Abstract
The most common side effect of angiotensin-converting enzyme inhibitor (ACEi) drugs is cough. We conducted a genome-wide association study (GWAS) of ACEi-induced cough among 7080 subjects of diverse ancestries in the Electronic Medical Records and Genomics (eMERGE) network. Cases were subjects diagnosed with ACEi-induced cough. Controls were subjects with at least 6 months of ACEi use and no cough. A GWAS (1595 cases and 5485 controls) identified associations on chromosome 4 in an intron of KCNIP4. The strongest association was at rs145489027 (minor allele frequency=0.33, odds ratio (OR)=1.3 (95% confidence interval (CI): 1.2–1.4), P=1.0 × 10−8). Replication for six single-nucleotide polymorphisms (SNPs) in KCNIP4 was tested in a second eMERGE population (n=926) and in the Genetics of Diabetes Audit and Research in Tayside, Scotland (GoDARTS) cohort (n=4309). Replication was observed at rs7675300 (OR=1.32 (1.01–1.70), P=0.04) in eMERGE and at rs16870989 and rs1495509 (OR=1.15 (1.01–1.30), P=0.03 for both) in GoDARTS. The combined association at rs1495509 was significant (OR=1.23 (1.15–1.32), P=1.9 × 10−9). These results indicate that SNPs in KCNIP4 may modulate ACEi-induced cough risk.
Introduction
Angiotensin-converting enzyme inhibitor drugs (ACEi) are frequently used in clinical medicine for the treatment of elevated blood pressure, heart failure and renal protection in chronic kidney disease.1 Although this class of medications is generally well tolerated, adverse reactions may prevent their use in some individuals. The most common side effect is a persistent, nonproductive cough that can start within days to months after initiating therapy and requires cessation of ACEi use.2 Women are 1.5–2 times more likely to develop a cough than men.3 Some epidemiological features of ACEi-induced cough suggest a genetic predisposition to this adverse effect. In particular, there is racial variation in the incidence of cough, with the highest rates observed in east Asian populations where rates are 20–45%,4 as compared with European American populations where rates are approximately 10%.5
The mechanism of ACEi-induced cough is not certain. ACEis block the proteolytic enzyme angiotensin I-converting enzyme (ACE), which cleaves a number of target proteins, including angiotensin I, the primary mediator of the blood pressure-lowering effects, and pro-inflammatory kinins.6 One suspected mechanism of cough is the accumulation of these inflammatory kinins, which may sensitize vagal afferent fibers leading to a neurogenically mediated cough.7 There have been multiple candidate gene studies testing for associations between genetic variation in ACE and bradykinin pathway members and ACEi-induced cough. There has been intense focus on the association of cough with a common 287-bp insertion–deletion in the ACE gene.3, 8, 9, 10, 11 A meta-analysis of 11 of these studies only found a consistent association in a subgroup analysis of subjects aged >60 years.12 Significant single-nucleotide polymorphism (SNP) associations have also been reported between mediators in the bradykinin pathway. including the bradykinin B2 receptor (BDKRB2), membrane metallo-endopeptidase (MME), prostaglandin receptor E (PTGER3), neurokinin 2 receptor (NK2R) and ACE.9, 13, 14, 15, 16 However, these findings have not been consistently observed across studies.3, 10, 17, 18 Additional studies also suggest a role for SNPs located within the ABO gene, which have been shown to regulate the plasma ACE levels and have also been associated with ACEi cough.10, 14, 19, 20
In the present study, we used a genome-wide association study (GWAS) approach to carry out a comprehensive analysis of genetic determinants of ACEi-induced cough. Subjects were identified through the Electronic Medical Records and Genomics (eMERGE) network, a consortium of medical centers that utilize electronic medical records (EMR) as a tool for genomic research.21 Candidate SNPs were first identified using a multi-racial discovery cohort. The most significant candidate SNPs were then evaluated in two independent replication cohorts. We report here a significant signal at the KCNIP4 locus indicating that variant potassium channel function or neuronal signaling contributes to the risk of a cough.
Materials and methods
Study population
The discovery study population comprised 7080 adult subjects collected from six sites participating in the phases I and II of eMERGE Network (Phase I: Vanderbilt University (VUMC), Marshfield Clinic, Northwestern University, Mayo Clinic and Group Health Research Institute; and Phase II: Geisinger Health System and Mount Sinai).21, 22 Subjects used for the discovery population were selected to maximize the representation across sites while minimizing the number of genotyping platforms represented. In addition, cases and controls genotyped on OMNI-QUAD platforms and that were part of the Vanderbilt Electronic Systems for Pharmacogenomic Assessment (VESPA) study that examined the genomics of drug response phenotypes were also included.23 For subgroup analyses, genetic ancestry assignment was determined using STRUCTURE24 in conjunction with 1917 ancestry informative markers (from the Illumina Test Panel25), with European ancestry defined as >90% probability of being in the CEU cluster and African ancestry defined as >70% probability of being in the YRI cluster, using a HapMap population as the reference (Supplementary Figures S1–S3). Site-specific subject counts are shown in Supplementary Table S1.
