Abstract
Spinal muscular atrophy is an autosomal-recessive pediatric neurodegenerative disease characterized by loss of spinal motor neurons. It is caused by mutation in the survival of motor neuron 1 gene (SMN1) leading to loss of function of the full-length SMN protein. SMN has a number of functions in neurons, including RNA-splicing and snRNP-biogenesis in the nucleus, and RNA-trafficking in neurites. The expression level of full-length SMN protein from the SMN2 locus modifies disease severity. Increasing full-length SMN protein by a small amount can lead to significant improvements in the neurological phenotype. Currently available interventions for spinal muscular atrophy my patients are physical therapy, orthopedic, nutritional, and pulmonary interventions; these are palliative or supportive measures and do not address the etiology of the disease. In the last decade there has been a push for developing therapeutics to improve motor phenotypes and increase lifespan of spinal muscular atrophy patients. These therapies are aimed primarily at restoration of full-length SMN protein levels; but other neuroprotective treatments have been investigated as well. Here, we discuss recent advances in basic and clinical studies towards finding safe and effective treatments of spinal muscular atrophy using gene therapy, antisense oligonucleotides, and other small molecule modulators of SMN expression.
Keywords: spinal muscular atrophy, antisense oligonucleotides, gene therapy, clinical trials
Introduction
Spinal muscular atrophy (SMA) is a severe neurodegenerative disease that results in degeneration and cell death of lower motor neurons in the spinal cord and is the leading genetic cause of infant mortality. It is an autosomal-recessive disorder with an incidence of 1:11,000 live births.1 SMA is caused by homozygous deletions or mutations involving the survival of motor neuron 1 gene (SMN1) that encodes the SMN protein.2 There are five clinical types of SMA defined by disease severity and age of onset, although the various types represent a continuum of severity with indistinct borders.3 Type I SMA, Werdnig-Hoffmann disease, is a severe infantile form diagnosed prior to six months of age with mortality occurring before two years of age. Type I patients are severely hypotonic, have denervation of muscles, and are never able sit up unaided. Type II SMA, Dubowitz disease, diagnosed between 6–18 months of age is less severe; patients can progress to sitting unsupported at some point, but they never gain the ability to walk unassisted and have a shortened lifespan. Type III SMA, Kugelberg–Welander disease, is diagnosed after 18 months of age; patients are able to stand and walk unaided, and have an approximately normal lifespan. Type 0 and type IV SMA are rare variants. Type 0, also referred to as type IA, is the most severe form of SMA; it is characterized by prenatal onset as determined by reduced fetal movement and arthrogryposis at birth, and mortality before 6 months of age.4 Type IV SMA is similar to type III, but with onset in adulthood and milder symptoms.
Humans possess an additional gene, SMN2, which is almost identical to SMN1 but produces only 10% of full-length SMN protein.5 SMN2 has a C-T substitution in exon 7 that leads to exclusion of exon 7 and formation of a truncated, unstable protein (SMNΔ7) that is rapidly degraded.5–7 In SMA patients, SMN2 is the primary modifier of the disease phenotype; the number of copies of SMN2 a patient carries is inversely correlated with disease severity. SMA type I patients typically carry only 1–2 copies of SMN2, while SMA type II and III patients carry between two and four copies of SMN2.8
To date there is no U.S. Food and Drug Association (FDA)-approved treatment for SMA, and only palliative therapies are available to patients. In recent years, several promising treatments have entered advanced clinical trials with promising results (Table 1). The therapeutic aim in the majority of these trials is to increase the amount of functional full-length SMN protein expressed, either by use of antisense oligonucleotide (ASO)–mediated exon 7 inclusion at the SMN2 locus or by exogenous expression of SMN1 by viral-mediated gene therapy vectors. Additionally, a number of other therapies that modulate SMN levels or function are being investigated. Here we describe current progress in cell culture, mouse models, and clinical trials towards an effective SMA treatment.
