SUMMARY
Salivary proteins modulate bacterial colonization in the oral cavity and interact with systemic pathogens that pass through the oropharynx. An interesting example is the opportunistic respiratory pathogen Streptococcus pneumoniae that normally resides in the nasopharynx, but belongs to the greater Mitis group of streptococci, most of which colonize the oral cavity. S. pneumoniae also expresses a serine-rich repeat (SRR) adhesin, PsrP, that is a homologue to oral Mitis group SRR adhesins, such as Hsa of S. gordonii and SrpA of S. sanguinis. Since the latter bind to salivary glycoproteins through recognition of terminal sialic acids, we wanted to determine whether S. pneumoniae also binds to salivary proteins through possibly the same mechanism. We found that only a capsule-free mutant of S. pneumoniae TIGR4 binds to salivary proteins, most prominently to mucin MUC7, but that this binding was not mediated through PsrP or recognition of sialic acid. We also found, however, that PsrP is involved in agglutination of human red blood cells (RBCs). After removal of PsrP, an additional previously masked lectin-like adhesin activity mediating agglutination of sialidase-treated RBCs becomes revealed. Using a custom-spotted glycoprotein and neoglycoprotein dot blot array, we identify candidate glycan motifs recognized by PsrP and by the putative S. pneumoniae adhesin that could perhaps be responsible for pneumococcal binding to salivary MUC7 and glycoproteins on RBCs.
Keywords: Saliva, salivary proteins, serine-rich repeat adhesins, bacterial lectins
INTRODUCTION
Saliva helps to maintain an ecological balance within the diverse oral biofilm microbiota through clearance of pathogens and fostering colonization of the mouth by a physiological commensal microflora (Ruhl, 2012, Scannapieco, 1994, Nobbs et al., 2011). Oral Mitis group streptococci are frequently found among the earliest colonizers of human teeth (Frandsen et al., 1991). Binding of these bacteria to the surface of teeth requires specific biochemical interactions of protein adhesins expressed on the bacteria with cognate receptor motifs on salivary proteins and glycoproteins that are adsorbed to the mineralized tooth surface as a thin film, the so-called acquired enamel pellicle (Lendenmann et al., 2000, Nobbs et al., 2011). On salivary glycoproteins, terminal sialic acids serve as important glycan receptor motifs recognized by corresponding sialic acid-binding adhesins of oral commensal Mitis group streptococci (Levine et al., 1978, Murray et al., 1992, Murray et al., 1982, Deng et al., 2014). One example for such an adhesin is Hsa, expressed on the surface of Streptococcus gordonii DL1 (Challis), that preferentially binds sialic acids in a terminal position α2-3-linked to O-linked glycan chains (Takahashi et al., 1997, Takahashi et al., 2002a, Deng et al., 2014). Major sialoglycoproteins in saliva that serve as counter-receptors for Hsa-mediated bacterial adhesion are the low-molecular weight salivary mucin-7 (MUC7) (Ruhl et al., 2004, Takamatsu et al., 2006) and the heavy-chain of secretory immunoglobulin A1 (Ruhl et al., 1996). Hsa structurally and genetically belongs to a wider family of bacterial serine-rich repeat protein (SRR) adhesins that all participate in bacterial adhesion, colonization, and opportunistic virulence (Lizcano et al., 2012, Löfling et al., 2011, Pyburn et al., 2011, Zhou & Wu, 2009, Deng et al., 2014, Turner et al., 2009). Homologues of SRR adhesins are expressed by different bacterial genera including a number of other streptococcal species (Wu et al., 1998, Bensing et al., 2004, Plummer et al., 2005, Obert et al., 2006). However, besides Hsa, binding to sialic acid on salivary glycoproteins by SRR adhesins has only been shown for GspB (Takamatsu et al., 2006, Deng et al., 2014), a close homologue of Hsa in S. gordonii M99, and SrpA of S. sanguinis SK36 (Plummer & Douglas, 2006, Deng et al., 2014).
One prominent non-oral Mitis group streptococcal species that also expresses an SRR adhesin is the opportunistic respiratory pathogen Streptococcus pneumoniae (Löfling et al., 2011, Rose et al., 2008, Obert et al., 2006, Kawamura et al., 1995, Kilian et al., 2014, Johnston et al., 2010). The SRR adhesin expressed by S. pneumoniae was identified on strain TIGR4 (Hava & Camilli, 2002, Tettelin et al., 2001) and termed pneumococcal serine-rich repeat protein (PsrP) (Obert et al., 2006). PsrP is not required for nasal colonization, but is rather a lung-specific virulence factor (Obert et al., 2006, Rose et al., 2008), which also mediates pneumococcal intra-species aggregation and biofilm formation (Sanchez et al., 2010, Blanchette-Cain et al., 2013). It has been shown that PsrP binds to keratin 10 present on human lung cells (Shivshankar et al., 2009) through a binding domain that resembles microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (Schulte et al., 2014). There exist, however, earlier reports suggesting a role of glycan binding for adherence of S. pneumoniae (Barthelson et al., 1998, Idänpään-Heikkila et al., 1997, Andersson et al., 1983, Cundell & Tuomanen, 1994, Krivan et al., 1988). Even though the binding of PsrP to keratin 10 was proven to be independent of lectin-carbohydrate interaction (Shivshankar et al., 2009), the possible binding of the PsrP adhesin to glycans on other host proteins has not been fully excluded and deserves further study, particularly in view of its close homology with the glycan-binding adhesins Hsa and SrpA. In this study, we aimed to determine by using the bacterial overlay technique (Deng et al., 2014, Walz et al., 2009, Prakobphol et al., 1987) if S. pneumoniae TIGR4 and, specifically, PsrP can bind to salivary glycoproteins through a similar mechanism as Hsa and SrpA. Because S. pneumoniae differs from the other Mitis group streptococci by the expression of a polysaccharide capsule that can vary in its thickness (Kim & Weiser, 1998, Lizcano et al., 2012, Löfling et al., 2011), we have also investigated the influence of this polysaccharide layer on adhesin activity.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The bacterial strains and mutants used in this study are listed in Table 1. All streptococcal strains were cultured overnight at 37°C as stationary cultures in a 5% CO2 environment. S. gordonii DL1 and isogenic Hsa-deficient mutant (kindly provided by Dr. Howard F. Jenkinson, University of Bristol, UK) as well as S. sanguinis SK36 and isogenic SrpA-deficient mutant (kindly provided by Dr. Hui Wu, University of Alabama at Birmingham) were grown in brain heart infusion (Becton Dickinson, Franklin Lakes, NJ). S. pneumoniae wild-type strain TIGR4, capsule-free mutant strain HR1001.1 and isogenic SRR adhesin-deficient mutants were grown as suspension cultures in Todd-Hewitt broth (Difco Laboratories, Detroit, MI) containing 0.5% yeast extract, or as plate cultures on tryptic soy agar supplemented with 5% sheep blood. Bacteria were washed three times in 0.02 M phosphate buffered saline containing 0.02% sodium azide (PBS, pH 7.2) before use in the assays. In assays involving S. pneumoniae, 2% choline chloride was included at all steps to prevent pneumococcal autolysis (Martner et al., 2009), which we found resulted in unreproducible bacterial binding results and also adhesin-independent agglutination of RBCs, as reported much earlier (Löffler, 1950). Antibiotic concentrations that were used for selection are as follows: kanamycin (Kan) was used at 250 μg/ml and 25 μg/ml for S. gordonii mutant strain UB1545 and S. sanguinis mutant strain VT1614, respectively; erythromycin (Erm) was used at 0.3 μg/ml and chloramphenicol (Cm) was used at 4 μg/ml for pneumococci where appropriate. All antibiotics were purchased from Sigma-Aldrich, St. Louis, MO.
