SUMMARY
Periodontitis is a polymicrobial inflammatory disease that results from the interaction between the oral microbiota and the host immunity. While the innate immune response is important for disease initiation and progression, the innate immune receptors that recognize both classical and putative periodontal pathogens that elicit an immune response have not been elucidated. By using the Human Oral Microbe Identification Microarray (HOMIM), we identified multiple predominant oral bacterial species in human plaque biofilm that strongly associate with severe periodontitis. Ten of the identified species were evaluated in greater depth, 6 being classical pathogens and 4 putative novel pathogens. Using human peripheral blood monocytes (HPBM) and murine bone marrow–derived macrophages (BMDM) from wild-type (WT) and toll-like receptor (TLR)-specific and MyD88 knockouts (KOs), we demonstrated that heat-killed Campylobacter concisus, Campylobacter rectus, Selenomonas infelix, Porphyromonas endodontalis, Porphyromonas gingivalis, and Tannerella forsythia mediate high immunostimulatory activity. C. concisus, C. rectus, and S. infelix exhibited robust TLR4 stimulatory activity. Studies using mesothelial cells from WT and NOD1-specific KOs and NOD2-expressing human embryonic kidney (HEK) cells demonstrated that Eubacterium saphenum, Eubacterium nodatum and Filifactor alocis exhibit robust NOD1 stimulatory activity, and that Porphyromonas endodontalis and Parvimonas micra have the highest NOD2-stimulatory activity. These studies allowed us to provide important evidence on newly-identified putative pathogens in periodontal disease pathogenesis showing that these bacteria exhibit different immunostimulatory activity via TLR4, NOD1, and NOD2 (Clinicaltrials.gov NCT01154855).
Keywords: Oral Microbiology, biofilms, innate immunity, periodontal disease, anaerobes, cell receptors
INTRODUCTION
Periodontitis is an immune-inflammatory infection of the tooth-supporting structures that is prevalent in 47% of the American adult population in mild, moderate, or severe forms (Eke et al., 2010). It is well established that periodontal disease development is based on a combination of factors: a susceptible host, environmental factors, and the presence of oral microorganisms. Over 600 bacterial species have been identified in the periodontal microbiota, both cultivable and not-yet-cultivable (Paster et al., 2006). Analysis of clinical plaque samples and disease correlation analysis have classically grouped microorganisms into 5 major complexes, with the 2 most relevant for chronic periodontal disease being the red complex with Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola and the orange complex with Fusobacterium nucleatum, Prevotella intermedia, Prevotella nigrescens, Parvimonas micra and other associated species (Socransky et al., 1998). Clinical studies have provided clues based on disease association and allowed for the development of in vitro and in vivo studies that have further contributed to explaining the mechanisms involved in periodontal disease development. Currently, the pathogen mostly explored in periodontal disease pathogenesis has been P. gingivalis. Investigation of this pathogen over the years has led to the novel and important keystone-pathogen hypothesis in which very low colonization levels of P. gingivalis lead to changes in the amount and composition of commensal microflora (dysbiosis) and further inflammatory periodontal bone loss (Hajishengallis et al., 2012, Hajishengallis & Lamont 2012, Hajishengallis et al., 2011). The development of novel methods for microbial identification in clinical plaque samples over the past 12 years has confirmed the importance of P. gingivalis and other classical periodontal bacterial species in disease and allowed for the detection of numerous novel potential commensal and pathogenic species (Kumar et al., 2005, Lourenco et al., 2014, Teles et al., 2011). Unfamiliar species such as Porphyromonas endodontalis, Eubacterium saphenum, and Filifactor alocis are now being proposed to play important roles in the development of disease (Belstrom et al., 2014, Colombo et al., 2012, Perez-Chaparro et al., 2014). These studies along with the concept of polymicrobial infection emphasize the importance of further investigating newly-identified pathogens in periodontal disease pathogenesis.
The host response that develops against microorganisms initiates with the innate immune response, including their recognition via pattern-recognition receptors (PRRs), such as toll-like receptors (TLR) and nucleotide-binding oligomerization domain (NOD) receptors (NLR) (Kawai & Akira 2009). Sensing by TLR2 and TLR4 (most extensively studied TLR) leads to production of inflammatory mediators via adaptors MyD88, while sensing by NOD1 and NOD2 (two well-characterized NLR) occurs mainly via receptor-interacting serine/threonine kinase (Alhawi et al.,) (Hasegawa et al., 2008). Some classical periodontal pathogens have been mechanistically explored in this context. Previous studies have reported that P. gingivalis (Burns et al., 2006, Jain et al., 2013) and T. forsythia (Myneni et al., 2011) are recognized by TLR2, Campylobacter rectus is recognized mainly by TLR4 (Arce et al., 2012), while multiple PRRs (including TLR2, TLR4, and NOD1) were suggested to mediate cytokine expression upon Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans stimulation (Park et al., 2014). Recent studies also indicate that NOD2 and TLR9 are also involved in P.gingivalis infection (Kim et al., 2015, Prates et al., 2014). Overall, these studies suggest that PRRs play important roles in the host response against periodontal pathogens and emphasize the importance of further characterizing the immune response to other newly-identified pathogens involved in periodontitis. However, the interaction between host PRRs and the majority of newly-identified periodontal pathogens remains unknown. In this study, we identified a group of predominant oral microorganisms, including classical and putative periodontal bacterial species, which are highly correlated with plaque biofilm derived from patients afflicted with severe periodontitis. Furthermore, we characterized the TLR2, TLR4, NOD1 and NOD2 stimulatory activity of these identified periodontal bacterial species. Our data support that these newly-identified bacterial species associated with periodontitis individually possess distinct immunostimulatory properties and suggests that TLR4, NOD1 and NOD2 are important in mediating the immune response to these pathogens.
METHODS
Study population
This descriptive study was approved by the University of Michigan Health Sciences Institutional Review Board and was registered with the NIH clinical trials registry (Clinicaltrials.gov NCT01154855) and conformed to STROBE guidelines for observational studies. After informed consent was provided, 40 individuals were recruited at the Michigan Center for Oral Health Research. Patients included in the study were 35 years old or older. All subjects possessed at least 20 teeth and had not received long-term (over 2 weeks) antibiotic-related therapy for medical or dental reasons within 3 months prior to study inclusion. Patients were also excluded if they received long-term use of medications known to affect periodontal status such as anti-inflammatory drugs, aspirin and ibuprofen. Patients on immunosuppressive therapies, including glucocorticoids or cyclosporines, were excluded from participation in this study (corticosteroid inhalers were allowed). The patients showed no history of metabolic bone diseases such as rheumatoid arthritis or post-menopausal osteoporosis. Pregnant women were excluded from participating in both groups due to x-ray exposure and possible harm to the fetus. All women were questioned regarding pregnancy status at the start of the study. Patients were enrolled into either healthy group or severe periodontitis group as previously described (Ramseier et al., 2009). Healthy category patients exhibited no probing depths (PD) >4 mm, possessed minimal to no radiographic bone loss, and < 20% of sites possessing bleeding on probing (BOP). Severe periodontitis subjects exhibited at least 8 sites with evidence of radiographic bone loss, had at least 8 sites with PD >4 mm, and >30% of sites with clinical attachment loss (Perez-Chaparro et al.,) >3 mm. Patients were excluded if they had any periodontal treatments in the 12 months prior to study initiation.