Table 1.
Current Open Clinical Trials for SMA Therapies. Open clinical trials enrolling patients as published on clinicaltrials.gov as of January 5th 2015.
| Treatment | SMA patients | Identifier/phase | Study design | Primary outcome measures |
|---|---|---|---|---|
| ISIS-SMN-Rx | Type I infants |
NCT02193074 Phase III |
Randomized, double-blind, sham- controlled safety and efficacy study Intrathecal administration |
Time to death or permanent ventilation |
| ISIS-SMN-Rx | Type II/III |
NCT02292537 Phase III |
Randomized, double-blind, sham- controlled safety and efficacy study Intrathecal administration |
Change from baseline HFMS |
| scAAV9.CB.SMN | Type I infants |
NCT02122952 Phase I |
Non-random, open label safety and efficacy study IV delivery escalating viral dose in three cohorts |
Grade three of higher treatment toxicity, mortality |
| LMI070 | Type I infants |
NCT02268552 Phase II |
Open-label multi-part safety and efficacy study Oral administration of escalating doses in three cohorts |
Change from baseline in physical exam, blood chemistry, urinalysis, vital signs, and cardiac function |
| RO6885247 | Type I/II/III |
NCT02240355 Phase I |
Randomized, double-blind, placebo- controlled, safety and efficacy study Oral administration |
Incidence of adverse evens (AEs) |
| Pyridostigmine bromide | Type III adults |
NCT02227823 Phase II |
Non-random, open-label Oral delivery of anti-cholinesterase for patients with abnormal NMJs |
Change in baseline distance in 6 minute walk test |
| 4-AP (dalfampridine) | Type III |
NCT01645787 PhaseII/III |
Randomized, double blind, placebo- controlled Oral administration |
Change in six-minute walk test Kinematic gait evaluation |
| Biomarker identification | Type I infants | NCT01736553 | Prospective observational study of SMA infants vs. healthy controls | SMN mRNA, SMN protein plasma proteins, CMAP, EIM, motor function |
Multifaceted functions of SMN
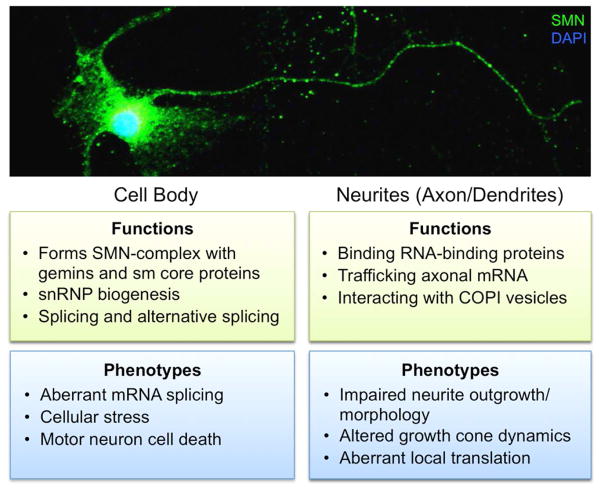
SMN protein is ubiquitously expressed very early in development and plays diverse roles in RNA processing and trafficking as summarized in Figure 1. In the cell body, SMN protein forms the SMN complex with eight other proteins: Gemin2–8 and unr-interacting protein (UNRIP) (reviewed in Ref. 9). This complex is involved in biogenesis of small nuclear ribonucleoproteins (snRNPs) via recruitment and binding of the seven Sm protein core to snRNA.10 SMN complex binding of Sm proteins confers specificity and efficiency of the Sm proteins to accurately bind specific snRNAs.11 SMN bound snRNPs localize to Cajal bodies where they are incorporated into the spliceosomal machinery. The SMN complex is also integral to U7 snRNP function and histone mRNA processing via interaction with a different subset of Sm proteins.12, 13 Furthermore, there is evidence that SMN expression levels are regulated in part by components of the spliceosome. UA1 protein, a component of the U1 snRNP complex, regulates SMN expression by binding to the 3′-UTR of SMN mRNA to inhibit polyadenylation and decrease SMN protein levels.14
Figure 1.