Phenotype data
The phenotype evaluated was cough attributable to the use of an ACEi. The algorithm identifying cases and controls was developed at VUMC and subsequently validated and deployed at the other eMERGE sites. The phenotype definition incorporated an iterative process whereby automated case and control assignment algorithms were validated against assignments made by manual review of the EMR.26 Phenotyping algorithms were refined until the positive predictive value reached the predesignated target of ⩾95%. In the final algorithms, cases were defined as subjects whose records contained either an ACEi drug name or ACEi class designator and ‘cough' on the same line within the structured ‘Allergy' section of the medical record. Hence, cases represent ACEi-induced cough recorded by a health-care provider. Controls were subjects who had an ACEi drug name or ACEi class on two medication listings with dates separated by at least 6 months and did not have a documented cough associated with ACEi use in the Allergy section. Complete details of the algorithm are available from PheKB (http://phekb.org/phenotypes). The mean positive predictive value for cases and controls in the VUMC and replication sites was 100% and 97.5%, respectively (Supplementary Table S2).
Birth decade and sex were also extracted in addition to diagnosis codes (International Classification of Diseases (ICD)-9 codes27) related to comorbidities potentially contributive to the risk of cough, including asthma, postnasal drip, gastroesophageal reflux disease, bronchitis, emphysema, bronchiectasis, allergic alveolitis and chronic obstructive pulmonary disease. Lists of the ICD-9 codes used to define each of these conditions are shown in Supplementary Table S3. Subjects with ⩾1 ICD-9 codes for any of the conditions were considered to have the comorbidity. Smoking status, categorized as ‘Ever', ‘Never' or ‘Missing', was also extracted from either structured EMR data or validated algorithms.28
Genotyping data
SNP genotype data were acquired on the Illumina HumanOmni1-Quad (Vanderbilt), HumanOmni5-Quad (VUMC), Human1M-Duov3_B (VUMC), HumanOmniExpress-12v1.0 (Geisinger), HumanOmniExpress (Mount Sinai) and Human660W-Quadv1_A (all other sites and VUMC) (Supplementary Table S1). Genotyping data for each platform were individually cleaned. Quality control (QC) steps included identifying sex mismatches, SNPs failing concordance with HapMap, Mendelian errors and duplicate removal. After QC, a merged data set was created that contained 267 485 SNPs present on all platforms and with a call rate >98%. Cryptic relatedness was assessed on the merged platforms by identical-by-descent analysis, and one of a pair of subjects (n=343 total) more closely related than half-siblings was randomly excluded. Imputation was performed on the merged intersection data set using IMPUTE229 in conjunction with the 1000 Genomes phase 3 reference panel for all populations. Prior to imputation, strand alignment between study and reference genotypes and prephasing of study genotypes into haplotypes was performed using SHAPEIT.30 Only those imputed SNPs with a genotype probability >90% were analyzed. SNPs with an Info score29 (measuring the average probabilities for a given SNP) <0.7 were excluded from analyses. The final analyses were restricted to 1 931 830 SNPs with a call rate >98%, a Hardy–Weinberg P>1 × 10−6 and minor allele frequency (MAF)>0.01. Principal components (PCs) fit to the preimputed SNP data set were computed using EIGENSTRAT31 to adjust for population structure.
The primary single SNP tests of association were performed using logistic regression assuming an additive genetic model, adjusting for 10 PCs, birth year and sex. A model that also incorporated binary covariates for smoking status (captured by two binary variables: ‘Ever smoked' and ‘Never smoked') and each of the comorbidities of asthma, postnatal drip, gastroesophageal reflux disease and lung disease (bronchitis, emphysema, bronchiectasis, allergic alveolitis and chronic obstructive pulmonary disease) was evaluated.
Replication cohorts
SNPs with an association P-value <5x10−6 were evaluated in two replication populations. The first replication set comprised additional subjects available through the eMERGE network who had SNP genotyping as part of eMERGE Phase II in addition to 53 additional cases and controls from VUMC that were not included in the original analyses. Covariate and phenotype extraction from the EMR was as described above. Subjects were genotyped on the Affymetrix Human SNP Array 6.0 (Mount Sinai, Marshfield), Human660W-Quad (Marshfield), Human610-Quad (Mayo) and HumanOmniExpress (Group Health, Northwestern) platforms (Supplementary Table S4). QC steps, as described above, were performed per the QC protocol established by the eMERGE Genomics Working Group.32 Each data set was then individually imputed by platform using IMPUTE229 and the 1000 Genomes cosmopolitan reference panel.33 Imputed data were extracted and merged using the same QC filters as described above. The replication analyses were conducted in 157 cases and 769 controls that had >80% probability of being in the CEU cluster by STRUCTURE analyses. Multivariable analyses adjusting for PCs and covariates were performed, as described above. A replication P-value of 0.05 was considered statistically significant.
The second replication set was derived from the GoDARTS (Genetics of Diabetes Audit and Research in Tayside, Scotland) 2011 cohort and contained prescription information between 1990 and 2011 for 17 601 self-reported Caucasian diabetics and non-diabetics from the Tayside area. Cases were defined as subjects who switched from an ACEi to an angiotensin receptor blocker, a drug that targets a downstream receptor in the angiotensin–renin–aldosterone pathway. The positive predictive values of a possible and probable ACEi adverse drug reaction using this case definition were 90.5% and 68.3%, respectively. The most frequent adverse reactions associated with this case definition were cough and the rare reaction of angioedema. Controls were defined as those who had filled ACEi prescriptions within 9 months of the study's censor date or their date of death. Subjects who were ever concurrently on ACEis and angiotensin receptor blockers were also categorized as controls. A total of 710 cases and 3599 controls were available for analysis. Subjects were genotyped on the Affymetrix 6.0 (Affymetrix, Santa Clara, CA, USA) (381 cases and 1924 controls) or Illumina HumanOmniExpress (Illumina, San Diego, CA, USA) (329 cases and 1675 controls) platforms. Both platforms were imputed using IMPUTE229 and the 1000 Genomes reference panel. SNPs deviating from Hardy–Weinberg equation (P<1 × 10−6) or with an Info Score<0.4 were excluded. An additive genetic model adjusting for age and sex was first computed separately for each genotyping platform and the results were meta-analyzed using GWAMA.34
Data analysis
All QC analyses and GWA analyses were performed using PLINK v1.07.35 GoDARTS data were also analyzed using SNPTEST v2.536 and GWAMA v2.1.34 All other analyses were performed using SAS v9.3 (SAS Institute, Cary, NC, USA). SNP data around the KCNIP4 gene were visualized using LocusZoom37. The forest plot was generated using the Metafor package.38 The publicly available GTEx39, HaploReg v340 and the NCBI expression quantitative trait locus (eQTL) databases were used to identify eQTLs and functional motifs associated with the most significant candidate SNPs.