Plaque collection and analysis by the Human Oral Microbe Identification Microarray (HOMIM)
Within two weeks of screening, a subgingival plaque/biofilm sample was collected from the most affected tooth and site in each sextant from patients in both groups as previously described (Beikler et al., 2006, Ramseier et al., 2009). Subgingival plaque was collected from the designated teeth, dried with a gentle blast of air prior to sampling, using a sterile Gracey curette by scraping with slight pressure against the tooth. The plaque was collected and immediately placed in a labeled vial containing 150 μl of TE buffer and stored at −20° for further analysis using HOMIM as previously described (Colombo et al., 2012, Lourenco et al., 2014). Briefly, a total of 400 16S rRNA-based reverse-capture oligonucleotide probes targeting ~300 bacterial taxa were utilized. Data were normalized by comparing individual signal intensities to the average of signals from universal probes.
Bacteria and culture conditions
E. saphenum ATCC 49989, F. alocis ATCC 35896, P. endodontalis ATCC 35406, S. infelix ATCC 700230, and C. concisus ATCC 33237 were purchased from ATCC. C. rectus ATCC 33238, T. forsythia ATCC 43037, E. nodatum ATCC 33099, and P. micra ATCC 33270 were a gift from Dr. Ricardo Teles of the Forsyth Institute. P. gingivalis W83 was a gift from the lab of Dr. Christopher Fenno of the University of Michigan. Bacterial identification was also confirmed by DNA sequencing using 16S ribosomal RNA gene consensus primers: 16s commonS1 CCAAACTCCTACGGGAGGCAGCAG and 16s commonA1 CATGGACTACCAGGGTATCTAATC. The 16S rDNA was amplified from a single bacterial colony - by 35 cycles of PCR at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min. PCR products were verified by agarose gel electrophoresis and followed by gel purification (QIAquick Gel Extraction Kit) and sequenced from both ends using the same primers. The sequences were subjected to an online BLASTN analysis (NCBI, National Institutes of Health) for confirmation of the bacterial identity.
F. alocis ATCC 35896, P. endodontalis ATCC 35406, S. infelix ATCC 700230, P. micra ATCC 33270, and P. gingivalis ATCC W83 were cultured on Brucella Blood Agar mono plates (AS-141, Anaerobe Systems). Eubacterium nodatum ATCC 33099 was cultured on Brucella Blood Agar mono plates supplemented with Arginine (0.5%) (Hill et al., 1987). E. saphenum ATCC 49989 was cultured on Brucella Blood Agar mono plates supplemented with Lysine (0.3%) (Uematsu et al., 2003). T. forsythia ATCC 43037 was cultured on TSA blood plates supplemented with 10mg/L N-acetyl muramic acid (NAM). All the above bacteria were cultured with anaerobic incubation (80% N2; 10% H2; 10% CO2). C. concisus ATCC 33237 and C. rectus ATCC 33238 were cultured on Trypticase soy agar (TSA) blood plates supplemented with 25mM formate and 50mM furmarate. Both Campylobacter species were cultured under microaerophilic conditions (80% N2, 6% O2, 8% CO2 and 6% H2).
A single bacterial colony on each plate was inoculated into broth to obtain a bacterial liquid culture P. endodontalis ATCC 35406 and P. gingivalis ATCC W83 were cultured in Brain heart infusion (BHI) supplemented with hemin (5 μg/ml) and vitamin K (0.5 μg/ml). F. alocis ATCC 35896 was cultured in BHI supplemented with hemin, vitamin K and 0.5% arginine and 0.1% cysteine. P. micra ATCC 33270 and S. infelix ATCC 700230 were cultured in Brucella broth supplemented with hemin and vitamin K. E. nodatum ATCC 33099 was cultured in Brucella broth supplemented with hemin, vitamin K and 1% Arginine. E. saphenum ATCC 49989 was cultured in Brucella broth supplemented with hemin, vitamin K and 0.3% Lysine. T. forsythia ATCC 43037 was cultured in Trypticase soy Broth (TSB) supplemented with hemin, vitamin K and 10mg/L NAM. C. concisus ATCC 33237 and C. rectus ATCC 33238 were cultured in TSB supplemented with hemin, vitamin K, 25mM formate and 50mM fumarate. The final concentrations of hemin and vitamin K in all liquid medium were 5μg/ml and 0.5μg/ml. All organisms were grown at 37°C to an early steady state. Numbers of bacteria were determined spectrophotometrically (optical density at 600 nm (OD600)). Bacterial cultures were inactivated by heating at 98°C for 10 min.
Human peripheral blood monocytes (HPBMs) isolation and bacterial stimulatory assay
HPBMs were purified from filter buffy coats obtained from a pool of healthy human donors from the blood bank at the University of Michigan. Human peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Hypaque (Meyer et al., 2005). Freshly isolated PBMCs were diluted into 1X106 cells/ml of Monocyte Attachment Medium (PromoCell, C-28051) and 200ul was seeded in 96 well plates. The cells were incubated at 37° under 5% CO2 without any further manipulation to allow monocyte attachment. After 1.5h incubation, the Monocyte Attachment Medium was changed into HPBMs culture medium (RPMI-1640 supplemented with 5% heat inactivated Human AB serum and 1% Penicillin-streptomycin (P/S) solution) and subsequently incubated overnight. The culture medium was changed after overnight culture, and the cells were then stimulated with heat-inactivated whole bacterial culture at a bacterial/HPBMs ratio of ~10:1, 1:1 and 0.1:1 for 12h. Presence of cytokines in the cell culture supernatants was measured by Human IL-6 ELISA Kit (BD OptEIA™) according to the manufacturer's recommended protocol.
Animal sources of cells used in the study
Mice deficient in MyD88, TLR2/TLR4, TLR2, TLR4, and NOD1 have been previously described (Hasegawa et al., 2010, Kim et al., 2013). All mice were crossed at least 5 times on a C57BL/6 background. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Michigan (Ann Arbor, MI).
BMDMs Cell culture and bacterial stimulatory assay
WT, MyD88, TLR2/TLR4, TLR2, and TLR4 KO macrophages were prepared from the femur and tibia of C57BL/6 mice and cultured for 3 to 7 days in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 10% fetal bovine serum (FBS, Invitrogen Life Technologies), 30% L-cell supernatant, non-essential amino acids, sodium pyruvate, 2-ME, and 1% P/S solution (Park et al., 2007). After 5 to 7 days of differentiation, BMDMs were detached by scraping cells in cold PBS and then seeded in 96-well plates at a density of 5×104 cells per well. After overnight culture, cells were stimulated with heat-inactivated whole bacterial culture at a bacteria/macrophage ratio of ~10:1 and 1:1 for 12h. The presence of cytokine in cell culture supernatants was measured by Mouse IL-6 DuoSet ELISA kits (R&D Systems, DY-406) according to the manufacturer's recommended protocol.
Preparation of primary mesothelial cells and stimulation with different bacterial species
Mesothelial cells from C57BL/6 WT and NOD1-KO mice were prepared from the peritoneum and external surface of the liver, spleen, and kidneys of adult mice as described (Park et al., 2007). Briefly, pieces of the peritoneum and intact organs were obtained from sacrificed mice and digested with 0.25%-trypsin-EDTA solution for 50 min at 37°C. Intact tissues and tissue debris were discarded and the cell suspension was centrifuged at 1000 rpm for 5 min. The pellet was resuspended in Dulbecco's modified eagle medium (DMEM) supplemented with 15% heat-inactivated FBS and 1% P/S and cultured overnight. The next day, non-adherent cells were removed and the resultant mesothelial cells were grown in the above culture medium for 1 week. Mesothelial cells were used between passages 2 and 4 for the stimulatory assay. The cells were stimulated with heat-inactivated bacteria at a bacteria/mesothelail cells ratio of ~10:1, 1:1 and 0.1 for 12h. Culture supernatants were collected and assayed for CXCL1 production by mouse CXCL1/KC DuoSet ELISA (R&D Systems, DY453).
