development and characterization of a new mouse model in which we targeted Msi1 expression. Our results reveal that Msi1 overexpression affects a network of genes and signaling pathways linked to cell cycle, proliferation and stemness. Our study also led to the identification of three new important MSI1 mRNA targets, Ccnd1, Cdk6 and Sox4, representing a step to expand its functional repertoire and define its multiple functions.
Supplementary Material
Figure S1. Generation and validation of v-Msi1 mice. A) Structure of the transgene and localization of the primers for genotyping. Msi1 cDNA has been tagged by a V5 epitope in C-terminal and cloned downstream of the Villin gene promoter. The construct allows the expression of Msi1 specifically in the gut endoderm and intestinal epithelium. B) Analysis of transgenic Msi1 DNA in animals from F5, F7 and F9 lines by PCR. The picture shows the presence of the transgene within the genomic DNA of v-Msi1 animals. CD8 is a positive control for genomic DNA template. C, D) The transgenic Msi1 mRNA is detectable in the intestine of transgenic animals from three founders, but not in the WT littermates (C) or in other organs (D). Ppib has been used as a positive control for the PCR reaction. -, negative control of PCR reaction; SI, small intestine. C, colon; Int, intestine; H, heart; Li, liver; Lu, lung; St, stomach; K, kidney. E) RT-qPCR analysis of Msi1 expression in WT or v-Msi1 intestines, as indicated. Values represent fold change ± SD, n=4, after normalization to WT. *: P<0.05, **: P<0.01, in comparison with WT, by Student’s t-test.
Figure S2. Morphological and proliferative properties of the v-Msi1 intestinal mucosa. A, C) Morphological analysis of the proximal small intestine (A) and colon (C) from WT and v-Msi1 mice as indicated. Graphs show the quantification, for each region, of the number of cells per crypt axis, as indicated by the black, dashed-line double-headed arrows. Approximately 40 axes were scored under the microscope from at least four mice per genotype; histograms represent the mean ± SD. **: P<0.01, in comparison with WT, by Student’s t-test. (B, D) Ki67 immunolabeling of proliferating cells in sections from proximal small intestine (B) and colon (D) of WT and v-Msi1 mice as indicated. Images show merged Ki67 immunolabeling (red) and nuclear staining (blue). Graphs show the quantification, for each region, of the percentage of Ki67 positive cells per crypt. Approximately 40 crypts were scored under the microscope from at least four mice per genotype; histograms represent the mean ± SD. **: P<0.01, in comparison with WT, by Student’s t-test. Bar=15μm. Solid double-headed black arrows in A and C define the length of the vertical epithelial axis. Dashed double-headed black arrows in A show the size of the crypts. White bars in B and D define the limit of the proliferative Ki67-positive cells and dashed double-headed white arrows show the size of the proliferating zone within the crypts.
Figure S3. Knockdown of Msi1 decreases cell proliferation in intestinal epithelial primary cultures. (A–C) Cells were infected with viral particles transducing control-GFP (Ctrl), control Sh-Scrambled (Scr), Sh1-1 or Sh1-4 (both targeting Msi1 mRNA). Results are representative of two independent experiments conducted in four replicates. A) RT-qPCR analysis of Msi1 expression in cells treated as indicated. Values represent mean ± SD, n=4. *: P<0.05, **: P<0.01, in comparison with control-GFP or with Sh-Scr conditions; #: P<0.05, ##: P<0.01, in comparison with Sh1-1, by Student’s t-test. B) Immunostaining for GFP, MSI1 and Ki67 on infected cells under the indicated experimental condition. Images show the merging of the nuclei (blue) and the specific labeling (green or red) as indicated. Bar: 10μm. C) Percentage of Ki67 positive nuclei in infected cells under different experimental conditions as indicated. The histograms represent the mean ± SD, n=4, obtained by counting the positive nuclei under the microscope (approximately 200 cells per experimental condition). *: P<0.05, **: P<0.01, in comparison with control-GFP or Sh-Scr conditions; #: P<0.05, ##: P<0.01, in comparison with Sh1-1, by Student’s t-test.