Ethics statement
The eMERGE study was approved by the Institutional Review Board at each site.21, 41 The GoDARTS study was approved by the Tayside Medical Ethics Committee and informed consent was obtained for all participants.
Results
A total of 1595 cases of ACEi-induced cough and 5485 controls were analyzed in the discovery cohort (Table 1). The majority of subjects were of European ancestry, with about 15% of subjects belonging to other racial/ethnic groups. As compared with controls, the cases had a higher proportion of females (P<0.001), were younger, as measured by birth decade (P<0.001), more likely to have been smokers (P<0.001) and differed with respect to frequencies of diagnoses of gastroesophageal reflux disease (P<0.001), postnatal drip (P<0.001) and structural lung disease (P<0.001), all of which may contribute to a chronic cough. However, the directions of associations for these variables were not consistent, as smoking and lung disease were associated with decreased rates of ACEi cough, whereas the other variables were associated with increased rates.
Table 1. Population characteristics of the discovery cohort.
| Cases, n (%) | Controls, n (%) | P-valuea | |
|---|---|---|---|
| Male | 626 (39.3) | 3029 (55.2) | <0.001 |
| Female | 969 (60.7) | 2456 (44.8) | |
| Genetic ancestry | |||
| European ancestry | 1346 (84.3) | 4661 (85.0) | <0.001 |
| African ancestry | 178 (11.2) | 701 (12.8) | |
| Other | 71 (4.5) | 123 (2.2) | |
| Birth decade (IQR) | 1930–1950 | 1920–1940 | <0.001 |
| Ever smoked | 515 (32.9) | 2403 (43.8) | <0.001b |
| History of | |||
| Asthma | 262 (16.4) | 882 (16.1) | 0.74 |
| GERD | 668 (41.9) | 2035 (37.1) | <0.001 |
| PND | 530 (33.2) | 1507 (27.5) | <0.001 |
| Other lung disease | 331 (20.8) | 1626 (29.7) | <0.001 |
Abbreviations: GERD, gastroesophageal reflux disease; IQR, interqquartile range; PND, postnasal drip.
P-value obtained from chi-square analysis.
Calculation excludes 776 subjects (184 cases, 592 controls) with an unknown smoking history.
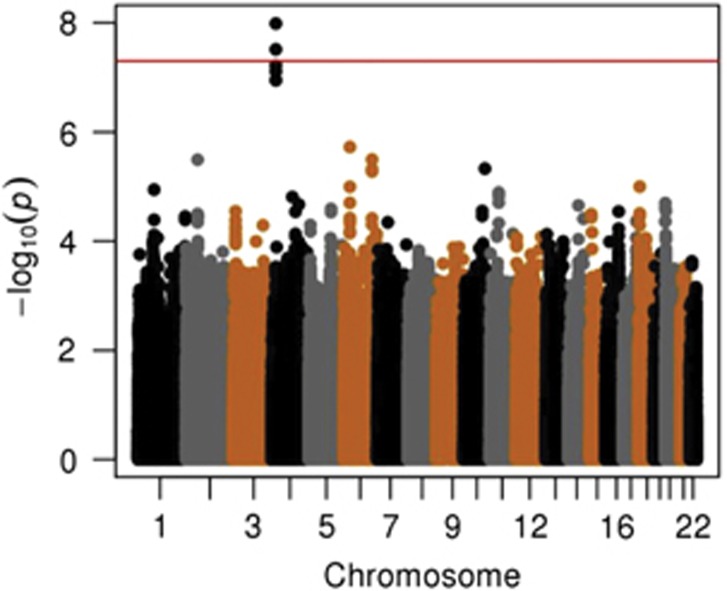
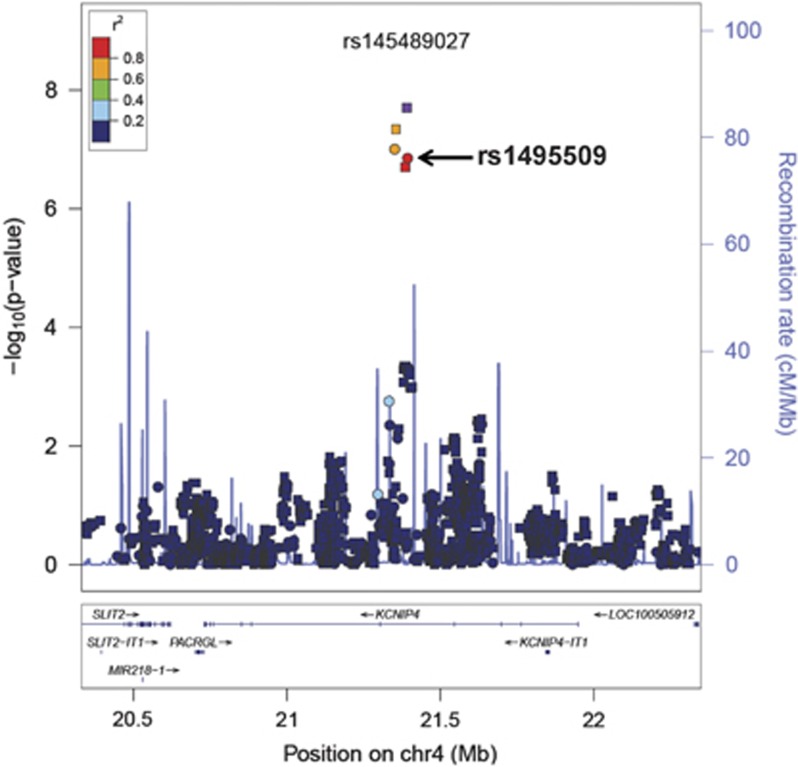
In multivariable regression analyses adjusting for PCs, sex and birth year, two SNPs located on chromosome 4 reached genome-wide significance (Figure 1, Supplementary Figure S4). All of the SNPs with an association P-value<5x10−6 were exclusively located in intron 4 in the gene ‘Kv Channel Interacting Protein 4' (KCNIP4) (Figure 2 and Supplementary Figure S5). The strongest association was in the imputed SNP rs145489027 (MAF=0.30, odds ratio (OR)=1.3 (95% confidence interval (CI): 1.2–1.4), P=1.0 × 10−8; Table 2). When results were stratified