Bioassay for NOD2 receptor
HEK 293 cells stably expressing human NOD2 (HEK293-NOD2) and an NF-κB luciferase reporter were cultured in DMEM with 10% FBS and 1% of P/S (Warner et al., 2013). HEK293-NOD2 cells were seeded in 48-well plates at a density of 5X104 cells and then cultured overnight. Whole bacterial cultures were added to each well giving a bacterial/HEK293-NOD2 cell ratio of ~10:1 and 1:1. Different concentrations of MDP (0, 0.1, 0.5, 0.75 and 1.5ng) were added to each well to establish a standard curve. After 16h stimulation, culture supernatants were removed and reporter lysis buffer (75ul) was added to each well to lyse cells. Thirty microliters of each sample of lysed cell supernatant was mixed with 30ul of substrate and luciferase assay. Each sample was carried using the fluorometer FluoroCount (PerkinElmer Life Sciences) with excitation at 485 nm and emission at 530 nm.
Statistical analysis
Demographical and clinical data were analyzed using the unpaired two-tailed Student's t-test. HOMIM data was evaluated using non-parametric Wilcoxon Rank Sum Test (adjusted with Benjamini-Hochberg). Statistical analyses among different bacterial species to HPBM, murine BMDM and HEK-293-NOD2 were analyzed by one-way analysis of variance (ANOVA) and the Bonferroni multiple-comparison test. Statistical analyses between WT and NOD1 group were performed using a two-tailed t test with unequal variance (Aspin-Welch's t test; Excel, Microsoft). Differences were considered significant at p≤0.05.
RESULTS
Demographics and clinical measurements
Fifty-eight patients were screened for study eligibility, of which 40 patients met all study criteria and were evenly distributed between the healthy and diseased groups. Demographic and clinical characteristics are described in Table 1. Thirty and fifty percent were male for the healthy and diseased group, respectively. The mean number of teeth differed between groups, with a mean of 26.8 teeth for the healthy group and 24.4 teeth for the diseased group. All the clinical parameters that classified each category in healthy or diseased were significantly different between groups, including percent sites with BOP (10.5% in healthy and 60.1% in disease), gingival redness (31.6% in healthy and 83.4% in disease), and sites with plaque (41.0% in healthy and 81.8% in disease). The mean PD was 1.8mm for healthy individuals and 3.3mm for individuals with the disease (p<0.001), with zero sites and 20.42% with PD>4mm in healthy and disease groups respectively. Healthy subjects had a mean CAL of 0.7mm, and diseased individuals had a mean 3.5mm of CAL (p<0.001).
Table 1.
Demographics and clinical measurements of periodontal disease in patient population
| Healthy (Magalhaes et al., ) | Periodontal disease (Magalhaes et al., ) | p values | |
|---|---|---|---|
| Subjects (n) | 20 | 20 | NA |
| Males (%) | 30 | 50 | 0.10 |
| Caucasian (%) | 95 | 85 | 0.15 |
| Current tobacco users (%) | 15 | 25 | 0.22 |
| Former tobacco users (%) | 35 | 70 | ≤0.05 |
| Mean number of teeth | 26.8 (24 – 28) | 24.45 (19 – 28) | ≤0.05 |
| Mean age (years) | 46.1 (24 – 74) | 53 (39 – 66) | ≤0.05 |
| Sites with BOP (%) | 10.46 (0 - 22.62) | 60.08 (32.10 – 86.31) | ≤0.001 |
| Sites with gingival redness (%) | 31.57 (0 – 82.10) | 83.42 (37.68 – 100) | ≤0.001 |
| Sites with plaque (%) | 41.04 (0 – 88.27) | 81.8 (51.45 – 100) | ≤0.001 |
| Mean PD (Beikler et al., ) | 1.83 (1.21 – 2.46) | 3.27 (2.19 – 4.13) | ≤0.001 |
| Sites with PD>4mm (%) | 0 | 20.42 | ≤0.001 |
| Mean CAL (Beikler et al., ) | 0.7 (0.06 – 1.93) | 3.45 (2.06 – 6.83) | ≤0.001 |
BOP = bleeding on probing; PD = probing depth; CAL = clinical attachment level. Healthy category patients exhibited no probing depths (PD) >4 mm, possessed minimal to no radiographic bone loss, and < 20 percent sites with bleeding on probing (BOP). Severe periodontitis subjects exhibited at least 8 sites with evidence of radiographic bone loss, had at least 8 sites with PD >4 mm, and >30% of sites with clinical attachment loss (Perez-Chaparro et al., ) >3 mm.
Microbial analysis of plaque samples
Based on the microbial analysis by HOMIM in plaque samples (Table S1), twenty-five microorganisms were significantly associated with the periodontitis group (Table S2). The periodontitis group included species from the red and orange complexes defined by Socransky (Socransky et al., 1998), including P. gingivalis, T. forsythia, P. micra, P. intermedia, E. nodatum and C. rectus. Other putative periodontal bacterial species were correlated to the diseased group, including F. alocis, E. saphenum, S. infelix, and P. endodontalis. The periodontal disease group was further evaluated based on the microorganism's predominance in the biofilm shown by the highest relative signal. The top 50 most predominant species found in the periodontitis group included again emerging periodontal pathogens P. micra with a 3.4 score, P. endodontalis with a 2.05 score, and F. alocis with a 1.45 score, and classic periodontal pathogens C. rectus, T. forsythia, P. gingivalis, and E. nodatum with scores 3.95, 2.95, 2.05, 1.9, respectively (Table S1 and 2). Based on these 2 analyses correlation to periodontal disease and predominance in the plaque, 15 bacterial species were initially selected to test for their immunostimulatory activity in vitro (Table 2). Peptostreptococcus stomatis was excluded due to the lack of commercial availability. Dialister pneumosintes, Mogibacterium timidum and Treponema maltophilum were excluded due to lack of bacterial growth in liquid media. Finally, we were able to obtain liquid culture for 10 species, comprised of 6 classical pathogens and 4 potential pathogens, and were selected for further evaluation (Table 2).
Table 2.
Microorganisms predominant in periodontal plaque samples and significantly correlated with human periodontitis
| Relative signal* | Bacterial species | Human Oral Taxon | Pathogen classification | p value*** |
|---|---|---|---|---|
| 3.95 | Campylobacter concisus / Campylobacter rectus |
HOT-575**
HOT 748** |
Green complex/Orange complex | 0.045 |
| 3.4 | Parvimonas micra | HOT-111** | Orange complex | 0.029 |
| 2.95 | Tannerella forsythia | HOT-613** | Red complex | 0.001 |
| 2.9 | Eubacterium [XI][G-5] saphenum | HOT-759** | Potential pathogen | 0.016 |
| 2.4 | Peptostreptococcus stomatis | HOT-112 | Potential pathogen | 0.005 |
| 2.35 | Porphyromonas endodontalis | HOT-273** | Potential pathogen | 0.004 |
| 2.3 | Porphyromonas gingivalis | HOT-619** | Red complex | 0.001 |
| 2.2 | Eubacterium [XI][G-6] nodatum | HOT-694** | Orange complex | 0.001 |
| 2.1 | Selenomonas infelix | HOT-639** | Potential pathogen | 0.045 |
| 2.1 | Desulfobulbus sp. | HOT-041 | Potential pathogen | 0.001 |
| 2.05 | Dialister pneumosintes | HOT-736 | Potential pathogen | 0.025 |
| 2.05 | Treponema maltophilum | HOT-664 | Potential pathogen | 0.015 |
| 1.95 | Filifactor alocis | HOT-539** | Potential pathogen | 0.029 |
| 1.9 | Mogibacterium timidum | HOT-042 | Potential pathogen | 0.001 |
Relative signal = mean signal intensity compared to the universal probe signal ranging from 0-5, categorized as 0=signal absence and 5=highest signal, as described by Colombo et al 2009.