Figure S4. Up- and down-regulated genes in v-Msi1 intestine in comparison to wild type mice. A) Differentially expressed genes were selected using three methods, Cuffdiff2, DESeq and EdgeR. Only coding genes defined as up- or down-regulated by at least two out of three methods were used (FDR < 0.05). In total, we selected 1,365 up-regulated genes and 1,018 down regulated genes. B) Heat map for differentially expressed genes in v-Msi1 vs. WT samples. C) Volcano plot; up- and down-regulated genes are represented in blue.
Figure S5. Enriched KEGG pathways for genes down-regulated in v-Msi1 samples. Only pathways with adjusted p-value < 0.05 were selected.
Figure S6. Genes down-regulated in Drug Metabolism - Cytochrome P450 pathway. Gene pathway data were obtained from DAVID (http://david.abcc.ncifcrf.gov/). Cytoscape (http://www.cytoscape.org/) was used for pathway visualization.
Figure S7. Analysis of the Wnt pathway in v-Msi1 intestinal mucosa. A) Up- and down-regulated genes present in the WNT pathway. Only 10 up-regulated genes (red cycles) and 4 down-regulated genes (blue cycles) in v-Msi1 intestine (in comparison to wild type mice) are present in the WNT pathway. WNT pathway (KEGG ID: mmu04310) contains 148 genes. B) Western blot analysis of Wnt-related genes. Images show specific protein levels in the intestine from WT and v-Msi1 animals, as indicated. Actin has been used as loading control.
Figure S8. RIP-RT-qPCR experiments show the association between MSI1 protein and Ccnd1, Cdk6 and Sox4 transcripts in the intestine of v-Msi1 mice. Tissue extracts were immuno-precipitated using either anti-MSI1 antibodies or IgG; isolated mRNAs were analyzed by RT-qPCR. In both cases we saw a large enrichment for the transcripts in the anti-MSI1 RIP experiments. In the case of Sox4 mRNA the transcript could not be detected in IgG immuno-precipitated samples in any of the experiments performed. In this case, we used ΔCT values to represent the results instead of RQ, as used for the other analyses.
Figure S9. Validation of differentially expressed genes in v-Msi1 intestinal mucosa and epithelial primary cultures. A) RT-qPCR analysis of indicated genes in animals of different genotype. Values represent fold change ± SD, n=4, after normalization to WT. *: P<0.05, **: P<0.01, in comparison with WT, by Student’s t-test. Note that the expression of the endogenous Msi1 mRNA (Msi1 UTR) is not affected by Msi1-overexpression. B, C) RTqPCR analysis of the indicated MSI1-target mRNAs (B) or stem cell marker mRNAs (C) in primary 2D or 3D cultures, as indicated, infected with control-GFP (Ctrl), Sh-Scr (Scr) or Sh1-1 and Sh1-4, both targeting Msi1 mRNA. Results are representative of two independent experiments conducted in three-four replicates. Values represent mean ± SD, n=3–4. *: P<0.05, **: P<0.01, ***: P<0.001 in comparison with control-GFP or with Sh-Scr conditions; #: P<0.05, ##: P<0.01, in comparison with Sh1-1, by Student’s t-test.
Figure S10. Western analysis shows an increase in expression levels of CCND1, CDK6 and SOX4 in 293T cells transiently transfected with a pcDNA3.1 plasmid expressing MSI1. A pcDNA3.1 plasmid expressing GST was used as a negative control. Tubulin was used as loading control.
List of the oligonucleotides used for the different approaches.
Table S2. Number of genes up- and down-regulated in v-Msi1 vs. wild type samples.
Table S3. Differentially expressed genes between v-Msi1 and wild type samples.
Table S4. Number of KEGG pathways for genes up- and down-regulated in v-Msi1 vs. wild type sample.
Table S5. Full data of enriched KEGG pathways annotation for differentially expressed genes.
Table S6. Differentially expressed genes in v-Msi1 animals matching to the intestinal stem cell signature genes defined by Muñoz et al. (EMBO J 2012, 31:3079-9).
Table S7. Differentially expressed genes in v-Msi1 animals matching to Sox4 targets defined by Scharer et al. (Cancer Res 2009, 69:709-17).
Significance Statement.