Subcellular localization of SMN function in motor neurons. The top image shows immunofluorescent staining of SMN protein throughout the cell body and processes of a primary rodent spinal motor neuron in culture. SMN has a number of different functions throughout the motor neuron cell body axon/neurites. Loss of SMN leads to perturbations in these cellular functions and therefore specific SMA phenotypes are associated with these subcellular functions.
An additional role for SMN was first reported by Bassell and colleagues, who described the presence and trafficking of SMN protein, in association with Gemin2 or Gemin3, in motor neuron axons and dendrites.15 In neuronal processes, SMN regulates RNA via direct interaction with several RNA binding proteins including FMRP,16 KSRP,17 HuD,18–20 and Imp1.21 Loss of SMN in neurons affects trafficking of SMN binding partners and target mRNAs.22 This leads to changes in axon and dendrite outgrowth and morphology, as well as aberrant growth cone dynamics and disrupted local translation of a number of axonal RNAs.18, 23, 24 In motor neuron axons, SMN-containing granules interact with the coatomer complex of coat protein I (COPI) vesicle family member α-COP, linking SMN function to the Golgi network.25 Disruption of this interaction by either knockdown of α-COP or inhibition of Golgi-mediated granule secretion decreases SMN in the cellular processes resulting in aberrant neurite outgrowth and morphology.26, 27 Further, in vitro evidence suggests SMN may associate with polyribosomes in the cytoplasm and act as a translational repressor in RNA granules.28 The relative contribution of each distinct role for SMA throughout the nucleus, cell body, and processes of motor neurons to the SMA phenotype is not yet known. However, understanding SMN protein functions and mechanisms of action in a subcellular context could shed light on additional pathways for therapeutic intervention and biomarkers.
Interestingly, aberrant RNA splicing and metabolism is common to other motor neuron diseases including amyotrophic lateral sclerosis (ALS). New evidence of a direct interaction between the SMN complex and FUS, an RNA binding protein mutated in familial ALS, is a functional link between the two disorders. Loss of nuclear FUS due to ALS-associated mutations leads to decreased levels of nuclear gems and cytoplasmic localization of SMN.29 As a result, snRNPs are unable to reenter the nucleus, thus leading to aberrant spliceosomal function.30 In ALS neurons, localization of SMN within cytoplasmic FUS granules leads to axonal SMN reduction, which can be rescued by SMN overexpression.31 Depletion of another ALS-associated protein, TDP-43, can increase SMN alternative splicing and decrease its expression,32 and may contribute to U12 minor spliceosome function.33 Overexpression of SMN in a mutant SOD1 mouse model of ALS alleviated motor deficits and inhibited neuron death, providing further evidence for SMN as a modifier of ALS phenotypes.34 Interactions between ALS and SMA disease pathways are indicative of common molecular mechanisms in motor neuron loss and may inform preclinical studies for other degenerative motor neuron diseases.
Models for development of SMA therapeutics
Mouse models
Complete loss of SMN expression is lethal very early in gestation,35 and therefore Smn−/− mice are not a viable model to study SMA disease progression and pathology. While humans with SMA have at least one copy of the disease-modifying SMN2, rodents do not carry the SMN2 gene. As a result, mouse models of SMA often include human SMN2 transgenes or modifications to the mouse Smn to mimic SMN2 exon 7 splicing. To date, a number of mouse models of SMA have been described that cover the spectrum of disease phenotypes from severe infantile onset to milder type III (reviewed in36). Of these models, two have emerged as the most widely used for studies of SMA therapeutics. The first, referred to as the Taiwanese model (Smnhung−/−;SMN2Hungtg/−) expresses a transgene with two copies of SMN2 and can be bred with Smn heterozygotes to obtain a range of phenotypes.37 The second model, SMNΔ7 (Smn−/−;SMN2tg/tg;SMNΔ7tg/tg), is homozygous for human SMN2 and SMNΔ7 and exhibits a severe motor phenotype, significant motor neuron loss, and mean lifespan of 15 days.38 These models allow for investigation of potential therapeutics with outcome measures ranging from increased lifespan and gross motor function improvement to neuromuscular junction (NMJ) morphology, motor neuron survival, and subcellular mechanistic readouts.