Microorganisms evaluated in this study
p values adjusted with Benjamini–Hochberg procedure
Immunostimulatory activity of various periodontal bacteria in human peripheral blood monocytes (HPBMs)
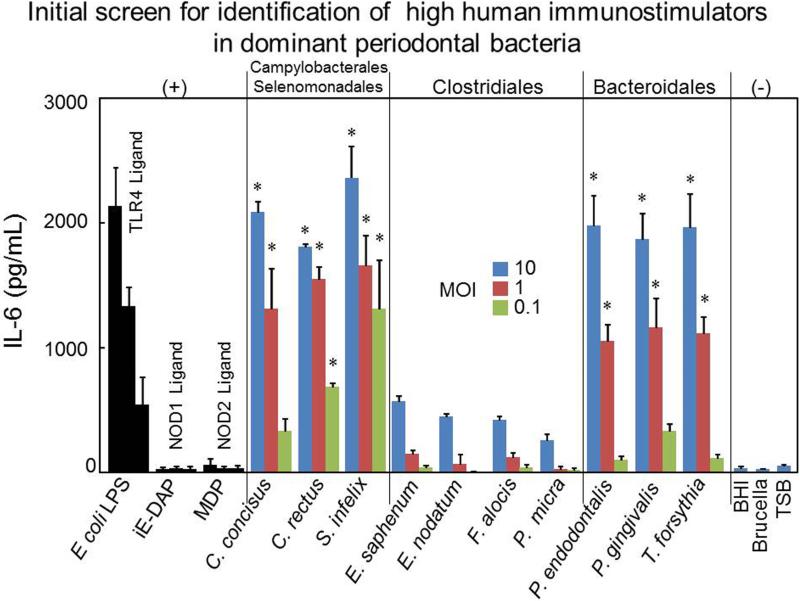
In order to determine the immunostimulatory activity of the identified periodontal bacterial species, HPBMs, a key component of the innate immune system, were stimulated with different doses of heat-inactivated periodontal bacteria for 12h. Culture supernatants were collected and IL-6 release was measured by ELISA. IL-6 is one of the major proinflammatory cytokines secreted by monocytes in response to specific microbial molecules. Three ratios of bacteria to HPBMs (10:1, 1:1 and 0.1:1) were chosen for the stimulatory experiments (Fig. 1). Following stimulation with different ratios of periodontal bacteria, HPBMs released IL-6 in a dose-related fashion. Consistent with previous studies (Jiang et al., 1996), P. gingivalis showed strong immunostimulatory activity to induce IL-6 in HPBMs, which is comparable to the positive control Escherichia coli lipopolysaccharide (LPS) (Fig. 1). C. concisus, C. rectus, S. infelix, P. endodontalis, and T. forsythia also released an amount of IL-6 that was comparable to that produced by the control E. coli LPS (Fig. 1). In contrast, E. nodatum, E. saphenum, F. alocis, and P. micra showed weak stimulation of IL-6 production in HPBMs (Fig. 1). Consistent with previous studies (Fritz et al., 2005), our results also indicate that human monocytes stimulated by NOD1 and NOD2 agonists produced significantly lower amounts of IL-6 compared with the same amount of LPS. We further assessed the IL-6 production by stimulating HPBMs with different concentration of TLR4, TLR2, NOD1 and NOD2 agonist with 12 h incubation, the results showed that LPS, Pam3CysSerLys4 (Pam3CSK4) and muramyl dipeptide (MDP) can induce robust IL-6 production, γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) cannot induce significant IL-6 production with high concentration (Fig S1). Overall, these results indicated HPBMs exhibited different IL-6 production to distinct innate receptor agonists, more importantly; these results indicated that different oral pathogens exhibited variable degrees of immunostimulatory activity in HPBMs. It strongly suggested that different oral pathogens may depend on different PAMPs to activate HPBMs (Fig. 1).
Figure 1. IL-6 production from Human peripheral blood monocytes (HPBMs) after exposure to periodontal bacteria.
HPBM were stimulated with the indicated ratios of bacteria to HPBM (MOI: 10:1, 1:1 and 0.1:1). After 12h stimulation, the supernatants were collected and pro-inflammatory IL-6 release was analyzed by ELISA. As positive controls, cells were incubated with 50 ng/ml E. coli O55B5 LPS, 50 ng/ml NOD1 ligand iE-DAP and 50 ng/ml NOD2 ligand MDP (with 10 times gradient dilution). Cells were stimulated with culture medium alone as a negative control. Results shown as mean ± SD of triplicate samples are representative of three independent experiments. Asterisks indicate statistically significant to culture medium negative control (P < 0.05) as determined by One-way ANOVA with Bonferroni post-test analysis.
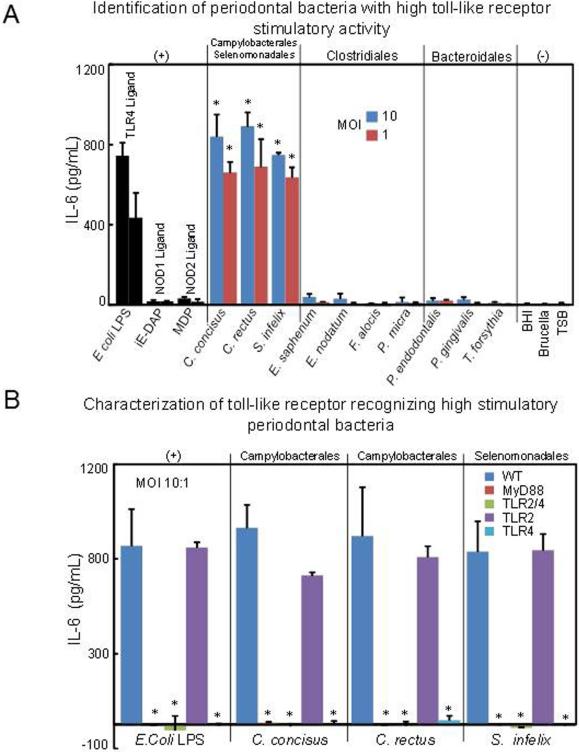
TLR4 is the major receptor for three periodontal microbes in murine BMDMs
The family of TLRs has been extensively studied as critical PRRs expressed at either the cell surface or the endosome membranes to recognize microorganisms (Kawai & Akira 2009). To investigate TLRs, essential for the immunologic recognition of periodontal bacteria, we first used BMDMs from wild type (WT) mice stimulated with various doses of oral bacteria for 12h (MOI 10:1 and 1:1), and the levels of IL-6 were measured from the culture supernatant. Consistent with HPBMs, E. coli LPS control, C. concisus, C. rectus, and S. infelix also induced robust IL-6 production in mice WT BMDMs (Fig. 2A). Meanwhile, iE-DAP, MDP, E. nodatum, E. saphenum, F. alocis, P.micra were also weak stimulators of IL-6 production in WT mice BMDMs (Fig. 2A). We also stimulated murine BMDM with different concentration of TLR4, TLR2, NOD1 and NOD2 agonist, the results indicated that LPS and Pam3 CSK4 induce robust IL-6 but ie-DAP and MDP cannot induce significant IL-6 production with high concentration (Fig S2). Unexpectedly, under MOI 10:1 and 12h stimulation, IL-6 production induced by P. gingivalis, P. endodontalis, and T. forsythia in murine WT BMDMs is not as robust as C. concisus, C. rectus, and S. infelix. Furthermore, IL-6 production induced by P. gingivalis, P. endodontalis, and T. forsythia is not in parallel with HPBMs in the same stimulatory condition (Fig. 2A). Coincidently, these three bacterial species all belong to the phylum Bacteroidetes. This result suggests murine immune cells may exhibit lower sensitivity to human Bacteroidetes as compared to other oral pathogens such as C. concisus, C. rectus, and S. infelix as well as human immune cells. TLR2 and TLR4 are two members of the TLR family which have been extensively studied. To further clarify the involvement of TLR2 and TLR4-induced cytokine production, oral bacteria with high immunostimulatory responses, including C. concisus, C. rectus and S. infelix, were used to stimulate BMDMs from MyD88, TLR2/4 double knock-out (DKO), TLR2 KO and TLR4 KO mice with the same doses of bacteria as WT BMDMs for 12h. Levels of IL-6 were measured from the culture supernatant. The production of IL-6 in response to these oral pathogens was completely impaired in MyD88, TLR2/4 DKO, and TLR4 KO macrophages in both MOI 10:1 (Fig. 2B) and 1:1 (Fig. S3). However, IL-6 production was as robust as WT in TLR2-deficient macrophages infected with these oral pathogens at MOIs of 10:1 (Fig. 2B) and 1:1 (Fig. S3). These results indicated that TLR4 is the major TLR that is responsive to C. concisus, C. rectus, and S. infelix in mice BMDMs, and suggested that these three periodontal bacterial species may induce inflammation during the development of periodontitis primarily via LPS.