The evolutionary conserved RNA-binding protein Musashi1 is a marker of adult stem cells, including those of the intestinal epithelium. With the aim to gain more insights into its function, we generated a mouse model overexpressing Musashi1 specifically in the intestinal epithelium. Detailed phenotypical analyses indicated that this targeted ectopic and over-expression in vivo affects crypt cell physiology, including increased proliferation rate. This is due to the modulation of a complex network of gene functions and pathways such as gut stem cell signature, drug metabolism, cell cycle and DNA synthesis and repair.
Acknowledgments
Grant support
MP’s lab was supported by the Institut National pour le Cancer (grant INCA-2009-175 and PLBIO14-292) and the Ligue contre le cancer Department du Rhone. FC was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) and the Association pour la Recherche sur le Cancer (ARC). LOFP lab was supported by the Voelcker Fund and NIH R01HG006015. BRC was supported by FAPESP (2013/07159-5).
We gratefully acknowledge Nadine Aguilera for animal handling and Julien Nadjar for the excellent technical help. We are grateful to Drs Araujo, Bishop and Frau for the critical reading of the manuscript.
Footnotes
Author’s contribution
Cambuli FM: Performed the studies on the v-Msi1 animals in vivo including histology, immunohistochemistry, RT-PCR and RT-qPCR; realized 3D crypt cultures and their analysis; contributed to the writing of the manuscript.
Correa BR: Conducted RNA-seq and most of the subsequent bioinformatics analyses, helped with RNA experiments and contributed to the writing of the manuscript.
Rezza A: Constructed the transgenesis vector to generate the v-Msi1 animals, helped establishing the different transgenic lines and contributed to the writing of the manuscript.
Burns SC: Executed sample preparation and RNA-seq experiments.
Qiao M: Performed western and RNA analyses.
Uren PJ: Contributed to the bioinformatics analysis, assisted in preparing the manuscript.
Kress E: Contributed to 3D crypt culture experiments and analysis.
Boussouar A: Contributed to primary cultures experiments and analysis.
Galante PAF: Coordinated RNA-seq and bioinformatics analyses and participated in the writing of the manuscript.
Penalva LOF: Led part of the study, coordinated data analysis, put together conclusions and coordinated the writing.
Plateroti M: Led part of the study, coordinated data analysis, put together conclusions and coordinated the writing.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
References
- 1.Stappenbeck TS, Wong MH, Saam JR, Mysorekar IU, Gordon JI. Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Curr Opin Cell Biol. 1998;10(6):702–709. doi: 10.1016/s0955-0674(98)80110-5. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136(5):903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 4.Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71(1):28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 5.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40(7):915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breault DT, Min IM, Carlone DL, Farilla LG, Ambruzs DM, Henderson DE, Algra S, Montgomery RK, Wagers AJ, Hole N. Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells. Proc Natl Acad Sci U S A. 2008;105(30):10420–10425. doi: 10.1073/pnas.0804800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149(1):146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MW, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14(4):401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14(1):106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31(14):3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535(1–3):131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- 12.Cambuli FM, Rezza A, Nadjar J, Plateroti M. Musashi1-Egfp Mice, a New Tool for Differential Isolation of the Intestinal Stem Cell Populations. Stem Cells. 2013;31(10):5. doi: 10.1002/stem.1428. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13(1):67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 14.Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A. 2006;103(22):8402–8407. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411(6833):94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Developmental neuroscience. 2000;22(1–2):139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 17.Sugiyama-Nakagiri Y, Akiyama M, Shibata S, Okano H, Shimizu H. Expression of RNA-binding protein Musashi in hair follicle development and hair cycle progression. Am J Pathol. 