Invertebrate models
Invertebrate models of SMA are integral to understanding aberrant development, axon outgrowth, and splicing. Both flies and worms have SMN homologs that are roughly 30% similarity to the human SMN1 gene. C. elegans has been used in RNAi-based screens for smn modifier genes.39 Drosophila are genetically tractable, have a short lifecycle, and are a good system for investigating motor behaviors. The generation of an allelic series of Drosophila mutants with SMA patient–specific point mutations allowed in-depth investigation of point-mutation effects on SMN binding, function, and phenotype.40 RNAi-mediated muscle-, neuron-, or motor neuron–specific SMN loss in the fly also has elucidated cell type–specific effects on neuromuscular junction morphology and electrophysiology.41
In zebrafish, the SMA axonal phenotype is recapitulated by administration of an antisense morpholino that knocks down Smn1 mRNA expression.42 Studies investigating compounds that modulate axon growth in zebrafish have used this morpholino model and found that kinesins,43 Nrxn2,44 and molecules involved in ubiquitin homeostasis45 modify the Smn1 axon phenotype. Zebrafish genetic models of SMA also have severe motor axon defects and can be used for rescue experiments to look at temporal effects of Smn1 loss on motor axons. 46, 47
Large animal models
Large animal and primate models of SMA have been used extensively for bioavailability and safety studies, particularly for antisense oligonucleotide (ASO) and viral vector–mediated gene therapy development. However, they lack the SMN2 gene and therefore cannot be used for efficacy studies of SMN2 modulators. Pigs lack the intron splicing silencer in exon 7 and have modified splicing of SMN1, so they are not a good model for splice modulation therapies.48 However, Burghes and colleagues have developed a porcine model using intrathecal delivery of scAAV9-mediated SMN mRNA knockdown that recapitulates the SMA phenotype of motor neuron loss and axon degeneration, as determined by assessment of motor unit function with electromyography (EMG), compound motor action potential (CMAP), and motor unit number estimation (MUNE).49 Further, the phenotype is rescued by administration of human SMN1 via scAAV9-SMN, providing evidence that this is a very useful pre-clinical model for gene therapy approaches.
In vitro human SMA stem-cell model
Induced pluripotent stem cells (iPSCs) are increasingly used to model CNS diseases including Alzheimer’s disease, Parkinson’s disease, and ALS (reviewed in Ref. 50). iPSCs derived from SMA patient fibroblasts can be differentiated into neural lineage cells and have low SMN expression and decreased nuclear gems.51 iPSCs, and the motor neurons derived from them, are an in vitro model of SMN loss that can be used to study cellular phenotypes and potential treatment compounds.51, 52 Wild-type mouse53 and human54 motor neuron progenitors injected into the cerebrospinal fluid (CSF) of SMA mouse models differentiate into neurons and provide trophic support for endogenous motor neurons to increase survival. Gene editing is used to correct mutations in iPSCs to directly compare the effect of a specific mutation in otherwise genetically identical cell lines. Correction of the mutation in SMA iPSC lines increases SMN expression and decreases motor neuron cell death in vitro, and injection in SMNΔ7 mice increases lifespan by forty percent.55
Antisense oligonucleotides
ASO technology and therapeutic approach