Figure 2. IL-6 production from wild-type (WT) and knock-out (KO) mice Bone marrow-derived macrophages (BMDMs) after exposure to periodontal bacteria.
(A) BMDMs from WT mice were stimulated with various periodontal bacteria at the indicated MOI of 10:1 and 1:1. At 12 h after stimulation, culture supernatants were collected, and IL-6 production was measured by ELISA. As positive controls, cells were incubated with 50 ng/ml E. coli LPS, 50 ng/ml NOD1 ligand iE-DAP and 50 ng/ml NOD2 ligand MDP (with 10 times gradient dilution). As negative controls, cells were stimulated with culture medium alone. Asterisks indicate statistically significant to culture medium negative control (P < 0.05) as determined by One-way ANOVA with Bonferroni post-test analysis. (B) BMDMs from MyD88 and different TLRs KO mice were stimulated with these three periodontal bacterial species which can induce robust IL-6 production to WT BMDMs at MOI 10:1. 50 ng/ml E. coli LPS were used as positive control for stimulation. Culture supernatants were also collected for IL-6 detection at 12h after stimulation. Data are shown as mean ± SD of triplicate samples from one representative of three independent experiments. Asterisks indicate statistically significant to WT BMDM control (P < 0.05) as determined by One-way ANOVA with Bonferroni post-test analysis.
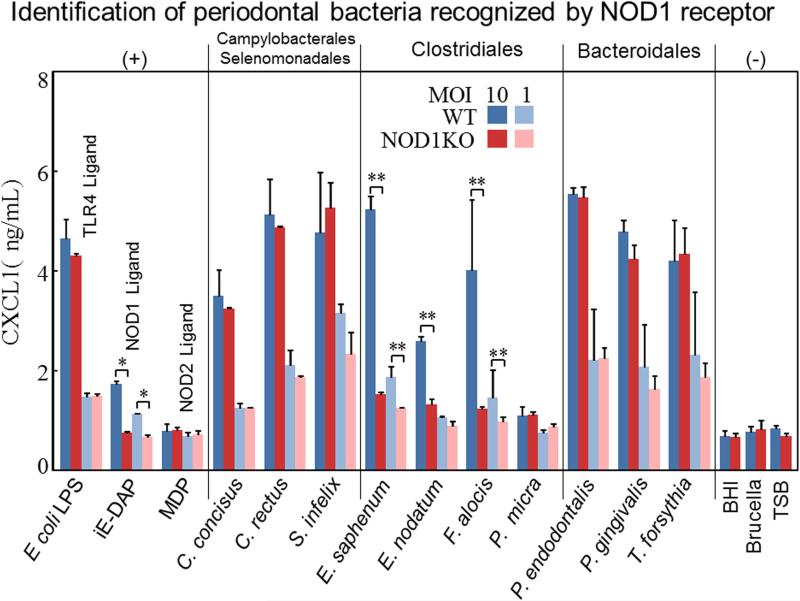
Diversity of NOD1 and NOD2 stimulatory activity among different periodontal bacterial species
In addition to TLRs, NOD1 and NOD2 represent 2 well-characterized PRRs of the NOD-like receptor (NLR) family and provide another level of microbial surveillance in the host cytosol to sense distinct peptidoglycan (PGN) products found in the cell wall of many types of bacteria (Caruso et al., 2014). NOD1 has been reported to be expressed by a variety of cells such as epithelial cells, stromal cells and endothelial cells (Caruso et al., 2014). Mesothelial cells are specialized epithelial cells that line the internal organs and the body wall of the peritoneal, pleural, and pericardial cavities that function monitor infection. Previous studies have shown that neutrophil-recruiting chemokines (CXCL1, CXCL2, and human IL-8) are secreted from mesothelial cells upon NOD1 stimulation (Park et al., 2007). Therefore, to determine the NOD1 stimulatory activity of periodontal bacteria, we stimulated WT and NOD1 KO mesothelial cells (Park et al., 2007) with various periodontal bacteria at the indicated ratio of 10:1 and 1:1 (Fig. 3) and 0.1:1 (Data not shown as no difference between WT and NOD1 KO was observed). The levels of CXCL1 in NOD1 KO mesothelial cells culture supernatant were compared with WT cells at 12h poststimulation. We found that E. nodatum, E. saphenum, and F. alocis showed significantly reduced levels of CXCL1 in NOD1 KO mesothelial cells (Fig. 3). However, the induction of CXCL1 secretion by C. concisus, C. rectus, S. infelix, P. gingivalis, P. endodontalis, T. forsythia, and P. micra was not dependent on NOD1 (Fig. 3). These data indicated that the periodontal bacteria E. nodatum, E. saphenum, and F. alocis display NOD1 stimulatory activity. These periodontal bacteria may potentially contribute to the pathogenesis of periodontal disease via NOD1 stimulatory ability. Surprisingly, unlike mice BMDMs, three members of Bacteroidetes (P. gingivalis, P. endodontalis, and T. forsythia) can induce strong CXCL1 production in mice mesothelial cells.
Figure 3. NOD1 stimulatory activities of various periodontal bacteria.
Mesothelial cells were prepared from both WT and NOD1 KO mice as described under “Material and Methods” and then stimulated with periodontal bacteria at defined ratios (MOI: 10:1, 1:1) for 12 h and the levels of CXCL1 secretion in cell culture supernatant was determined by ELISA. As positive controls, cells were incubated with 50 ng/ml E. coli LPS, 50ng/ml Nod1 ligand iE-DAP and 50 ng/ml NOD2 ligand MDP (with 10 times gradient dilution). Culture medium also was used as negative control for stimulation. The data shown represent the means ± SD from triplicate wells from one representative experiment of at least three separate experiments.