2006;168(1):80–92. doi: 10.2353/ajpath.2006.050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke RB, Spence K, Anderson E, Howell A, Okano H, Potten CS. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277(2):443–456. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 19.Glazer RI, Wang XY, Yuan H, Yin Y. Musashi1: a stem cell marker no longer in search of a function. Cell Cycle. 2008;7(17):2635–2639. doi: 10.4161/cc.7.17.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang XY, Yu H, Linnoila RI, Li L, Li D, Mo B, Okano H, Penalva LO, Glazer RI. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget. 2013;4(5):739–750. doi: 10.18632/oncotarget.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 24.Muto J, Imai T, Ogawa D, Nishimoto Y, Okada Y, Mabuchi Y, Kawase T, Iwanami A, Mischel PS, Saya H, et al. RNA-binding protein Musashi1 modulates glioma cell growth through the post-transcriptional regulation of Notch and PI3 kinase/Akt signaling pathways. PLoS One. 2012;7(3):e33431. doi: 10.1371/journal.pone.0033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vo DT, Subramaniam D, Remke M, Burton TL, Uren PJ, Gelfond JA, de Sousa Abreu R, Burns SC, Qiao M, Suresh U, et al. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol. 2012;181(5):1762–1772. doi: 10.1016/j.ajpath.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu HJ, Saito T, Watanabe H, Ito JI, Takeda H, Okano H, Kawata S. Expression of the Musashi1 gene encoding the RNA-binding protein in human hepatoma cell lines. Biochem Biophys Res Commun. 2002;293(1):150–154. doi: 10.1016/S0006-291X(02)00175-4. [DOI] [PubMed] [Google Scholar]
- 27.Kanai R, Eguchi K, Takahashi M, Goldman S, Okano H, Kawase T, Yazaki T. Enhanced therapeutic efficacy of oncolytic herpes vector G207 against human non-small cell lung cancer--expression of an RNA-binding protein, Musashi1, as a marker for the tailored gene therapy. The journal of gene medicine. 2006;8(11):1329–1340. doi: 10.1002/jgm.965. [DOI] [PubMed] [Google Scholar]
- 28.Kanai R, Tomita H, Shinoda A, Takahashi M, Goldman S, Okano H, Kawase T, Yazaki T. Enhanced therapeutic efficacy of G207 for the treatment of glioma through Musashi1 promoter retargeting of gamma34. 5-mediated virulence. Gene therapy. 2006;13(2):106–116. doi: 10.1038/sj.gt.3302636. [DOI] [PubMed] [Google Scholar]
- 29.Seigel GM. Differentiation potential of human retinoblastoma cells. Current pharmaceutical biotechnology. 2011;12(2):213–216. doi: 10.2174/138920111794295846. [DOI] [PubMed] [Google Scholar]
- 30.Ye F, Zhou C, Cheng Q, Shen J, Chen H. Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer. 2008;8:108. doi: 10.1186/1471-2407-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotte M, Wolf M, Staebler A, Buchweitz O, Kelsch R, Schuring AN, Kiesel L. Increased expression of the adult stem cell marker Musashi-1 in endometriosis and endometrial carcinoma. The Journal of pathology. 2008;215(3):317–329. doi: 10.1002/path.2364. [DOI] [PubMed] [Google Scholar]
- 32.Schiapparelli P, Enguita-German M, Balbuena J, Rey JA, Lazcoz P, Castresana JS. Analysis of stemness gene expression and CD133 abnormal methylation in neuroblastoma cell lines. Oncology reports. 2010;24(5):1355–1362. doi: 10.3892/or_00000993. [DOI] [PubMed] [Google Scholar]
- 33.Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, Gelfond JA, Brenner AJ, Patel V, Trageser D, et al. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res. 2012;10(1):143–155. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmuck R, Warneke V, Behrens HM, Simon E, Weichert W, Rocken C. Genotypic and phenotypic characterization of side population of gastric cancer cell lines. Am J Pathol. 2011;178(4):1792–1804. doi: 10.1016/j.ajpath.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan LF, Dong WG, Jiang CQ, Xia D, Liao F, Yu QF. Expression of putative stem cell genes Musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. International journal of colorectal disease. 2010;25(1):17–23. doi: 10.1007/s00384-009-0791-2. [DOI] [PubMed] [Google Scholar]
- 36.Li D, Peng X, Yan D, Tang H, Huang F, Yang Y, Peng Z. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Annals of surgical oncology. 2011;18(7):2074–2083. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 37.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, Matsumoto T, Tamura K, Imai T, Ohnishi T, et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation. 2001;68(2–3):141–152. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu DC, Yang ZL, Jiang S. Identification of musashi-1 and ALDH1 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Cancer biomarkers: section A of Disease markers. 2010;8(3):113–121. doi: 10.3233/DMA-2011-0812. [DOI] [PubMed] [Google Scholar]