by site, there was a consistent direction of association across sites, with ORs ranging from 1.1 to 1.5 (Supplementary Table S5). When the analyses were additionally adjusted for cough risk factors, the association at the SNP remained significant (OR=1.3 (95% CI: 1.2–1.4), P=1.0 × 10−8; Supplementary Figure S6). Similar results were observed when subjects with lung disease were excluded (OR=1.3 (95% CI: 1.2–1.5), P=3.0x10−8; Supplementary Figure S7). A regression model adjusting for the rs145489027 SNP eliminated the associations (P>0.05) for the other most significant SNPs, indicating that the SNPs represent a common signal (the r2 between this SNP and other most significant SNPs was 0.68 for rs7661530 and rs6838116 and 0.98 for rs7675300, rs16870989 and rs1495509 in a 1000G European ancestry population). In a subset analysis of African Americans and European Americans, this SNP was most strongly associated with cough in European Americans (MAF=0.33, OR=1.3 (95% CI: 1.2–1.4), P=2.5 × 10−7; Table 3). Although the direction of the effect was similar in African Americans, these associations were not significant in this smaller subset of subjects (n=879). ACEi-induced cough has been observed to occur more frequently in women3. In analyses stratified by sex, ORs were similar for men (OR=1.3 (95% CI: 1.2–1.5), P=1.4 × 10−4) and women (OR=1.3 (95% CI: 1.1–1.5), P=1.1 × 10−5). A search of publicly available data sets did not identify eQTLs or functional motifs associated the most significant SNPs. We specifically examined the associations using all SNPs located in 16 previously reported candidate genes (see Supplementary Materials). No significant associations (P<0.05) were observed after applying a Bonferroni correction adjusting for multiple testing or in a subset analysis of older subjects born before 1960.
Figure 1.
Manhattan plot of genotyped single-nucleotide polymorphisms associated with angiotensin-converting enzyme inhibitor-induced cough using an additive model adjusted for 10 principal components, age, sex, history of asthma, reflux, postnasal drip and lung disease. The red line indicates the genome-wide significance threshold of alpha=5 × 10−8.
Figure 2.
LocusZoom plot of most strongly associated single-nucleotide polymorphisms (SNPs) from genome-wide association study located in the region of KCNIP4 (chr4:20330238–22350374), centered around SNP rs145489027 (shown in purple). Linkage disequilibrium (based on r2 values) with respect to rs145489027 are based on the CEU reference population. Imputed SNPs are denoted by squares and genotyped SNPs by circles.
Table 2. Most significantly associated SNPs.