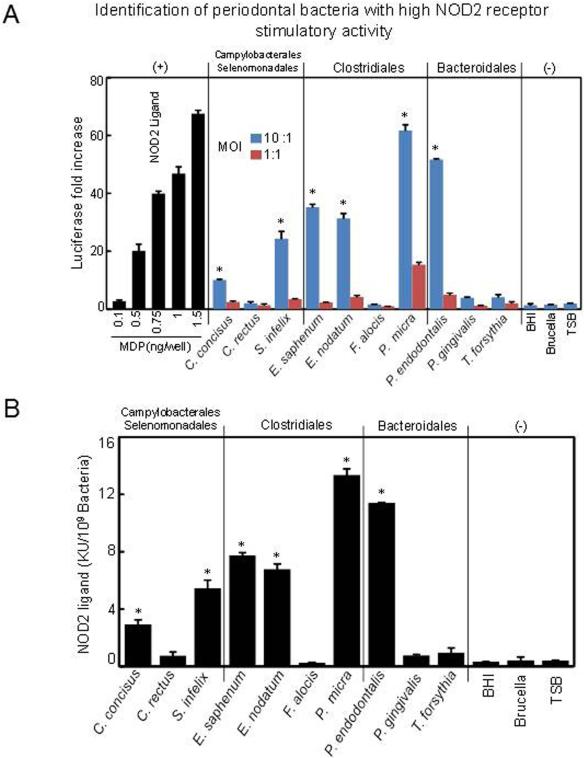
We next tested the NOD2 stimulatory activity of different periodontal bacteria by using human embryonic kidney (HEK) 293 cells with permanent transfections of NOD2 receptor (Warner et al., 2013). In this experiment, HEK293-NOD2 cells were stimulated with periodontal bacteria for 16h at the ratio of 10:1 and 1:1; then the luciferase assay was used to measure the NOD2 stimulatory activity. The data demonstrated that P. micra and P. endodontalis had the highest NOD2-stimulatory activity in HEK293-NOD2 cells (Fig. 4A and B). Meanwhile, S. infelix, E. saphenum, and E. nodatum also exhibited considerable NOD2 stimulatory activity, but to a lower extent (Fig. 4A and B). In contrast, C. concisus, C. rectus, F. alocis, P. gingivalis, and T. forsythia exhibited very weak NOD2 stimulatory activity (Fig. 4A and B). These results suggest that specific periodontal bacteria with high NOD2-stimulatory activity may induce the innate immune response by the NOD2 pathway.
Figure 4. NOD2 stimulatory activities of specific periodontal bacteria.
The NOD2 stimulatory activities were determined by the human embryonic kidney (HEK)-NOD2 cells. Defined ratios of periodontal bacteria (MOI: 10:1 and 1:1) were added to the cells for 16 h, the cells were lysed, and the amount of luciferase produced was determined. (A) Values are reported as the fold increase of relative luciferase compared to the nonstimulated control response, which was set at 1. Different concentrations of MDP were used as positive control. Culture medium was also used to stimulate cells as negative control. (B) The activity of bacterial culture is also given as KU/109 bacteria of the original culture according to the standard curve of MDP. One unit of the NOD2-stimulatory activity is equivalent to those of 1 ng of MDP. The data shown represent the means ± SD from triplicate wells from one experiment of at least three separate experiments. Asterisks indicate statistically significant to culture medium negative control (P < 0.05) as determined by One-way ANOVA with Bonferroni post-test analysis.
DISCUSSION
Periodontal bacteria are regarded as the causative agents that induce an excessive inflammatory response in a susceptible host. Recent studies demonstrated that the periodontal bacteria of health and disease are dramatically shifted from a symbiotic microbial community to a dysbiotic microbial community (Hajishengallis et al., 2011). A current study monitored the bacterial composition and diversity of plaque samples between healthy and periodontitis patients using HOMIM (Colombo et al., 2012). Consistent with previous studies (Griffen et al., 2012), our data also show that the oral microbiome in periodontitis patients exhibits greater disparities from healthy subjects (Table S1). It implies that these shifts in oral microbiota may be associated with the development of periodontal disease. As specific species indicate relative abundance in the bacterial community, the predominant composition of the oral microbiota in subjects with periodontitis may be relatively critical to the pathogenesis of periodontal disease. Based on the above two standards, we identified 15 bacterial species that not only significantly correlate with periodontitis, but also indicate predominance in the microbial community of periodontitis afflicted individuals. Our analysis showed that several classic pathogens and putative pathogens, including E. saphenum, F. alocis, P. endodontalis and S. infelix, were strongly associated with periodontal disease. This is in accordance to previous studies demonstrating that subjects with chronic periodontitis or general periodontitis are infected with these bacterial species more frequently and at higher levels as compared to healthy subjects (Belstrom et al., 2014). The presence of P. endodontalis was recently shown an a risk indicator for periodontal disease along with T. forsythia (Lourenco et al., 2014). Interestingly, two of these potential periodontal pathogens F. alocis and P. endodontalis were shown to persist or increase along with classic periodontal pathogens after mechanical detriment in refractory periodontitis patients (Colombo et al., 2012). A recent systematic review identified E. saphenum, F. alocis, and P. endodontalis as microorganisms with moderate evidence to be set as newly described periodontal pathogens (Perez-Chaparro et al., 2014). Together, the data indicate that the pathogenesis of these emerging periodontal pathogens requires further exploration.
As periodontal bacteria are the trigger of inflammatory immune response in periodontitis, their immunostimulatory activity to induce a host immune response is tightly related to the pathogenesis of periodontal disease. Recent studies have provided evidence that oral bacterial species possess different levels of immunostimulatory activities (Jiao et al., 2014). In this study, we observed a robust IL-6 production in HPBMs after stimulation by C. concisus, C. rectus, S. infelix, P. endodontalis, P. gingivalis and T. forsythia. However, four Clostridia including E. saphenum, E. nodatum, F. alocis and P. micra showed weak ability to induce cytokine in HPBMs. We also observed robust IL-6 production in WT mouse macrophages after stimulation by Campylobacter concisus, C. rectus, and S. infelix. As expected, the specific Clostridia remained unable to induce cytokine production in WT murine macrophages. Surprisingly, inconsistent with HPBMs, WT murine macrophages showed weak responses to three Bacteroidetes including P. endodontalis, P. gingivalis and T. forsythia compared to Campylobacter concisus, C. rectus, and S. infelix at MOI 10:1. These data may reflect the difference in immune response sensitivity to Bacteroidetes between human and mouse immune cells. Immunological differences between murine and human cells have been previously acknowledged. Interestingly, the expression of TLR2 in murine peripheral blood leukocytes has been reported to be low compared to the constitutive expression in human leukocytes (Mestas & Hughes 2004). This finding could partly explain our results of low IL-6 expression in murine macrophage compared to human monocyte in response to human Bacteroidetes since several human Bacteroidetes such as P. gingivalis, T. forsythia and Bacteroides fragilis have been reported to induce responses mainly via TLR2 (Alhawi et al., 2009, Hajishengallis et al., 2006, Onishi et al., 2008). Further studies are necessary to more completely elucidate the mechanisms why murine BMDMs exhibit lower inflammatory responses to human Bacteroidetes than human monocyte. In contrast to our current data, previous studies demonstrate that P. gingivalis showed stimulatory activity in murine macrophages, expressing IL-6 after stimulation (Kim et al., 2015, Shaik-Dasthagirisaheb et al., 2010). This difference is attributed to the fact that our study applied lower MOI and shorter stimulation time compared with previous studies. All of these studies support that the P.gingivalis stimulates murine BMDM to induce the immune response but significantly lower than the same challenger of Campylobacter concisus, C. rectus, or S. infelix.
Previous studies have reported that bacterial component such as lipoteichoic acid, lipoarabinomannan, LPS, and muropeptides degraded from peptidoglycan are highly heat-stable (Grunfeld et al., 1999, Hasegawa et al., 2006, Magalhaes et al., 2007). This finding indicates that the heat-killed bacteria will maintain immunostimulatory activity for TLR2, TLR4, NOD1 and NOD2, but limited the potential for other virulence factors such as proteinases. Since the goal of this study was to evaluate the innate immune response elicited via TLR2, TLR4, NOD1 and NOD2 to periodontal pathogens, we selected heat–killed bacteria for the assays. Interestingly, unlike murine macrophages, murine mesothelial cells were found to be sensitive to these three human oral Bacteroidetes (Table 3). Similar to our findings, previous studies have indicated that murine peritoneal mesothelial cells can mediate significant immune responses in response to B. fragilis (Kim et al., 2012). These results strongly suggest that murine mesothelial cells have great potential as a valuable murine cell type to study the immunostimulatory property of human Bacteroidetes. Further studies using murine mesothelial cells with specific gene-deficiency may assist in characterizing the pathogen-associated molecular patterns (PAMPs) of human oral Bacteroidetes.