Table 1.
Bacterial strains used in this study.
| Strain | Relevant characteristics | References |
|---|---|---|
| DL1 (Challis) | Streptococcus gordonii blood isolate | (Kilian et al., 1989) |
| UB1545 | DL1, hsa::aphA3; Kanr | (Jakubovics et al., 2005) |
| SK36 | S. sanguinis human oral plaque isolate | (Plummer et al., 2005) |
| VT1614 | SK36, srpA::aphA3; Kanr | (Plummer et al., 2005) |
| TIGR4 | S. pneumoniae serotype 4 clinical isolate | (Tettelin et al., 2001) |
| STM199 | Signature-tagged mutagenesis of psrP gene done in TIGR4; Cmr | (Hava & Camilli, 2002) |
| HR1001.1 | Capsule-free TIGR4 | This study |
| HR1001.1ΔpsrP | Capsule-free and ΔpsrP | This study |
Kanr, Kanamycin resistant; Cmr, Chloramphenicol resistant.
Construction of capsule-free TIGR4 and psrP-deficient mutants
A capsule-free S. pneumoniae TIGR4 mutant was generated by transforming encapsulated TIGR4 bacteria with chromosomal DNA from strain JD908 (a capsule-free serotype 3 strain carrying an Erm cassette in the capsule synthase gene wchE (cpsS) required for capsule production and having homology to the TIGR4 capsule locus (Dillard & Yother, 1994)) in the presence of competence signal peptide 2 (CSP2). Unencapsulated clones were selected on blood agar plates containing 0.3 μg/ml erythromycin after overnight grown at 37°C in a 5% CO2 environment. Two independent erythromycin-resistant clones were selected and were transformed with chromosomal DNA from the TIGR4 wild-type parent strain (back-crossed) three times and selected on erythromycin agar plates to retain the capsule mutation, yet ensure that the remaining genetic background consisted purely of TIGR4 chromosomal DNA, as described (Kelly et al., 1994). The final capsule-free S. pneumoniae TIGR4 mutant used in this study was named HR1001.1. This mutant was shown to lack capsule production by ELISA, according to the method of Bender and Yother (Bender & Yother, 2001). Briefly, pneumococci were grown to an OD600 of 0.5, washed and resuspended in PBS and heat killed at 56°C. Bacterial suspension adjusted to an OD of 0.2 with PBS (100 μL) was added in a two-fold dilution series to ELISA plates and left at 4°C overnight. After washing the plates three times with PBS containing 0.05% Tween 20, a capsule type 4-specific rabbit anti-serum (Statens Serum Institute, Copenhagen, Denmark) was added as the primary antibody (1:10,000 dilution in PBS). After 1 h of incubation at room temperature, the plates were washed and HRP-conjugated goat anti-rabbit antibodies (1:1,000 in PBS-T) were added for 1 h. The plates were washed and developed using o-phenylenediamine dihydrochloride substrate, and the absorbance was read at 492 nm in a Multiskan MS microtitre plate reader (Labsystems). Results of the HR1001.1 mutant were compared to wild-type TIGR4 using JD908 as a negative control for background absorbance. The HR1001.1 showed no absorbance above the JD908 background.
For deletion of the psrP gene, HR1001.1 was transformed with chromosomal DNA from STM199, a chloramphenicol-resistant TIGR4 transposon-mutant lacking expression of the psrP gene (Hava & Camilli, 2002), kindly provided by Dr. Andrew Camilli, Tufts University School of Medicine, Boston, MA. Transformants were selected on blood agar plates containing chloramphenicol (4 μg/ml) and erythromycin (0.3 μg/ml). The resulting strain used in this study was named HR1001.1ΔpsrP. All mutants were verified by PCR and sequencing of amplicons, using primers upstream of the insertion of each construct and from within the inserted resistance cassette.
Bacterial-mediated hemagglutination
Human blood (group 0) was collected in Vacutainers containing 3.2% sodium citrate as anticoagulant (Becton Dickenson) according to guidelines approved by the Health Science Institutional Review Board (#ORB0511008E), University at Buffalo. Red blood cells (RBCs) were washed and resuspended to a final concentration of 0.5 % (v/v) in PBS containing 2 mg/ml bovine serum albumin (Sigma-Aldrich, St. Louis, MO) (PBS-BSA). For removal of sialic acids, RBCs were treated for 1 h at 37°C with sialidase from Clostridium perfringens (Type X, Sigma-Aldrich) at a final concentration of 0.05 U/ml in PBS and washed three times with PBS thereafter. To demonstrate glycan-dependency of binding, periodate treatment was performed with either 2 mM or 10 mM sodium periodate in PBS (pH 6.5) to oxidize sialic acids or general glycans, respectively (Fukuda, 2001). The reaction was carried out for 1 h at 4°C in the dark with constant agitation and was stopped thereafter by the addition of 100 mM sodium borohydride for 10 min at room temperature. The volume ratio of sodium periodate to sodium borohydride was 4 parts to 1 part. After treatment, RBCs were washed three times with sodium acetate buffer (50 mM sodium acetate, 100 mM sodium chloride, pH 5.5), followed by washing with PBS twice more before resuspension. Both sialidase- and periodate-treated RBCs were resuspended in PBS-BSA to a final concentration of 0.5% (v/v) for later use in the hemagglutination assay. Hemagglutination was performed as previously described (Takahashi et al., 1997). Two-fold serial dilutions of 75 μl bacterial suspensions at an optical density of 1.0 (~ 109 streptococci per ml) in PBS-BSA were mixed with 25 μl untreated, sialidase-treated, or periodate-treated RBCs in round-bottom 96-well microtiter plates (Costar, Corning Incorporated, Corning, NY). As a control for the presence of sialic acids on RBCs, serial two-fold dilutions of Maackia amurensis lectin (MAL II, Vector Laboratories, Burlingame, CA) starting at 0.01 mg/ml were substituted for bacteria. Hemagglutination titers were scored after overnight stationary incubation of plates at 4°C and recorded using a digital camera.