- 39.Rezza A, Skah S, Roche C, Nadjar J, Samarut J, Plateroti M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci. 2010;123(Pt 19):3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 40.Pinto D, Robine S, Jaisser F, El Marjou FE, Louvard D. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J Biol Chem. 1999;274(10):6476–6482. doi: 10.1074/jbc.274.10.6476. [DOI] [PubMed] [Google Scholar]
- 41.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci. 1992;101(Pt1):219–231. doi: 10.1242/jcs.101.1.219. [DOI] [PubMed] [Google Scholar]
- 42.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306(2):349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 50.Spears E, Neufeld KL. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J Biol Chem. 2011;286(7):4946–50. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang XY, Yin Y, Yuan H, Sakamaki T, Okano H, Glazer RI. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol Cell Biol. 2008;28(11):3589–3599. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uren PJ, Vo DT, Rosa de Araujo P, Pötschke R, Burns SC, Bahrami-Samani E, Qiao M, de Sousa Abreu R, Nakaya HI, Correa BR, et al. The RNA-binding protein Musashi1 is a central regulator of adhesion pathways in glioblastoma. Mol Cell Biol. 2015 Jun 22; doi: 10.1128/MCB.00410-15. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP--transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp. 2011 Apr;30(50) doi: 10.3791/2638. pii:2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun B, Mallampati S, Gong Y, Wang D, Lefebvre V, Sun X. Sox4 is required for the survival of pro-B cells. Journal of immunology. 2013;190(5):2080–2089. doi: 10.4049/jimmunol.1202736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku JL, Shin YK, Kim DW, Kim KH, Choi JS, Hong SH, Jeon YK, Kim SH, Kim HS, Park JH, et al. Establishment and characterization of 13 human colorectal carcinoma cell lines: mutations of genes and expressions of drug-sensitivity genes and cancer stem cell markers. Carcinogenesis. 2010;31(6):1003–1009. doi: 10.1093/carcin/bgq043. [DOI] [PubMed] [Google Scholar]
- 56.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69(2):709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plateroti M, de Araujo PR, da Silva AE, Penalva LO. The RNA-Binding Protein Musashi1: A Major Player in Intestinal Epithelium Renewal and Colon Cancer Development. Curr Colorectal Cancer Rep. 2012;8(4):290–297. doi: 10.1007/s11888-012-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A. 2002;99(23):15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuqi L, Chengtang W, Ying W, Shangtong L, Kangxiong L. The expression of Msi-1 and its significance in small intestinal mucosa severely damaged by high-dose 5-FU. Dig Dis Sci. 2008;53(9):2436–2442. doi: 10.1007/s10620-007-0155-0. [DOI] [PubMed] [Google Scholar]
- 60.Todaro M, Perez Alea M, Scopelliti A, Medema JP, Stassi G. IL-4-mediated drug resistance in colon cancer stem cells. Cell Cycle. 2008;7(3):309–313. doi: 10.4161/cc.7.3.5389. [DOI] [PubMed] [Google Scholar]
- 61.Sharom FJ. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9(1):105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 62.Guengerich FP. Cytochrome p450 and chemical toxicology. Chemical research in toxicology. 2008;21(1):70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 63.Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22(47):7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forsburg SL. The MCM helicase: linking checkpoints to the replication fork. Biochem Soc Trans. 2008;36(Pt 1):114–119. doi: 10.1042/BST0360114. [DOI] [PubMed] [Google Scholar]
- 65.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Advances in cancer research. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 66.Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129(5):1567–1580. doi: 10.1053/j.gastro.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Deneault E, Cellot S, Faubert A, Laverdure JP, Frechette M, Chagraoui J, Mayotte N, Sauvageau M, Ting SB, Sauvageau G. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137(2):369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4(2):155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hunt SM, Clarke CL. Expression and hormonal regulation of the Sox4 gene in mouse female reproductive tissues. Biology of reproduction. 1999;61(2):476–481. doi: 10.1095/biolreprod61.2.476. [DOI] [PubMed] [Google Scholar]