| SNP | Minor allele | MAF | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| Discoverya | |||||
| rs7661530 | T | 0.33 | 1.27 | (1.17–1.39) | 6.1 × 10−8 |
| rs6838116 | A | 0.33 | 1.28 | (1.17–1.40) | 3.1 × 10−8 |
| rs7675300 | A | 0.31 | 1.27 | (1.16–1.39) | 1.1 × 10−7 |
| rs16870989 | A | 0.31 | 1.27 | (1.16–1.39) | 1.1 × 10−7 |
| rs145489027 | A | 0.30 | 1.30 | (1.19–1.42) | 1.0 × 10−8 |
| rs1495509 | C | 0.31 | 1.27 | (1.17–1.39) | 7.8 × 10−8 |
| Replication | |||||
| eMERGEb | |||||
| rs7661530 | T | 0.35 | 1.24 | (0.96–1.59) | 0.09 |
| rs7675300 | A | 0.33 | 1.32 | (1.01–1.70) | 0.04 |
| rs16870989 | A | 0.33 | 1.28 | (0.98–1.65) | 0.06 |
| rs145489027 | A | 0.32 | 1.28 | (0.98–1.68) | 0.07 |
| rs1495509 | C | 0.33 | 1.28 | (0.98–1.65) | 0.06 |
| GoDARTSc | |||||
| rs7661530 | T | 0.37 | 1.05 | (0.89–1.23) | 0.84 |
| rs6838116 | A | 0.37 | 1.08 | (0.95–1.20) | 0.57 |
| rs16870989 | A | 0.34 | 1.15 | (1.01–1.30) | 0.03 |
| rs145489027 | A | 0.33 | 1.12 | (0.99–1.27) | 0.08 |
| rs1495509 | C | 0.34 | 1.15 | (1.01–1.30) | 0.03 |
| Meta-analysis | |||||
| rs7661530 | T | — | 1.22 | (1.13–1.31) | 9.6 × 10−8 |
| rs6838116d | A | — | 1.17 | (1.10–1.24) | 4.2 × 10−7 |
| rs7675300d | A | — | 1.27 | (1.17–1.39) | 1.4 × 10−8 |
| rs16870989 | A | — | 1.23 | (1.15–1.32) | 4.9 × 10−9 |
| rs145489027 | A | — | 1.24 | (1.16–1.33) | 2 × 10−9 |
| rs1495509 | C | — | 1.23 | (1.15–1.32) | 1.9 × 10−9 |
Abbreviations: CI, confidence interval; eMERGE, Electronic Medical Records and Genomics; GoDARTS, Genetics of Diabetes Audit and Research in Tayside, Scotland; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism.
From a multivariable logistic regression assuming an additive genetic model and adjusted for 10 principal components (PCs), birth year and sex.
From a multivariable logistic regression (157 cases and 769 controls) assuming an additive genetic model and adjusted for 3 PCs, birth year and sex.
From a meta-analysis (710 cases and 3,599 controls) adjusted for age and sex. The P-value for the Q-score P-value was >0.05 for each SNP.
Based on only two data sets.
Table 3. Association analyses by racial groups for SNP rs145489027.
| Racial group | Cases | Controls | MAF cases | MAF controls | OR | 95% CI | P-valuea |
|---|---|---|---|---|---|---|---|
| European ancestry | 1346 | 4661 | 0.37 | 0.32 | 1.3 | (1.2–1.4) | 2.5 × 10−7 |
| African ancestry | 178 | 701 | 0.13 | 0.10 | 1.2 | (0.9–1.8) | 0.24 |
| Otherb | 71 | 123 | 0.29 | 0.20 | 1.4 | (0.8–2.6) | 0.25 |
Abbreviations: CI, confidence interval; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism.
From a multivariable logistic regression assuming an additive genetic model and adjusting for 10 principal components, birth year and sex.
Includes subjects not meeting criteria for inclusion with African ancestry and European ancestry.
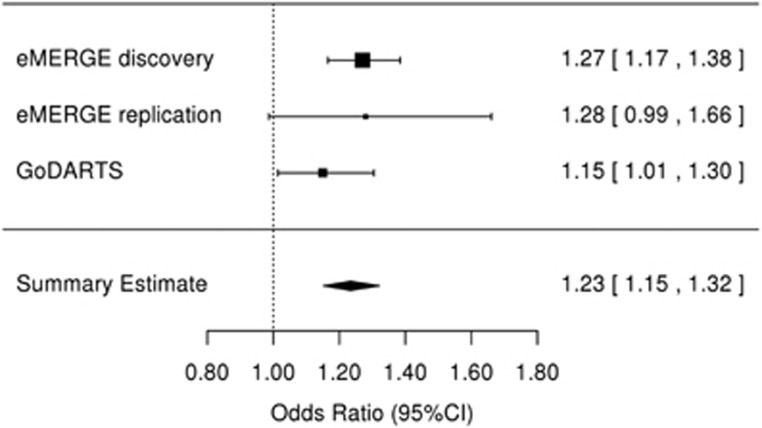
We sought replication for the SNPs in KCNIP4 with an association P-value <5 × 10−6 in two independent data sets (Supplementary Tables S6 and S7). Of the six SNPs identified in the discovery cohort, five were available in each of the final QC'ed eMERGE and GoDARTS replication sets. In both replication sets, the MAFs and association statistics were similar to those observed in the European ancestry subjects in the discovery population (Table 2). The most significantly associated SNP from the discovery analysis (rs145489027) did not reach statistical significance in either replication population (Table 2). One SNP (rs7675300) was significantly associated with ACEi cough in the eMERGE set, (OR=1.32 (1.01–1.70), P=0.04). Two SNPs were significantly associated in the GoDARTS set: rs16870989 (OR=1.15 (1.01–1.30), P=0.03) and rs1495509 (OR=1.15 (1.01–1.30), P=0.03). In a meta-analysis performed using both replication sets, three of the four SNPs present in both sets had an association at P<0.05 (rs16870989: OR=1.17 (1.03–1.34), P=0.009; rs145489027: OR=1.15 (1.01–1.31), P=0.02); and rs1495509: OR=1.17 (1.03–1.34), P=0.009); Supplementary Table S8). In a meta-analysis across all discovery and replication populations, four of the most significant SNPs reached genome-wide significance (Table 2) with the non-imputed SNP rs1495509 (highlighted in Figure 2), showing the strongest association (Figure 3). The combined association statistic across the replication sets at this SNP was (OR=1.23 (1.15–1.32), P=1.9 × 10−9) and did not demonstrate significant heterogeneity across studies (P=0.42).
Figure 3.