Dot blot preparations, SDS PAGE, and electroblot transfer
Dot blots for bacterial overlay were prepared by immobilizing human salivary samples, naturally occurring glycoproteins (Table S1) and neoglycoproteins (Table S2) as dots containing 1 μg of protein on nitrocellulose (0.45 μm pore size, Whatman Protran BA 85, Fisher Scientific). Salivary samples that were collected on ice as previously described (Heo et al., 2013) according to the protocol approved by the University at Buffalo’s Health Sciences Institutional Review Board (HSIRB #ORB0511008E) included whole saliva (WS), parotid saliva (PAR) and submandibular/sublingual saliva (SMSL). Unstimulated WS samples were centrifuged at 12,000 x g for 15 min at 4°C to remove unwanted particulates. The resultant clarified saliva supernatant was transferred into a separate polypropylene microtube and stored at −80°C until further use. Samples treated with sialidase from Clostridium perfringens (Type X, Sigma-Aldrich) at a final concentration of 0.05 U/ml at 37°C for 30 min were also included in the dot blots where indicated. Proteins and glycoproteins of WS, PAR and SMSL secretions, both untreated and sialidase-treated, were denatured under reducing conditions and separated by 8–16% gradient Tris-glycine mini gels (Novex, Invitrogen, Carlsbad, CA) as previously described (Walz et al., 2009). Separated salivary glycoproteins (15 μg of total protein per lane) were visualized in gels by periodic acid-Schiff (PAS) stain (Heo et al., 2013) and imaged using a flat-bed scanner (ImageScanner III, GE Healthcare, Piscataway, NJ) in the transparent mode. In parallel, proteins and glycoproteins (15 μg of total protein per lane) were transferred to nitrocellulose (0.45 μm pore size, Whatman Protran BA 85, Fisher Scientific, Fair Lawn, NJ) in a semi-dry transfer unit (Amersham Biosciences, GE Healthcare, Piscataway, NJ) for 3 h at room temperature using transfer buffer (25 mM Tris, 192 mM glycine, 20% v/v methanol) under constant current of 45 mA per gel. Transfer membranes were briefly rinsed in Tris-buffered saline (0.15 M NaCl, 20 mM Tris HCl and 0.02% NaN3) (TBS) prior to performing immunoblotting or bacterial overlay.
Immunoblotting
Immunoblotting was performed as previously described (Heo et al., 2013). In brief, membranes were blocked with 2% non-fat dry milk (Carnation, Nestlé, Solon, OH) in TBS containing 0.1% Tween-20 (TBST) and subsequently incubated with 1:2,000 diluted rabbit polyclonal anti-human mucin-7 (MUC7) antiserum (AB-3, a gift from Dr. Libuse A. Bobek at the University at Buffalo). After washing with TBST, bound antibodies were detected with 1:1,000 diluted AlexaFluor 488 IgG (H+L) secondary antibodies (Molecular Probes, Invitrogen). All incubations were done for 1 h at room temperature. Signals of bound antibodies were detected using a fluorescence laser scanner (Typhoon 9400, GE Healthcare).
Bacterial overlay
Fluorochrome-labeling of bacteria and overlay method were performed as previously described (Walz et al., 2005). In brief, dot blots or transfers were blocked in 5% BSA and subsequently overlaid with suspensions of fluorescein-5-isothiocyanate (FITC)-labeled bacteria for 2 h to allow for binding. Membranes were then washed at 4°C to remove unbound bacteria and dried. Unless otherwise indicated, 1 mM CaCl2 and 1 mM MgCl2 were included in buffers, while 5 mM EDTA was used for conditions where the overlay was performed in the absence of divalent cations. Fluorescence signals of bound bacteria were detected using a fluorescence laser scanner (Typhoon 9400) and were analyzed by densitometry using the ImageQuant software (Version 5.2, Molecular Dynamics, Sunnyvale, CA).
RESULTS
S. pneumoniae binds to salivary proteins only when unencapsulated
Binding of the streptococcal strains to saliva was first tested by overlaying dot blots of immobilized salivary secretions with suspensions of fluorochrome-labeled bacteria (Fig. 1). Wild-type strain S. gordonii DL1 bound strongly to whole saliva (WS) and submandibular-sublingual (SMSL) secretion, but only weakly to parotid (PAR) secretion. This binding was abrogated or, in the case of SMSL secretion, much reduced when the salivary samples were treated with sialidase prior to immobilizing them on the membrane. Together with the fact that strain DL1 bound to fetuin but not to its de-sialylated form, these findings confirm that binding of S. gordonii DL1 to these substrates is mediated by recognition of sialic acid. Furthermore, because the Hsa adhesin-deficient mutant of strain DL1 did not bind to the same substrates, it can be concluded that binding was mediated through the SRR adhesin Hsa, which is in agreement with previous studies (Takahashi et al., 1997, Deng et al., 2014). It has to be noted that the binding experiment was performed in the presence of EDTA to focus on the sialic acid binding activity and to abrogate other adhesin activities on strain DL1 (Takahashi et al., 2002b, Ruhl et al., 2004, Deng et al., 2014) that require the presence of divalent cations (Takahashi et al., 1997).
Fig. 1.
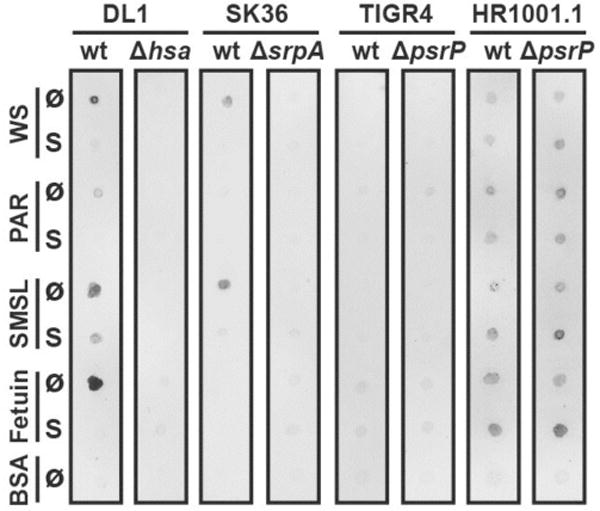
Streptococcal adhesion to immobilized whole saliva, parotid saliva and submandibular/sublingual saliva. Suspensions of fluorochrome-labeled S. gordonii DL1, S. sanguinis SK36, S. pneumoniae wild-type strain TIGR4, capsule-free mutant strain HR1001.1, and the respective isogenic SRR adhesin-deficient mutants were overlaid on nitrocellulose membranes carrying immobilized untreated (Ø) or sialidase-treated (S) dots (1 μg of total protein per dot) of whole saliva (WS), parotid saliva (PAR), and submandibular/sublingual saliva (SMSL). Fetuin and bovine serum albumin (BSA) were included as controls. Fluorescent signals of bound bacteria were detected by use of a fluorescence scanner.
Wild-type strain S. sanguinis SK36 bound to WS and SMSL secretions similar to strain DL1, and no binding was seen to sialidase-treated samples. Binding of strain SK36 was clearly mediated by the SRR adhesin SrpA, since the adhesin-deficient mutant did not bind. This finding also agrees with previous reports (Plummer & Douglas, 2006). In contrast to DL1, strain SK36 did not bind to fetuin. This could be due to overall weaker binding of SK36 or to the fact that this strain requires a certain subtype, linkage, or sterical presentation of sialic acid for binding that is present on salivary proteins, but not on fetuin (Deng et al., 2014).