Table 3.
Periodontal bacteria show recognition via different pathogen-recognition receptors
| Bacteria | Order | Family | Genus | Gram Staining | HPBM | Mice Mø | Mice Mesothelial | TLR4 | NOD1 | NOD2 |
|---|---|---|---|---|---|---|---|---|---|---|
| C.concicus | Campylobacterales | Campylobacteraceae | Campylobacter | Negative | Strong | Strong | Strong | + | - | + |
| C.rectus | Campylobacterales | Campylobacteraceae | Campylobacter | Negative | Strong | Strong | Strong | + | - | - |
| S.infelix* | Selenomonadales | Veillonellaceae | Selenomonas | Negative | Strong | Strong | Strong | + | - | + |
| E.saphenum | Clostridiales | Eubacteriaceae | Eubacterium | Positive | Weak | Weak | Strong | - | + | + |
| E.nodatum | Clostridiales | Eubacteriaceae | Eubacterium | Positive | Weak | Weak | Strong | - | + | + |
| F.alocis | Clostridiales | Peptostreptococcaceae | Filifactor | Positive | Weak | Weak | Strong | - | + | - |
| P.micra | Clostridiales | Clostridiaceae | Peptostreptococcus | Positive | Weak | Weak | Weak | - | - | + |
| P.endodontalis | Bacteroidales | Porphyromonadaceae | Porphyromonas | Negative | Strong | Weak | Strong | - | - | + |
| P.gingivalis | Bacteroidales | Porphyromonadaceae | Porphyromonas | Negative | Strong | Weak | Strong | - | - | - |
| T.forsythia | Bacteroidiales | Porphyromonadaceae | Tannerella | Negative | Strong | Weak | Strong | - | - | - |
Gram-negative wall but belongs within the Phylum Firmicutes.
- no stimulatory activity compared to the negative control, + significant stimulatory activity compared to the negative control (P<0.05),
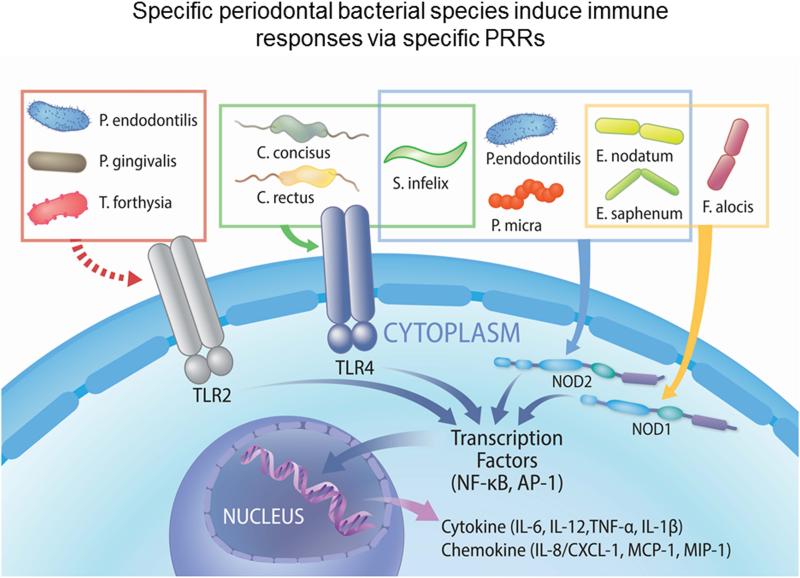
Although the function of PRRs in response to several periodontal pathogens has been widely studied (Burns et al., 2006, Jain et al., 2013, Myneni et al., 2011, Park et al., 2014), there is also a paucity of information on the interaction between host PRRs and periodontal microbes, especially specific newly-identified periodontal pathogens. We used assays from BMDMs, mesothelial cells, and NOD2-HEK-293 cells to estimate the immunostimulatory activities of TLR2, TLR4, NOD1 and NOD2 ligands in the present study. Consistent with previous findings (Arce et al., 2012), we noted that C. concisus, C. rectus, and S. infelix showed strong TLR4-stimulatory activity. E. saphenum, E. nodantum, and F. alocis showed considerable NOD1-stimulatory activity. P. micra and P. endodontalis had the highest NOD2-stimulatory activity. Our studies support that these periodontal bacterial species may contribute to the pathogenesis of periodontitis relying on TLR4, NOD1 and NOD2 (Table 3 and Fig. 5). It will be important to further determine whether these newly-identified putative pathogens induce periodontitis in vivo. Recent exploration of infection with F. alocis in a murine subcutaneous chamber model showed that this microorganism can establish a proinflammatory and proapoptotic local infection with neutrophil influx (Wang et al., 2014). One of the potential mechanisms that F. alocis induced neutrophil recruitment is by CXCL1 expression via NOD1, as we observed in our in vitro assay.
Figure 5. Specific periodontal bacterial species induce specific cytokine production via specific pattern recognition receptors (PRRs).
Three gram-negative bacteria including C. concisus, C. rectus and S. infelix can stimulate host to induce dramatic immune response via TLR4. Three gram-positive bacteria including E. nodatum, E. saphenum and F. alocis can be recognized by host via NOD1. Meanwhile, P. micra and P. endodontilis are predominantly sensed by host through NOD2. S. infelix, C. concisus and C. rectus also show considerable ability to induce host response through NOD2. According to previous studies, the host can sense two Bacteroidales including P. gingivalis and T. forsythia through TLR2. P. endodontilis also belongs to Bacteroidales, so it may also have TLR2 stimulatory activity. These events indicated that specific periodontal pathogens exhibit different properties to activate innate immune activation. It also suggested that the host may recognize these bacteria to initiate responses for defending against pathogens.
In this study, we evaluated IL-6 and CXCL-1 production since these are classical inflammatory cytokines and chemokines subsequent to bacterial stimulation. Similar parallel responses for TNF-α and IL-1β expression will be expected according to previous studies (Kim et al., 2015, Shaik-Dasthagirisaheb et al., 2010). However, future studies will be necessary to assist in better understanding the inflammatory response of these individual oral pathogens.
As mentioned above, in order to focus on the innate immune responses that are elicited via TLR2, TLR4, NOD1 and NOD2, we utilized heat-killed bacteria, which can potentially inactivate other pathogenic virulence factors, such as proteinases, toxins and bacterial invasion. However, we acknowledge that the expression of other virulence factors in these new-identified putative periodontal pathogens may also be critical for affecting the host immune response. Further investigation using live bacteria with the presence of other potential virulence factors will be important for future studies to better understand the virulence of these periodontal pathogens. In addition, some PAMPs for other TLRs and NLRs may be heat-labile (Akira et al., 2006). Therefore, the immunostimulatory activity of these heat-labile PAMPs may need to be tested by live bacteria or by non-heat inactivation methods such as antibiotics, sonication and paraformaldehyde fixation.