Forest plot for the association between angiotensin-converting enzyme inhibitor cough and KCNIP4 intronic single-nucleotide polymorphism rs1495509 for the discovery (eMERGE (Electronic Medical Records and Genomics) Network) and replication (eMERGE and GoDARTS (Genetics of Diabetes Audit and Research in Tayside, Scotland)) data sets. There was no heterogeneity across studies (P=0.43). CI, confidence interval.
Discussion
We describe the first large GWAS investigating SNP variants associated with clinically diagnosed ACEi-associated cough. All phenotype and genotype data for this study were derived from clinical research settings that incorporate an EMR data system. We found significant associations in a set of intronic SNPs located within the gene KCNIP4. Several of these significant associations were independently replicated in two European ancestry populations. In summary, these analyses identify a novel candidate gene that may have a role in this common adverse reaction.
The SNPs showing the strongest association with the cough phenotype are located exclusively in intron 4 of KCNIP4. KCNIP4 is a member of the KChIP family of EF hand (helix-loop-helix)-containing calcium-binding proteins. A major function attributed to KCNIP4 is the regulation of Kv4 potassium channels, which are significant contributors to action potential activity in neurons and cardiac myocytes.42 The amino terminus of the KCNIP4 gene product undergoes extensive alternative splicing and at least six splice variants have been described.43 Of note, several of these splice variants remove the flanking exons surrounding the intronic region identified in these analyses. Alternative splicing has been shown to have functional significance and can result in modulation of Kv4 channel functions and their subcellular location.42, 44 KCNIP4 isoforms are predominantly expressed in neuronal structures in the brain and spinal cord, though some isoforms are found in the kidney, stomach and small intestine.43 Little-to-no expression of the gene has been observed in lung extracts.45, 46
The most strongly associated SNPs are located within a single intron of KCNIP4. These SNPs are not in linkage disequilibrium with an amino-acid changing variant that would alter the primary structure of the protein. Hence, it is more likely that these SNPs have a regulatory role likely related to mRNA splicing or expression. A leading hypothesized mechanism of the ACEi-induced cough is stimulation of sensory nerve afferents within the lung resulting from the accumulation of inflammatory mediators that are normally cleaved by the ACE enzyme.47 This hypothesis has served as the basis for candidate gene studies that have focused on variation in inflammatory pathways within the lung. Our results would suggest that the important source of this variation may be directly related to the sensory nerves themselves, as KCNIP4 has been found to be in both central and peripheral neuronal structures. Indeed, if KCNIP4 expression in the lung were restricted to sensory nerves, its protein and mRNA levels would be expected to be low in samples derived from lung whole-cell homogenates, especially if samples were taken from the lung periphery. This restricted expression pattern could account for why KCNIP4 has not been detected in the lung. In further support of a role for KCNIP4 in lung physiology, a GWAS in mice identified an association between KCNIP4 and airway hyper-responsiveness, which was confirmed in studies of human asthma and airway hyper-reactivity.48
Epidemiological studies have identified several factors associated with an increased risk of ACEi-induced cough, including sex and race. In particular, the prevalence of cough is higher among women and east Asian populations.3, 4 In our replication set, the allele frequencies and effect sizes in subjects of European ancestry were generally comparable to those observed in the discovery sets. The ORs were weaker in the GoDARTS replication set, which is likely attributable to the lower positive predictive value and specificity of the case definition (switching from an ACEi medication to an angiotensin receptor blocker) used in this cohort.49, 50 Among African Americans, the associated alleles had lower MAFs. Although the association statistics trended in the same direction, they did not reach statistical significance. However, there was only 5% power to replicate the association in this group owing to small numbers of subjects and lower allele frequencies. Analyses across races suggest that this genomic region around KCNIP4 might be a trans-population risk factor and allele frequencies may contribute to prevalence differences in ACEi-induced cough. However, a comparison of the MAFs for the most significantly associated SNPs among HapMap European and Asian populations show that the minor alleles are very similar in these groups. Hence, racial differences in the frequencies of these SNPs would not account for the prevalence differences between these populations and would suggest that other independent or modifying genetic factors may be contributing to racial differences.
There are several limitations to this study. Cases and controls were identified using the EMR data sources. A limitation of EMR data is that data collection is not systematic and can be incomplete. These limitations can lead to both differential and nondifferential misclassification, which can skew or weaken associations. For instance, subjects who had been switched from ACEi therapy due to cough prior to the time period captured by the EMR could be inappropriately assigned to the control group. In addition, information pertaining to ACEi dosage, treatment duration and indication could not be systematically extracted for analysis. Hence, the contributions of these factors to ACEi cough could not be evaluated. We were also unable to incorporate elements of ACEi cough phenotype definitions that have been used in some studies, such as evaluating that effect of cessation of an ACEi on cough at a fixed time interval,18 as these protocols are not standard in clinical practice. The study also had limited power to detect associations for SNPs with an MAF<10%. Hence, the contribution of low frequency variants to the phenotype could not be quantified. Genotyping was performed using a number of commercial SNP-genotyping platforms. Imputation across multiple genotyping platforms can give rise to systematic frequency differences, which can lead to inflated type I error in GWAS studies.51 This problem is exacerbated when there are case and control imbalances across platforms. To attenuate this bias, the genotype data in the discovery set were imputed from an intersection of SNPs across each platform being evaluated, an approach that has been shown to decrease type I error rates.52 We did not demonstrate a functional role for KCNIP4 in the cough phenotype. Hence, it is possible the SNPs we identified are involved in the regulation of a nearby gene unrelated to KCNIP4, as has been reported for a number of other SNPs located within or in close proximity of a gene.53, 54 Epidemiological studies have observed higher rates of ACEi-induced cough in some Asian populations. This study did not assess the risks of the candidate SNPs in this population.