- 70.Lien WH, Polak L, Lin M, Lay K, Zheng D, Fuchs E. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat Cell Biol. 2014;16(2):179–190. doi: 10.1038/ncb2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19(13):1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. European journal of immunology. 1997;27(5):1292–1295. doi: 10.1002/eji.1830270534. [DOI] [PubMed] [Google Scholar]
- 73.Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G, et al. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132(2):628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 74.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54(12):3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Generation and validation of v-Msi1 mice. A) Structure of the transgene and localization of the primers for genotyping. Msi1 cDNA has been tagged by a V5 epitope in C-terminal and cloned downstream of the Villin gene promoter. The construct allows the expression of Msi1 specifically in the gut endoderm and intestinal epithelium. B) Analysis of transgenic Msi1 DNA in animals from F5, F7 and F9 lines by PCR. The picture shows the presence of the transgene within the genomic DNA of v-Msi1 animals. CD8 is a positive control for genomic DNA template. C, D) The transgenic Msi1 mRNA is detectable in the intestine of transgenic animals from three founders, but not in the WT littermates (C) or in other organs (D). Ppib has been used as a positive control for the PCR reaction. -, negative control of PCR reaction; SI, small intestine. C, colon; Int, intestine; H, heart; Li, liver; Lu, lung; St, stomach; K, kidney. E) RT-qPCR analysis of Msi1 expression in WT or v-Msi1 intestines, as indicated. Values represent fold change ± SD, n=4, after normalization to WT. *: P<0.05, **: P<0.01, in comparison with WT, by Student’s t-test.
Figure S2. Morphological and proliferative properties of the v-Msi1 intestinal mucosa. A, C) Morphological analysis of the proximal small intestine (A) and colon (C) from WT and v-Msi1 mice as indicated. Graphs show the quantification, for each region, of the number of cells per crypt axis, as indicated by the black, dashed-line double-headed arrows. Approximately 40 axes were scored under the microscope from at least four mice per genotype; histograms represent the mean ± SD. **: P<0.01, in comparison with WT, by Student’s t-test. (B, D) Ki67 immunolabeling of proliferating cells in sections from proximal small intestine (B) and colon (D) of WT and v-Msi1 mice as indicated. Images show merged Ki67 immunolabeling (red) and nuclear staining (blue). Graphs show the quantification, for each region, of the percentage of Ki67 positive cells per crypt. Approximately 40 crypts were scored under the microscope from at least four mice per genotype; histograms represent the mean ± SD. **: P<0.01, in comparison with WT, by Student’s t-test. Bar=15μm. Solid double-headed black arrows in A and C define the length of the vertical epithelial axis. Dashed double-headed black arrows in A show the size of the crypts. White bars in B and D define the limit of the proliferative Ki67-positive cells and dashed double-headed white arrows show the size of the proliferating zone within the crypts.
Figure S3. Knockdown of Msi1 decreases cell proliferation in intestinal epithelial primary cultures. (A–C) Cells were infected with viral particles transducing control-GFP (Ctrl), control Sh-Scrambled (Scr), Sh1-1 or Sh1-4 (both targeting Msi1 mRNA). Results are representative of two independent experiments conducted in four replicates. A) RT-qPCR analysis of Msi1 expression in cells treated as indicated. Values represent mean ± SD, n=4. *: P<0.05, **: P<0.01, in comparison with control-GFP or with Sh-Scr conditions; #: P<0.05, ##: P<0.01, in comparison with Sh1-1, by Student’s t-test. B) Immunostaining for GFP, MSI1 and Ki67 on infected cells under the indicated experimental condition. Images show the merging of the nuclei (blue) and the specific labeling (green or red) as indicated. Bar: 10μm. C) Percentage of Ki67 positive nuclei in infected cells under different experimental conditions as indicated. The histograms represent the mean ± SD, n=4, obtained by counting the positive nuclei under the microscope (approximately 200 cells per experimental condition). *: P<0.05, **: P<0.01, in comparison with control-GFP or Sh-Scr conditions; #: P<0.05, ##: P<0.01, in comparison with Sh1-1, by Student’s t-test.