Neither S. pneumoniae TIGR4 wild-type nor the PsrP-deficient mutant strain bound to the immobilized saliva samples. However, the capsule-free mutant strain HR1001.1 showed binding. Interestingly, strain HR1001.1 also bound to fetuin. However, neither treatment of the samples by sialidase nor removal of PsrP affected binding of strain HR1001.1. Notably, in the case of fetuin and to a certain degree also for SMSL-secretion, binding by the PsrP-deficient HR1001.1 mutant strain was stronger to the sialidase-treated samples. These findings show that after removal of their capsule, pneumococci are able to bind to salivary compounds but suggest that neither the recognition of sialic acid nor the adhesin PsrP play a dominant role in binding to saliva.
Unencapsulated S. pneumoniae binds to salivary mucin MUC7
To find out which salivary proteins serve as counter-receptors for lectin-like bacterial adhesin-mediated binding, transfer blots of SDS-PAGE-separated salivary secretions were overlaid with fluorochrome-labeled streptococcal strains and respective SRR adhesin-deficient mutants (Fig. 2). Both S. gordonii DL1 and S. sanguinis SK36 bound predominantly to a glycosylated protein band of ~150 kDa, present in WS and SMSL secretion, which was identified by periodic acid-Schiff glycan stain (Fig. 2 A) and immunoblotting (Fig. 2 B) as the low-molecular weight salivary mucin-7 (MUC7). Binding of DL1 occurred only to the sialylated form of MUC7. When sialic acid was removed by treating the samples with sialidase prior to electrophoretic separation, binding was abolished (Fig. 2 C). The loss of sialic acids on MUC7 became also evident from the glycoprotein stain and the immunoblot (Fig. 2 A, B) in that the sialylated form of MUC7 migrated faster in the gel than the de-sialylated form due to the overall loss of negative charge of the molecule after removal of sialic acids (Kirkbride et al., 2001). Binding of both strains to MUC7 was also clearly SRR adhesin-dependent, as seen with loss of binding in the adhesin-deficient mutants. For DL1 and to a lesser degree for SK36, binding to a lower 130 kDa band was observed. This band represents most likely a subtype of MUC7 that can be detected by various MUC7-specific antibodies (Soares et al., 2012), but was apparently not recognized by the anti-MUC7 antibody used in this study.
Fig. 2.
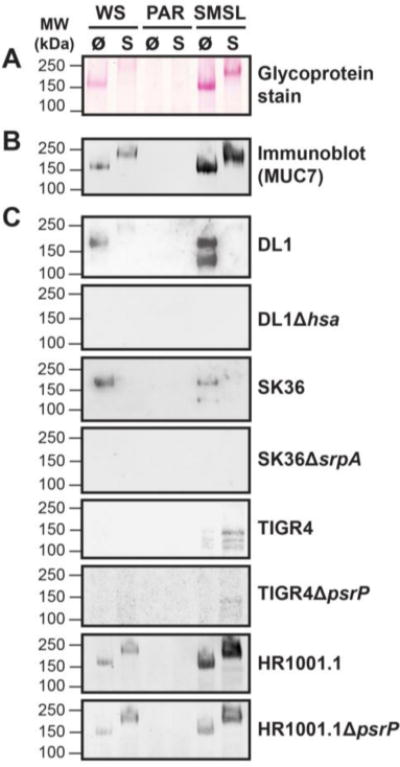
Streptococcal binding to salivary mucin MUC7. Untreated (Ø) or sialidase-treated (S) whole saliva (WS) as well as parotid (PAR), and submandibular/sublingual glandular secretions (SMSL) were separated by SDS-PAGE under reducing conditions, transferred to nitrocellulose and overlaid with fluorochrome-labeled bacteria. A, Periodic acid-Schiff stain to reveal the glycosylated mucin bands. B, Immunoblot with anti-MUC7 antibody. C, Bacterial overlay with fluorochrome-labeled S. gordonii DL1, S. sanguinis SK36, encapsulated S. pneumoniae wild-type strain TIGR4, capsule-free mutant strain HR1001.1, and respective isogenic SRR adhesin-deficient mutants. Fluorescent signals of bound antibodies or bound bacteria were detected using a fluorescence scanner.
For S. pneumoniae, encapsulated strain TIGR4 did not bind to any secretions and only weak binding was observed to a triplet of bands ranging between ~100 – 150 kDa in sialidase-treated SMSL secretion that could not be identified. However, the capsule-deficient mutant strain HR1001.1 bound to MUC7, but binding was neither dependent on the presence of sialic acid nor on the expression of the PsrP adhesin on the pneumococci (Fig. 2 C). This shows that the PsrP adhesin, similar to the results obtained in the dot blot experiment (Fig. 1), is not required for binding to salivary proteins and suggests that another putative adhesin might possibly be responsible for binding to MUC7.
Unencapsulated S. pneumoniae agglutinates RBCs in a glycan-dependent but sialic acid-independent manner
To further scrutinize whether S. pneumoniae can bind to sialic acids, we tested for hemagglutination ability of these bacteria using untreated and sialidase-treated human RBCs (Fig. 3 A). S. gordonii DL1 hemagglutinated human RBCs to a similar extent as lectin Maackia amurensis (MAL II), which specifically binds to α2,3-linked sialic acids. Hemagglutination caused by DL1 was sialic acid-dependent since it could be abolished by sialidase treatment of the RBCs. Hemagglutination activity was also entirely dependent on the presence of the Hsa adhesin on strain DL1. S. sanguinis SK36 displayed much weaker hemagglutination activity than S. gordonii DL1, but its activity was clearly sialic acid- and SrpA adhesin-dependent.
Fig. 3.