CONCLUSIONS
In summary, evidence is provided to support the concept of potential novel periodontal pathogens in periodontitis. Our study identified a group of oral microbes including classical and putative pathogens in periodontitis patients. Specifically, this is the first report that characterizes the TLR2, TLR4, NOD1 and NOD2 stimulatory features of newly-identified putative pathogens. Finally, we demonstrate that specific bacterial species exhibit distinct immunostimulatory properties that may be involved in the pathogenesis of periodontitis by distinct innate immune receptor activation.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by Colgate Palmolive Co, NIH DE13397 (to W.V.G) and NIH F32DE021934 (to JTM). The authors would like to thank Dr. Gabriel Nunez (University of Michigan) for providing MyD88 and TLR2/4 double KO mice mouse BMDM and NOD2-HEK293, Drs. R. Teles (Forsyth Institute) and C. Fenno (University of Michigan) for providing bacteria, Dr. Robertson Davenport (University of Michigan) for providing filter buffy coat. The authors would also thank Dr. Thomas Braun (University of Michigan School of Public Health) for the biostatistics discussion. The authors would also like to thank James Sugai, Christina Huffman, Anna Galloro, Hilye Pittman, Amy Collins for clinical and laboratory support.
Footnotes
All the authors declare no conflict of interest.
REFERENCES
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alhawi M, Stewart J, Erridge C, Patrick S, Poxton IR. Bacteroides fragilis signals through Toll-like receptor (TLR) 2 and not through TLR4. J Med Microbiol. 2009;58:1015–1022. doi: 10.1099/jmm.0.009936-0. [DOI] [PubMed] [Google Scholar]
- Arce RM, Caron KM, Barros SP, Offenbacher S. Toll-like receptor 4 mediates intrauterine growth restriction after systemic Campylobacter rectus infection in mice. Mol Oral Microbiol. 2012;27:373–381. doi: 10.1111/j.2041-1014.2012.00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beikler T, Schnitzer S, Abdeen G, Ehmke B, Eisenacher M, Flemmig TF. Sampling strategy for intraoral detection of periodontal pathogens before and following periodontal therapy. J Periodontol. 2006;77:1323–1332. doi: 10.1902/jop.2006.050204. [DOI] [PubMed] [Google Scholar]
- Belstrom D, Fiehn NE, Nielsen CH, et al. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41:104–112. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AP, Bennet S, Cotton SL, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–1287. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Thornton-Evans GO, Wei L, Borgnakke WS, Dye BA. Accuracy of NHANES periodontal examination protocols. J Dent Res. 2010;89:1208–1213. doi: 10.1177/0022034510377793. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Girardin SE, Fitting C, et al. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005;35:2459–2470. doi: 10.1002/eji.200526286. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunfeld C, Marshall M, Shigenaga JK, Moser AH, Tobias P, Feingold KR. Lipoproteins inhibit macrophage activation by lipoteichoic acid. J Lipid Res. 1999;40:245–252. [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Tapping RI, Harokopakis E, et al. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell Microbiol. 2006;8:1557–1570. doi: 10.1111/j.1462-5822.2006.00730.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Osaka T, Tawaratsumida K, et al. Transitions in oral and intestinal microflora composition and innate immune receptor-dependent stimulation during mouse development. Infect Immun. 2010;78:639–650. doi: 10.1128/IAI.01043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Yang K, Hashimoto M, et al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281:29054–29063. doi: 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- Hill GB, Ayers OM, Kohan AP. Characteristics and sites of infection of Eubacterium nodatum, Eubacterium timidum, Eubacterium brachy, and other asaccharolytic eubacteria. J Clin Microbiol. 1987;25:1540–1545. doi: 10.1128/jcm.25.8.1540-1545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Coats SR, Chang AM, Darveau RP. A novel class of lipoprotein lipase-sensitive molecules mediates Toll-like receptor 2 activation by Porphyromonas gingivalis. Infect Immun. 2013;81:1277–1286. doi: 10.1128/IAI.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Russell TR, Graves DT, Cheng H, Nong SH, Levitz SM. Monocyte chemoattractant protein 1 and interleukin-8 production in mononuclear cells stimulated by oral microorganisms. Infect Immun. 1996;64:4450–4455. doi: 10.1128/iai.64.11.4450-4455.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Hasegawa M, Inohara N. Emerging roles of immunostimulatory oral bacteria in periodontitis development. Trends Microbiol. 2014;22:157–163. doi: 10.1016/j.tim.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Park JH, Franchi L, Backert S, Nunez G. The Cag pathogenicity island and interaction between TLR2/NOD2 and NLRP3 regulate IL-1beta production in Helicobacter pylori infected dendritic cells. Eur J Immunol. 2013;43:2650–2658. doi: 10.1002/eji.201243281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PD, Xia-Juan X, Crump KE, Abe T, Hajishengallis G, Sahingur SE. Toll-Like Receptor 9-Mediated Inflammation Triggers Alveolar Bone Loss in Experimental Murine Periodontitis. Infect Immun. 2015;83:2992–3002. doi: 10.1128/IAI.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Lee KB, Kang MJ, Park JH. Critical role of Toll-like receptor 2 in Bacteroides fragilis-mediated immune responses in murine peritoneal mesothelial cells. Microbiol Immunol. 2012;56:782–788. doi: 10.1111/j.1348-0421.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco TG, Heller D, Silva-Boghossian CM, Cotton SL, Paster BJ, Colombo AP. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol. 2014;41:1027–1036. doi: 10.1111/jcpe.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes PO, Lopes AM, Mazzola PG, Rangel-Yagui C, Penna TC, Pessoa A., Jr. Methods of endotoxin removal from biological preparations: a review. J Pharm Pharm Sci. 2007;10:388–404. [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Meyer TP, Zehnter I, Hofmann B, et al. Filter Buffy Coats (FBC): a source of peripheral blood leukocytes recovered from leukocyte depletion filters. J Immunol Methods. 2005;307:150–166. doi: 10.1016/j.jim.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Myneni SR, Settem RP, Connell TD, Keegan AD, Gaffen SL, Sharma A. TLR2 signaling and Th2 responses drive Tannerella forsythia-induced periodontal bone loss. J Immunol. 2011;187:501–509. doi: 10.4049/jimmunol.1100683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi S, Honma K, Liang S, et al. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, Shaw M, et al. Nod1/RICK and TLR signaling regulate chemokine and antimicrobial innate immune responses in mesothelial cells. J Immunol. 2007;179:514–521. doi: 10.4049/jimmunol.179.1.514. [DOI] [PubMed] [Google Scholar]
- Park SR, Kim DJ, Han SH, et al. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect Immun. 2014;82:1914–1920. doi: 10.1128/IAI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Perez-Chaparro PJ, Goncalves C, Figueiredo LC, et al. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 2014;93:846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prates TP, Taira TM, Holanda MC, et al. NOD2 contributes to Porphyromonas gingivalis-induced bone resorption. J Dent Res. 2014;93:1155–1162. doi: 10.1177/0022034514551770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramseier CA, Kinney JS, Herr AE, et al. Identification of pathogen and host-response markers correlated with periodontal disease. J Periodontol. 2009;80:436–446. doi: 10.1902/jop.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik-Dasthagirisaheb YB, Kantarci A, Gibson FC., 3rd Immune response of macrophages from young and aged mice to the oral pathogenic bacterium Porphyromonas gingivalis. Immun Ageing. 2010;7:15. doi: 10.1186/1742-4933-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Teles FR, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Mol Oral Microbiol. 2011;26:127–139. doi: 10.1111/j.2041-1014.2010.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu H, Sato N, Hossain MZ, Ikeda T, Hoshino E. Degradation of arginine and other amino acids by butyrate-producing asaccharolytic anaerobic Gram-positive rods in periodontal pockets. Arch Oral Biol. 2003;48:423–429. doi: 10.1016/s0003-9969(03)00031-1. [DOI] [PubMed] [Google Scholar]
- Wang Q, Jotwani R, Le J, et al. Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect Immun. 2014;82:1205–1212. doi: 10.1128/IAI.01434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner N, Burberry A, Franchi L, et al. A genome-wide siRNA screen reveals positive and negative regulators of the NOD2 and NF-kappaB signaling pathways. Sci Signal. 2013;6:rs3. doi: 10.1126/scisignal.2003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.