Figure S4. Up- and down-regulated genes in v-Msi1 intestine in comparison to wild type mice. A) Differentially expressed genes were selected using three methods, Cuffdiff2, DESeq and EdgeR. Only coding genes defined as up- or down-regulated by at least two out of three methods were used (FDR < 0.05). In total, we selected 1,365 up-regulated genes and 1,018 down regulated genes. B) Heat map for differentially expressed genes in v-Msi1 vs. WT samples. C) Volcano plot; up- and down-regulated genes are represented in blue.
Figure S5. Enriched KEGG pathways for genes down-regulated in v-Msi1 samples. Only pathways with adjusted p-value < 0.05 were selected.
Figure S6. Genes down-regulated in Drug Metabolism - Cytochrome P450 pathway. Gene pathway data were obtained from DAVID (http://david.abcc.ncifcrf.gov/). Cytoscape (http://www.cytoscape.org/) was used for pathway visualization.
Figure S7. Analysis of the Wnt pathway in v-Msi1 intestinal mucosa. A) Up- and down-regulated genes present in the WNT pathway. Only 10 up-regulated genes (red cycles) and 4 down-regulated genes (blue cycles) in v-Msi1 intestine (in comparison to wild type mice) are present in the WNT pathway. WNT pathway (KEGG ID: mmu04310) contains 148 genes. B) Western blot analysis of Wnt-related genes. Images show specific protein levels in the intestine from WT and v-Msi1 animals, as indicated. Actin has been used as loading control.
Figure S8. RIP-RT-qPCR experiments show the association between MSI1 protein and Ccnd1, Cdk6 and Sox4 transcripts in the intestine of v-Msi1 mice. Tissue extracts were immuno-precipitated using either anti-MSI1 antibodies or IgG; isolated mRNAs were analyzed by RT-qPCR. In both cases we saw a large enrichment for the transcripts in the anti-MSI1 RIP experiments. In the case of Sox4 mRNA the transcript could not be detected in IgG immuno-precipitated samples in any of the experiments performed. In this case, we used ΔCT values to represent the results instead of RQ, as used for the other analyses.
Figure S9. Validation of differentially expressed genes in v-Msi1 intestinal mucosa and epithelial primary cultures. A) RT-qPCR analysis of indicated genes in animals of different genotype. Values represent fold change ± SD, n=4, after normalization to WT. *: P<0.05, **: P<0.01, in comparison with WT, by Student’s t-test. Note that the expression of the endogenous Msi1 mRNA (Msi1 UTR) is not affected by Msi1-overexpression. B, C) RTqPCR analysis of the indicated MSI1-target mRNAs (B) or stem cell marker mRNAs (C) in primary 2D or 3D cultures, as indicated, infected with control-GFP (Ctrl), Sh-Scr (Scr) or Sh1-1 and Sh1-4, both targeting Msi1 mRNA. Results are representative of two independent experiments conducted in three-four replicates. Values represent mean ± SD, n=3–4. *: P<0.05, **: P<0.01, ***: P<0.001 in comparison with control-GFP or with Sh-Scr conditions; #: P<0.05, ##: P<0.01, in comparison with Sh1-1, by Student’s t-test.
Figure S10. Western analysis shows an increase in expression levels of CCND1, CDK6 and SOX4 in 293T cells transiently transfected with a pcDNA3.1 plasmid expressing MSI1. A pcDNA3.1 plasmid expressing GST was used as a negative control. Tubulin was used as loading control.
List of the oligonucleotides used for the different approaches.
Table S2. Number of genes up- and down-regulated in v-Msi1 vs. wild type samples.
Table S3. Differentially expressed genes between v-Msi1 and wild type samples.
Table S4. Number of KEGG pathways for genes up- and down-regulated in v-Msi1 vs. wild type sample.
Table S5. Full data of enriched KEGG pathways annotation for differentially expressed genes.
Table S6. Differentially expressed genes in v-Msi1 animals matching to the intestinal stem cell signature genes defined by Muñoz et al. (EMBO J 2012, 31:3079-9).
Table S7. Differentially expressed genes in v-Msi1 animals matching to Sox4 targets defined by Scharer et al. (Cancer Res 2009, 69:709-17).