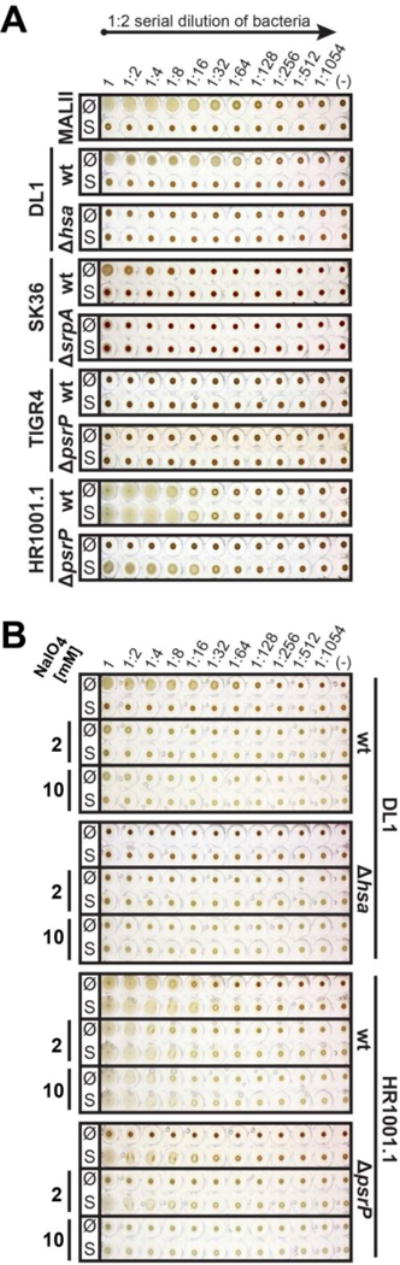
Streptococcal-mediated hemagglutination of human red blood cells. Two-fold serial dilutions of S. gordonii DL1, S. sanguinis SK36, S. pneumoniae wild-type strain TIGR4, capsule-free mutant strain HR1001.1, and respective isogenic SRR adhesin-deficient mutants were mixed and incubated with human red blood cells. The sialic acid specific lectin from Maackia amurensis (MAL II) was used as a control for the presence or loss of sialic acid residues. A, Hemagglutination done with untreated (Ø) or sialidase-treated (S) RBCs. B, Treatment of RBCs with 2 mM or 10 mM sodium periodate to oxidize sialic acids or general glycans, respectively, prior to hemagglutination.
For S. pneumoniae, hemagglutination occurred only with the capsule-free mutant strain HR1001.1, which agrees with its ability to bind salivary proteins (Fig. 2). Sialidase treatment of RBCs did not reduce, but rather improved hemagglutination by strain HR1001.1. This indicates that sialic acid may not be a receptor on RBCs but that subterminal glycans, revealed after removal of terminal sialic acids, may serve as receptor motifs for pneumococcal binding. Remarkably, agglutinating activity was abolished in the PsrP adhesin-deficient mutant of strain HR1001.1 which suggests that PsrP may exhibit lectin-like binding. However, the PsrP adhesin-deficient mutant still agglutinated sialidase-treated RBCs. Taken together, these observations support the assumption that S. pneumoniae may express two glycan-binding adhesins.
To determine glycan dependency of S. pneumoniae binding to RBCs, we performed periodate treatment of both untreated and sialidase-treated RBCs prior to hemagglutination with 2 mM or 10 mM sodium periodate to oxidize sialic acids or general glycans on the RBCs, respectively (Fig. 3 B). Treatment with 2 mM sodium periodate (mild periodate treatment) oxidizes vicinal hydroxyl groups of terminal sialic acid into permanent aldehydes, thereby “destroying” terminal sialic acid. Dissimilar to the treatment with sialidase, where terminal-end sialic acids are completely cleaved off, mild periodate treatment allows for the terminal oxidized sialic acid moiety to be retained on the glycan chain. Treatment with 10 mM sodium periodate (strong periodate treatment) can oxidize all vicinal hydroxyl groups of monosaccharide moieties within the glycan chain, thereby “destroying” the entire glycan chain without cleaving it from the surface of RBCs (Fukuda, 2001). As expected, sialic acid-dependent hemagglutination by S. gordonii DL1 was markedly reduced by mild periodate treatment of RBCs (Fig. 3 B) confirming that the binding of S. gordonii DL1 to RBCs is mediated by the recognition of sialic acid. For S. pneumoniae HR1001.1, only a partial reduction in hemagglutination titer was seen after periodate treatment of RBCs. Interestingly, when the PsrP-deficient mutant of HR1001.1 was tested, only strong periodate treatment of RBCs completely abolished hemagglutination activity. Taken together, these results suggest that S. pneumoniae strain HR1001.1 possesses an additional adhesin besides PsrP which binds to glycans other than sialic acids, and that its glycan-mediated binding activity might be partially masked by the presence of PsrP.
Unencapsulated S. pneumoniae binds to glycans via PsrP and another lectin-like adhesin
To further assess the specificity of glycan-mediated binding of capsule-free S. pneumoniae strain HR1001.1, custom-spotted glycan array dot blots containing immobilized salivary proteins, naturally occurring glycoproteins, and related neoglycoproteins were overlaid with fluorochrome-labeled bacteria (Fig. 4). Wild-type S. gordonii strain DL1 bound strongly to SMSL (A3) and considerably to both WS (A1) and PAR (A5) secretions, but not to their corresponding de-sialylated salivary samples (A2, A4 and A6). Strain DL1 also bound to a number of naturally occurring sialylated glycoproteins, which include salivary mucins MUC5B (B1) and MUC7 (B3), red blood cell membrane-associated glycoprotein glycophorin A (C1) and blood glycoprotein fetuin (C3). Binding activities to these substrates were abrogated when these samples were de-sialylated (B2, B4, C2 and C4). Strain DL1 bound explicitly to neoglycoconjugates that presented sialylated glycans (D1, E1, E5, and F1), but not to the desialylated glycans (D2, D3, E2, E6, and F2). Taken together, these findings confirmed that the binding of S. gordonii DL1 to these immobilized substrates was solely mediated by the recognition of sialic acid. Furthermore, all mentioned binding activities were no longer observed when the adhesin Hsa-deficient mutant strain of DL1 was used under the same conditions, signifying that binding activity was mediated through the SRR adhesin Hsa. These observations are in agreement with previous results in the study and previous studies (Deng et al., 2014, Takahashi et al., 1997, Takamatsu et al., 2005).
Fig. 4.

Streptococcal adhesion to immobilized WS and ductal glandular secretions (SMSL, PAR), naturally occurring glycoproteins and related neoglycoproteins. Suspensions of fluorochrome-labeled S. gordonii DL1, capsule-free S. pneumoniae TIGR4 mutant strain HR1001.1, and the respective isogenic SRR adhesin-deficient mutants were overlaid on dot blots of immobilized naturally occurring glycoproteins (Table S1) and related neoglycoproteins (Table S2). Fluorescent signals of bound bacteria were detected by use of a fluorescence scanner.
Capsule-free S. pneumoniae HR1001.1 displayed a much broader binding profile. Strain HR1001.1 bound to all immobilized saliva samples (A1, A3 and A5) with no influence of sialidase treatment (A2, A4 and A6). In the case of MUC7 (B3), glycophorin A (C1) and fetuin (C3), binding was even stronger to the sialidase-treated samples (B4, C2 and C4), supporting our previous observations (see Fig. 1). Binding to glycophorin and fetuin was markedly reduced or abrogated when these glycoproteins were treated with 10 mM periodate prior to immobilization on the blot (data not shown), further consolidating the glycan-mediated mechanism of binding. Strain HR1001.1 bound to neoglycoproteins carrying Galβ/α1-3GalNAc-(D2, D3 and D4), lacto-N-tetraose (E4), N-acetyllactosamine (E6), lacto-N-neotetraose (F4), lactose (F5), globo-N-tetrasose (G1), gangliotetraose (G2), and to a lesser extent to GalNAcβ1-3/4-Galα-(D5 and D6). Binding to these glycans was not lost when the PsrP-deficient mutant strain of HR1001.1 was used. The differences in pneumococcal binding to glycoconjugates on the dot blot array were quantified by densitometry (Figure S1). In summary, these results provide strong evidence for the existence of an additional lectin-like adhesin on S. pneumoniae other than PsrP that is involved in the recognition of these Gal- or GalNAc containing glycans, and perhaps salivary secretions as well.