In conclusion, we used a GWAS to identify SNP variants associated with ACEi-induced cough using data derived from eMERGE, a network of medical centers that utilize electronic medical records as a tool for genomic research. We identified SNPs in the intron of the gene KCNIP4 as potential candidates for this adverse reaction. The mechanisms by which KCNIP4 may contribute to the cough are not known, and functional studies are needed to further elucidate the pathophysiological mechanisms.
Acknowledgments
This work was supported by the VUMC Clinical Pharmacology Training grant (T32 GM07569), the electronic MEdical Records and GEnomics (eMERGE) Network U01-HG-04603 (Vanderbilt), 1U02HG004608-01, 1U01HG006389 and NCATS/NIH grant UL1TR000427 (Marshfield/EIRH/Penn State), U01HG006375 and U01AG06781 (Group Health and University of Washington), U01-HG04599 (Mayo Clinic), U01-HG004609 (Northwestern University), U01HG006382 (Geisinger), an ARRA grant RC2 GM092618, R01 LM 010685, a Vanderbilt PGRN grant U19 HL065962 and the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH. At Geisinger, the sample collection was supported by NIH (P30DK072488, R01DK088231 and R01DK091601), Pennsylvania Commonwealth Universal Research Enhancement Program, the Ben Franklin Technology Development Fund of PA, the Geisinger Clinical Research Fund and a Grant-In-Aid from the American Heart Association. GoDARTS was funded by the Wellcome Trust (084727/Z/08/Z, 085475/Z/08/Z, 085475/B/08/Z) and as part of the EU IMI-SUMMIT program.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Zaman MA, Oparil S, Calhoun DA. Drugs targeting the renin-angiotensin-aldosterone system. Nat Rev Drug Discov 2002; 1: 621–636. [DOI] [PubMed] [Google Scholar]
- Overlack A. ACE inhibitor-induced cough and bronchospasm. Incidence, mechanisms and management. Drug Saf 1996; 15: 72–78. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Tsai JC. Angiotensin-converting enzyme gene insertion/deletion, not bradykinin B2 receptor -58 T/C gene polymorphism, associated with angiotensin-converting enzyme inhibitor-related cough in Chinese female patients with non-insulin-dependent diabetes mellitus. Metab Clin Exp 2001; 50: 1346–1350. [DOI] [PubMed] [Google Scholar]
- Woo KS, Nicholls MG. High prevalence of persistent cough with angiotensin converting enzyme inhibitors in Chinese. Br J Clin Pharmacol 1995; 40: 141–144. [PMC free article] [PubMed] [Google Scholar]
- Wood R. Bronchospasm and cough as adverse reactions to the ACE inhibitors captopril, enalapril and lisinopril. A controlled retrospective cohort study. Br J Clin Pharmacol 1995; 39: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KE, Ong FS, Blackwell W-LB, Shah KH, Giani JF, Gonzalez-Villalobos RA et al. A modern understanding of the traditional and nontraditional biological functions of angiotensin-converting enzyme. Pharmacol Rev 2013; 65: 1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AJ, Lalloo UG, Belvisi MG, Bernareggi M, Chung KF, Barnes PJ. Bradykinin-evoked sensitization of airway sensory nerves: a mechanism for ACE-inhibitor cough. Nat Med 1996; 2: 814–817. [DOI] [PubMed] [Google Scholar]
- Furuya K, Yamaguchi E, Hirabayashi T, Itoh A, Hizawa N, Ohnuma N et al. Angiotensin-I-converting enzyme gene polymorphism and susceptibility to cough. Lancet 1994; 343: 354. [DOI] [PubMed] [Google Scholar]
- Mukae S, Itoh S, Aoki S, Iwata T, Nishio K, Sato R et al. Association of polymorphisms of the renin-angiotensin system and bradykinin B2 receptor with ACE-inhibitor-related cough. J Hum Hypertens 2002; 16: 857–863. [DOI] [PubMed] [Google Scholar]
- Woo SW, Bang S, Chung MW, Jin SK, Kim YS, Lee SH. Lack of association between ACE and bradykinin B2 receptor gene polymorphisms and ACE inhibitor-induced coughing in hypertensive Koreans. J Clin Pharm Ther 2009; 34: 561–567. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Yamaguchi E, Furuya K, Kawakami Y. The ACE gene polymorphism and cough threshold for capsaicin after cilazapril usage. Respir Med 2001; 95: 130–135. [DOI] [PubMed] [Google Scholar]
- Li Y-F, Zhu X-M, Liu F, Xiao C-S, Bian Y-F, Li H et al. Angiotensin-converting enzyme (ACE) gene insertion/deletion polymorphism and ACE inhibitor-related cough: a meta-analysis. PLoS One 2012; 7: e37396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilo A, Sáez-Rosas MP, Santos-Morano J, Sánchez E, Moreno-Rey C, Real LM et al. Identification of genetic factors associated with susceptibility to angiotensin-converting enzyme inhibitors-induced cough. Pharmacogenet Genomics 2011; 21: 10–17. [DOI] [PubMed] [Google Scholar]
- Mas S, Gassò P, Alvarez S, Ortiz J, Sotoca JM, Francino A et al. Pharmacogenetic predictors of angiotensin-converting enzyme inhibitor-induced cough: the role of ACE, ABO, and BDKRB2 genes. Pharmacogenet Genomics 2011; 21: 531–538. [DOI] [PubMed] [Google Scholar]
- Mukae S, Aoki S, Itoh S, Iwata T, Ueda H, Katagiri T. Bradykinin B(2) receptor gene polymorphism is associated with angiotensin-converting enzyme inhibitor-related cough. Hypertension 2000; 36: 127–131. [DOI] [PubMed] [Google Scholar]
- Kim T-B, Oh S-Y, Park H-K, Jeon S-G, Chang Y-S, Lee K-Y et al. Polymorphisms in the neurokinin-2 receptor gene are associated with angiotensin-converting enzyme inhibitor-induced cough. J Clin Pharm Ther 2009; 34: 457–464. [DOI] [PubMed] [Google Scholar]