Notably however, after deletion of PsrP, binding to the Lewis blood group-related glycans (E1 – E3 and F1 – F3) was lost. Furthermore, binding to GalNAcβ1-3/4-Galα- (D5 and D6), lacto-N-tetraose (E4), lacto-N-neotetraose (F4), globo-N-tetraose (G1), gangliotetraose (G2), N-acetyllactosamine (E6) and lactose (F5), as well as all monosaccharide-containing neoglycoconjugates (G3 – G6 and H1 – H5) was reduced when PsrP was not expressed. Taken together, these results suggest that PsrP might exhibit an as of yet undetermined, but sialic acid independent, glycan binding activity. It is perplexing that binding of strain HR1001.1 was equally strong to all the monosaccharide conjugates (G3 – G6 and H1 – H5), and was attenuated after removal of PsrP. One possible explanation could be that all the monosaccharides used for this array were linked to the carrier protein by the chemical spacer arm p-aminophenyl (PAP), whereas all other neoglycoproteins on this array were linked by different spacers (Table S2). Thus, the binding results to the monosaccharide units have to interpreted with reservation, because of possible sterical interference of the chemical linker. Another explanation could be that binding by PsrP is more complex and may require additional unknown structural motifs. Perhaps the true high-affinity counter-receptor for PsrP was not included among the neoglycoconjugates on the present array.
DISCUSSION
Here we showed that, when its capsule is absent, S. pneumoniae binds to low molecular weight salivary mucin MUC7 through presumably a glycan-mediated mechanism. Different to related oral Mitis group streptococci, binding of MUC7 by the pneumococci was not dependent on the expression of the SRR adhesin PsrP and recognition of sialic acid. The adhesin PsrP, however, was involved in agglutination of RBCs. Using a dot blot glycan array, we obtained evidence for glycan binding by PsrP and also showed that an additional glycan-binding adhesin exists on S. pneumoniae that becomes exposed only after the removal of the polysaccharide capsule and requires the removal of terminal sialic acids for its counter-receptors to become revealed.
We observed that S. pneumoniae TIGR4 bound to salivary MUC7 and hemagglutinated RBCs only when its capsule was absent. This suggests that the capsule hinders adhesive interactions with host receptors. Others have also shown that the presence of a capsular polysaccharide layer prevents pneumococcal biofilm formation (Muñoz-Elías et al., 2008) and hinders adherence to epithelial cells (Hammerschmidt et al., 2005). In fact, it was proposed that S. pneumoniae down-regulate their capsule expression in the presence of host mucosal surfaces thereby exposing previously buried surface adhesins (Hammerschmidt et al., 2005, Sanchez et al., 2011, Marks et al., 2012). S. pneumoniae are also known to switch from a highly encapsulated phenotype (opaque colonies) to a reduced capsule-expressing phenotype (transparent colonies) as a response to environmental cues (Kim & Weiser, 1998). Although exposure of the keratin 10 binding region of PsrP was shown not to be hindered by the capsule, and adherence to lung epithelial cells was not inhibited by the presence of a capsule (Shivshankar et al., 2009, Rose et al., 2008), it still could be that the putative domain responsible for binding to MUC7 and RBCs remains buried in the capsular polysaccharide layer and becomes exposed only when the capsule is removed.
Deletion of the PsrP adhesin in the unencapsulated mutant strain abolished hemagglutination of untreated RBCs. This opens up the possibility that PsrP possesses a lectin activity recognizing a glycan counter-receptor on the RBCs. There are earlier reports on sialic acid recognition by S. pneumoniae (Barthelson et al., 1998, Idänpään-Heikkila et al., 1997), but in our study neither hemagglutination nor binding to MUC7, glycophorin A and fetuin by S. pneumoniae were reduced after sialidase treatment. Even though this clearly speaks against recognition of sialic acids by PsrP, the possibility that this adhesin might bind to an unusual sialidase-resistant subtype of sialic acid (Powell & Varki, 2001) still remains. A more likely explanation, however, is that PsrP binds to glycans other than sialic acids. By using a custom-spotted array of natural glycoproteins and neoglycoproteins, evidence was obtained that PsrP could possibly recognize Lewis blood group-related glycans containing Galβ1-3/4GlcNAc. Binding of S. pneumoniae to Galβ1-3/4GlcNAc-containing glycan motifs, has been reported in earlier studies (Barthelson et al., 1998, Idänpään-Heikkila et al., 1997). Further binding studies involving larger glycan arrays and sugar inhibition experiments will have to narrow down on the preferred glycan motif recognized by PsrP.
When PsrP was deleted, unencapsulated S. pneumoniae still retained its binding to sialidase-treated RBCs, MUC7, glycophorin A, fetuin, as well as GalNAcβ1-3/4-Galα-, lacto-N-tetraose, N-acetyllactosamine, lacto-N-neotetraose, lactose, globo-N-tetraose, and gangliotetraose. This suggests that S. pneumoniae must possess at least one additional lectin-like adhesin besides PsrP. There are earlier reports by different groups demonstrating evidence for lectin-mediated binding by S. pneumoniae to GlcNAc-, GalNAc-, or Gal-containing glycans (Andersson et al., 1983, Krivan et al., 1988, Cundell & Tuomanen, 1994, Barthelson et al., 1998). Presently, lectin-mediated binding by S. pneumoniae is still poorly understood (Voß et al., 2012), although some glycan-binding proteins on S. pneumoniae have been described (Limoli et al., 2011, King et al., 2006, Dalia et al., 2010, King, 2010). From our current hemagglutination data using periodate-treated RBCs as well as the binding results with our custom-spotted glycan array, it can now be concluded that S. pneumoniae TIGR4 possesses an additional glycan-binding adhesin, other than PsrP with a specificity for Gal/GalNAc-containing glycan motifs. It is likely that related glycan motifs are present among the highly diverse oligosaccharides found on salivary mucin MUC7 (Karlsson & Thomsson, 2009, Prakobphol et al., 1982) and may explain S. pneumoniae binding to this molecule.
At this stage, we can only speculate what the binding of unencapsulated pneumococci to salivary MUC7 could mean in terms of physiology or pathology. On the one hand, binding to this mucin could mediate adhesion of pneumococci to saliva-coated surfaces in the oral cavity. However, S. pneumoniae is rarely found as an early colonizer in the oral cavity, much in contrast to other members of the Mitis group of streptococci (Whitmore & Lamont, 2011, Li et al., 2004). On the other hand, binding to MUC7 could agglutinate pneumococci and, thus, constitute a mechanism of clearance of these bacteria from the mouth. Notably, MUC7 is also produced by the mucous glands lining the nasopharynx and upper respiratory tract (Sharma et al., 1998), the natural habitat of the pneumococcus (Kadioglu et al., 2010, Kilian et al., 2014, Marks et al., 2012). As such, MUC7 could either mediate pneumococcal binding or contribute to the clearance of these bacteria from respiratory mucous surfaces (King et al., 2004). Alternative explanations are that the pneumococci bind MUC7 for nutritional purposes (Yesilkaya et al., 2008) or as a stealth strategy to evade mucosal host defense through decoration with host-derived molecules (Heo et al., 2013). As all these hypothetical scenarios cause quite opposing biological implications, further work will be required to better understand the role of MUC7 and glycan-mediated binding in pneumococcal colonization.
Supplementary Material
Acknowledgments
The authors are grateful to Roberta Anderson for excellent technical help. We are thankful to Frank Scannapieco, Elaine Haase, and Vera Dovirak for their thoughtful comments and critical review of the manuscript. The work was supported by grants NIDCR DE019807 (SR) and NIDCD DC013554 (AH).
References
- Andersson B, Dahmén J, Frejd T, et al. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J Exp Med. 1983;158:559–570. doi: 10.1084/jem.158.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelson R, Mobasseri A, Zopf D, Simon P. Adherence of Streptococcus pneumoniae to respiratory epithelial cells is inhibited by sialylated oligosaccharides. Infect Immun. 1998;66:1439–1444. doi: 10.1128/iai.66.4.1439-1444.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MH, Yother J. CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J Biol Chem. 2001;276:47966–47974. doi: 10.1074/jbc.M105448200. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Lopez JA, Sullam PM. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha. Infect Immun. 2004;72:6528–6537. doi: 10.1128/IAI.72.11.6528-6537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette-Cain K, Hinojosa CA, Akula Suresh Babu R, et al. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. MBio. 2013;4:e00745–00713. doi: 10.1128/mBio.00745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell DR, Tuomanen EI. Receptor specificity of adherence of Streptococcus pneumoniae to human type-II pneumocytes and vascular endothelial cells in vitro. Microb Pathog. 1994;17:361–374. doi: 10.1006/mpat.1994.1082. [DOI] [PubMed] [Google Scholar]
- Dalia AB, Standish AJ, Weiser JN. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect Immun. 2010;78:2108–2116. doi: 10.1128/IAI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Bensing BA, Thamadilok S, et al. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 2014;10:e1004540. doi: 10.1371/journal.ppat.1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP, Yother J. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol Microbiol. 1994;12:959–972. doi: 10.1111/j.1365-2958.1994.tb01084.x. [DOI] [PubMed] [Google Scholar]
- Frandsen EV, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6:129–133. doi: 10.1111/j.1399-302x.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- Fukuda M. Chemical labeling of carbohydrates by oxidation and sodium borohydride reduction. Curr Protoc Mol Biol Wiley Interscience. 2001:17.15.11–17.15.18. doi: 10.1002/0471142727.mb1705s26. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Muller E, Rohde M. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun. 2005;73:4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- Heo SM, Choi KS, Kazim LA, et al. Host defense proteins derived from human saliva bind to Staphylococcus aureus. Infect Immun. 2013;81:1364–1373. doi: 10.1128/IAI.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idänpään-Heikkila I, Simon PM, Zopf D, et al. Oligosaccharides interfere with the establishment and progression of experimental pneumococcal pneumonia. J Infect Dis. 1997;176:704–712. doi: 10.1086/514094. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Kerrigan SW, Nobbs AH, et al. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect Immun. 2005;73:6629–6638. doi: 10.1128/IAI.73.10.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Hinds J, Smith A, van der Linden M, Van Eldere J, Mitchell TJ. Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J Clin Microbiol. 2010;48:2762–2769. doi: 10.1128/JCM.01746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadioglu A, Brewin H, Härtel T, et al. Pneumococcal protein PavA is important for nasopharyngeal carriage and development of sepsis. Mol Oral Microbiol. 2010;25:50–60. doi: 10.1111/j.2041-1014.2009.00561.x. [DOI] [PubMed] [Google Scholar]
- Karlsson NG, Thomsson KA. Salivary MUC7 is a major carrier of blood group I type O-linked oligosaccharides serving as the scaffold for sialyl Lewis x. Glycobiology. 2009;19:288–300. doi: 10.1093/glycob/cwn136. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- Kelly T, Dillard JP, Yother J. Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infect Immun. 1994;62:1813–1819. doi: 10.1128/iai.62.5.1813-1819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906) Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- Kilian M, Riley DR, Jensen A, Brüggemann H, Tettelin H. Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. MBio. 2014;5:e01490–01414. doi: 10.1128/mBio.01490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JO, Weiser JN. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis. 1998;177:368–377. doi: 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- King SJ. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol. 2010;25:15–24. doi: 10.1111/j.2041-1014.2009.00564.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Gould JM, et al. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol. 2004;54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- Kirkbride HJ, Bolscher JG, Nazmi K, et al. Genetic polymorphism of MUC7: allele frequencies and association with asthma. Eur J Hum Genet. 2001;9:347–354. doi: 10.1038/sj.ejhg.5200642. [DOI] [PubMed] [Google Scholar]
- Krivan HC, Roberts DD, Ginsburg V. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc b1-4Gal found in some glycolipids. Proc Natl Acad Sci U S A. 1988;85:6157–6161. doi: 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendenmann U, Grogan J, Oppenheim FG. Saliva and dental pellicle - a review. Adv Dent Res. 2000;14:22–28. doi: 10.1177/08959374000140010301. [DOI] [PubMed] [Google Scholar]
- Levine MJ, Herzberg MC, Levine MS, et al. Specificity of salivary-bacterial interactions: role of terminal sialic acid residues in the interaction of salivary glycoproteins with Streptococcus sanguis and Streptococcus mutans. Infect Immun. 1978;19:107–115. doi: 10.1128/iai.19.1.107-115.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Helmerhorst EJ, Leone CW, et al. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol. 2004;97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- Limoli DH, Sladek JA, Fuller LA, Singh AK, King SJ. BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells. Microbiology. 2011;157:2369–2381. doi: 10.1099/mic.0.045609-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano A, Sanchez CJ, Orihuela CJ. A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol Oral Microbiol. 2012;27:257–269. doi: 10.1111/j.2041-1014.2012.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler H. Hemagglutination by pneumococcus. Schweiz Z Pathol Bakteriol. 1950;13:605–608. [PubMed] [Google Scholar]