- Mahmoudpour SH, Leusink M, Putten L, van der, Terreehorst I, Asselbergs FW, de Boer A et al. Pharmacogenetics of ACE inhibitor-induced angioedema and cough: a systematic review and meta-analysis. Pharmacogenomics 2013; 14: 249–260. [DOI] [PubMed] [Google Scholar]
- Zee RY, Rao VS, Paster RZ, Sweet CS, Lindpaintner K. Three candidate genes and angiotensin-converting enzyme inhibitor-related cough: a pharmacogenetic analysis. Hypertension 1998; 31: 925–928. [DOI] [PubMed] [Google Scholar]
- Terao C, Bayoumi N, McKenzie CA, Zelenika D, Muro S, Mishima M et al. Quantitative variation in plasma angiotensin-I converting enzyme activity shows allelic heterogeneity in the ABO blood group locus. Ann Hum Genet 2013; 77: 465–471. [DOI] [PubMed] [Google Scholar]
- Luo J-Q, He F-Z, Luo Z-Y, Wen J-G, Wang L-Y, Sun N-L et al. Rs495828 polymorphism of the ABO gene is a predictor of enalapril-induced cough in Chinese patients with essential hypertension. Pharmacogenet Genomics 2014; 24: 306–313. [DOI] [PubMed] [Google Scholar]
- Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 2013; 15: 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG et al. eMERGEing progress in genomics-the first seven years. Front Genet 2014; 5: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowton E, Field JR, Wang S, Schildcrout JS, Van Driest SL, Delaney JT et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med 2014; 6: 234cm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L, Ritchie MD, Brown-Gentry K, Pulley JM, Basford M, Denny JC et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med 2010; 12: 648–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011; 3: 79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel RA. The International Classification of Disease. Two hundred years of development. Public Health Rep 1978; 93: 150–152. [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shah A, Jiang M, Peterson NB, Dai Q, Aldrich MC et al. A study of transportability of an existing smoking status detection module across institutions. AMIA Annu Symp Proc 2012; 2012: 577–586. [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 2013; 10: 5–6. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet 2006; 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol 2011; 35: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SS, de Andrade M, Tromp G, Kuivaniemi H, Pugh E, Namjou B et al. Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet 2014; 5: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010; 11: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw 2010; 36: 1–48. [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84: 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Misonou H, Campomanes CR, Anderson AE, Schrader LA, Doliveira LC et al. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J Biol Chem 2003; 278: 36445–36454. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Timmusk T. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics 2005; 86: 581–593. [DOI] [PubMed] [Google Scholar]
- Massone S, Vassallo I, Castelnuovo M, Fiorino G, Gatta E, Robello M et al. RNA polymerase III drives alternative splicing of the potassium channel-interacting protein contributing to brain complexity and neurodegeneration. J Cell Biol 2011; 193: 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Kovacs I, Zhang Z. Differential distribution of KChIPs mRNAs in adult mouse brain. Brain Res Mol Brain Res 2004; 128: 103–111. [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Hatano N, Ohya S, Takikawa R, Watabiki T, Takasugi N et al. Molecular cloning and characterization of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit Kv4. J Biol Chem 2002; 277: 14965–14975. [DOI] [PubMed] [Google Scholar]
- Takahama K, Araki T, Fuchikami J, Kohjimoto Y, Miyata T. Studies on the magnitude and the mechanism of cough potentiation by angiotensin-converting enzyme inhibitors in guinea-pigs: involvement of bradykinin in the potentiation. J Pharm Pharmacol 1996; 48: 1027–1033. [DOI] [PubMed] [Google Scholar]
- Himes BE, Sheppard K, Berndt A, Leme AS, Myers RA, Gignoux CR et al. Integration of mouse and human genome-wide association data identifies KCNIP4 as an asthma gene. PLoS One 2013; 8: e56179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojczynski MK, Tiwari HK. Definition of phenotype. Adv Genet 2008; 60: 75–105. [DOI] [PubMed] [Google Scholar]
- Rice JP, Saccone NL, Rasmussen E. Definition of the phenotype. Adv Genet 2001; 42: 69–76. [DOI] [PubMed] [Google Scholar]
- Sinnott JA, Kraft P. Artifact due to differential error when cases and controls are imputed from different platforms. Hum Genet 2012; 131: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EO, Hancock DB, Levy JL, Gaddis NC, Saccone NL, Bierut LJ et al. Imputation across genotyping arrays for genome-wide association studies: assessment of bias and a correction strategy. Hum Genet 2013; 132: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 2014; 507: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJG, Klous P et al. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Invest 2014; 124: 1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.