- Löfling J, Vimberg V, Battig P, Henriques-Normark B. Cellular interactions by LPxTG-anchored pneumococcal adhesins and their streptococcal homologues. Cell Microbiol. 2011;13:186–197. doi: 10.1111/j.1462-5822.2010.01560.x. [DOI] [PubMed] [Google Scholar]
- Marks LR, Parameswaran GI, Håkansson AP. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect Immun. 2012;80:2744–2760. doi: 10.1128/IAI.00488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martner A, Skovbjerg S, Paton JC, Wold AE. Streptococcus pneumoniae autolysis prevents phagocytosis and production of phagocyte-activating cytokines. Infect Immun. 2009;77:3826–3837. doi: 10.1128/IAI.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Elías EJ, Marcano J, Camilli A. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun. 2008;76:5049–5061. doi: 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PA, Levine MJ, Tabak LA, Reddy MS. Specificity of salivary-bacterial interactions: II. Evidence for a lectin on Streptococcus sanguis with specificity for a NeuAc a2, 3Ga1 b1, 3Ga1NAc sequence. Biochem Biophys Res Commun. 1982;106:390–396. doi: 10.1016/0006-291x(82)91122-6. [DOI] [PubMed] [Google Scholar]
- Murray PA, Prakobphol A, Lee T, Hoover CI, Fisher SJ. Adherence of oral streptococci to salivary glycoproteins. 1992;60:31–38. doi: 10.1128/iai.60.1.31-38.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF, Jakubovics NS. Stick to your gums: mechanisms of oral microbial adherence. J Dent Res. 2011;90:1271–1278. doi: 10.1177/0022034511399096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obert C, Sublett J, Kaushal D, et al. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–4777. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer C, Douglas CW. Relationship between the ability of oral streptococci to interact with platelet glycoprotein Iba and with the salivary low-molecular-weight mucin, MG2. FEMS Immunol Med Microbiol. 2006;48:390–399. doi: 10.1111/j.1574-695X.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- Plummer C, Wu H, Kerrigan SW, Meade G, Cox D, Ian Douglas CW. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br J Haematol. 2005;129:101–109. doi: 10.1111/j.1365-2141.2005.05421.x. [DOI] [PubMed] [Google Scholar]
- Powell LD, Varki AP. Sialidases. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb1712s27. Chapter 17: Unit17.12. [DOI] [PubMed] [Google Scholar]
- Prakobphol A, Levine MJ, Tabak LA, Reddy MS. Purification of a low-molecular-weight, mucin-type glycoprotein from human submandibular-sublingual saliva. Carbohydr Res. 1982;108:111–122. doi: 10.1016/s0008-6215(00)81896-0. [DOI] [PubMed] [Google Scholar]
- Prakobphol A, Murray PA, Fisher SJ. Bacterial adherence on replicas of sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1987;164:5–11. doi: 10.1016/0003-2697(87)90359-9. [DOI] [PubMed] [Google Scholar]
- Pyburn TM, Bensing BA, Xiong YQ, et al. A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 2011;7:e1002112. doi: 10.1371/journal.ppat.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubbu N, Reddy MS, Bergey EJ, Haraszthy GG, Soni SD, Levine MJ. Large-scale purification and characterization of the major phosphoproteins and mucins of human submandibular-sublingual saliva. Biochem J. 1991;280(Pt 2):341–352. doi: 10.1042/bj2800341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Shivshankar P, Hinojosa E, Rodriguez A, Sanchez CJ, Orihuela CJ. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis. 2008;198:375–383. doi: 10.1086/589775. [DOI] [PubMed] [Google Scholar]
- Ruhl S. The scientific exploration of saliva in the post-proteomic era: from database back to basic function. Expert Rev Proteomics. 2012;9:85–96. doi: 10.1586/epr.11.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl S, Sandberg AL, Cisar JO. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J Dent Res. 2004;83:505–510. doi: 10.1177/154405910408300614. [DOI] [PubMed] [Google Scholar]
- Ruhl S, Sandberg AL, Cole MF, Cisar JO. Recognition of immunoglobulin A1 by oral actinomyces and streptococcal lectins. Infect Immun. 1996;64:5421–5424. doi: 10.1128/iai.64.12.5421-5424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CJ, Kumar N, Lizcano A, et al. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS One. 2011;6:e28738. doi: 10.1371/journal.pone.0028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CJ, Shivshankar P, Stol K, et al. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 2010;6:e1001044. doi: 10.1371/journal.ppat.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannapieco FA. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- Schulte T, Löfling J, Mikaelsson C, et al. The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold. Open Biol. 2014;4:130090. doi: 10.1098/rsob.130090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Dudus L, Nielsen PA, et al. MUC5B and MUC7 are differentially expressed in mucous and serous cells of submucosal glands in human bronchial airways. Am J Respir Cell Mol Biol. 1998;19:30–37. doi: 10.1165/ajrcmb.19.1.3054. [DOI] [PubMed] [Google Scholar]
- Shivshankar P, Sanchez C, Rose LF, Orihuela CJ. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol. 2009;73:663–679. doi: 10.1111/j.1365-2958.2009.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares RV, Offner GD, Assis MA, Silva KC, Zenobio EG. An unusual glycoform of human salivary mucin MG2. Clin Oral Investig. 2012;16:761–766. doi: 10.1007/s00784-011-0556-5. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun. 2002a;70:1209–1218. doi: 10.1128/IAI.70.3.1209-1218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ruhl S, Yoon JW, Sandberg AL, Cisar JO. Adhesion of viridans group streptococci to sialic acid-, galactose- and N-acetylgalactosamine-containing receptors. Oral Microbiol Immunol. 2002b;17:257–262. doi: 10.1034/j.1399-302x.2002.170409.x. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Sandberg AL, Ruhl S, Muller J, Cisar JO. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to a2-3-linked sialic acid-containing receptors. Infect Immun. 1997;65:5042–5051. doi: 10.1128/iai.65.12.5042-5051.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Cheng H, et al. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Iba. Mol Microbiol. 2005;58:380–392. doi: 10.1111/j.1365-2958.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 2006;74:1933–1940. doi: 10.1128/IAI.74.3.1933-1940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Turner LS, Kanamoto T, Unoki T, Munro CL, Wu H, Kitten T. Comprehensive evaluation of Streptococcus sanguinis cell wall-anchored proteins in early infective endocarditis. Infect Immun. 2009;77:4966–4975. doi: 10.1128/IAI.00760-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voß S, Gámez G, Hammerschmidt S. Impact of pneumococcal microbial surface components recognizing adhesive matrix molecules on colonization. Mol Oral Microbiol. 2012;27:246–256. doi: 10.1111/j.2041-1014.2012.00654.x. [DOI] [PubMed] [Google Scholar]
- Walz A, Odenbreit S, Mahdavi J, Borén T, Ruhl S. Identification and characterization of binding properties of Helicobacter pylori by glycoconjugate arrays. Glycobiology. 2005;15:700–708. doi: 10.1093/glycob/cwi049. [DOI] [PubMed] [Google Scholar]
- Walz A, Odenbreit S, Stühler K, et al. Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay. Proteomics. 2009;9:1582–1592. doi: 10.1002/pmic.200700808. [DOI] [PubMed] [Google Scholar]
- Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- Yesilkaya H, Manco S, Kadioglu A, Terra VS, Andrew PW. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol Lett. 2008;278:231–235. doi: 10.1111/j.1574-6968.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 2009;155:317–327. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