ASOs are single-strand, DNA-like molecules 15–35 nucleotides long that can modulate target gene expression by hybridization to the pre-mRNA. Targets can be regulated by several different mechanisms including restoration of the correct open reading frame, decreasing gene expression, alternative splicing, or, in the case of SMA, exon inclusion. ASOs are being developed as therapeutics for a number of rare genetic diseases caused by aberrant mRNA splicing, including Duchenne muscular dystrophy, fibrodysplasia ossificans progressiva, Leber congenitial amaurosis, and tauopathies.56 While a growing number of ASOs are being tested in clinical trials, to date only two have been FDA-approved, both reduce target gene expression: fomivirsen sodium for cytomegalovirus retinitis in immune-compromised AIDS patients and mipomersen sodium for familial hypercholesterolemia.57
The pharmaceutical properties of ASOs can be altered by the chemical structure of the nucleotide backbone or sugar ring. Each modification of the backbone confers slightly different stability, toxicity, and function to the ASO. In addition, bifunctional ASOs have added high-affinity splice enhancer sequences that promote binding of SR proteins and increase splicing.58–60 One ASO modification extensively used in the clinical application to SMA for SMN2 alternative splicing is 2′-OME, which provides enhanced target RNA binding affinity and resistance to endonuclease cleavage conferring stability to the ASO.61, 62
SMN is an attractive candidate for ASO therapy as patients have a functional copy of SMN2 that is an ideal target for modulation. As compared to SMN1, a single C-T substitution in exon7 of SMN2 weakens a binding site for the splicing activator SRSF1 leading to exon skipping and a 90% decrease in full-length SMN protein. Early studies of ASOs in SMA patient fibroblasts proved that they could be used to promote the inclusion of SMN2 exon 7 (Ref. 63) and identified the intronic splicing silencer N1 (ISS-N1) site in SMN2 exon 7 as a particularly potent target for ASO binding. Adjacent to the exon 7 5′-splice site, ISS-N1 contains a binding site for the A1-dependent ISS hnRNP-A1, which decreases exon 7 inclusion. ASOs targeted to the ISS-N1 sequence competitively bind the mRNA and prevent repressor binding by hnRNP to increase exon 7–containing SMN2 expression, in both in vitro and in vivo in mouse models of SMA.64–66
Development of ASO for clinical trials in SMA
Preclinical studies of the specific oligonucleotide ISIS 396442 have shown that either peripheral67 or intraventricular (ICV) injection of this ASO into either mouse model (SMNΔ7 or Taiwanese) results in stable in vivo rescue of full-length SMN in the spinal cord.68, 69 In utero ICV injection at E15.5 and postnatal injection partially rescued ear and tail necrosis in a milder SMA type III mouse model.68 Intrathecal infusion of the ASO into cynomolgus monkeys was sufficient for ASO distribution at therapeutic levels throughout the spinal cord.69 In comparison to the other chemically-modified ASOs, ISIS 396443 not only is more potent, but also yields more efficient SMN2 splicing correction throughout the CNS, both in the spinal cord and brain. Furthermore, administration of a single intrathecal bolus injection leads to better distribution and accumulation than infusion, which has important clinical significance for dosing.70 These studies laid the foundation for clinical trials of ASO 396643 (also known as ISIS-SMNRx). In a Phase I clinical trial, ISIS-SMNRx was administered by intrathecal injection into type II/III SMA patients and seemed well tolerated (NCT01494701). In a Phase II trial with type II/III SMA patients, multiple injections were well tolerated, and patients showed improvements in the Expanded Hammersmith Functional Motor Scale (HFMSE) scores at all doses (NCT01703988). Two double blind sham-controlled Phase III clinical trials are currently enrolling infants with type I (NCT02193074) and children with type II (NCT02292537) for intrathecal administration of ISIS-SMNRx, and will run through 2017 (Table 1).
Other ASOs with different chemistries and binding sites are also being developed and tested in clinical trials. Short 8-mer ASOs were developed to target a GC-rich site adjacent to the ISS-NI.66, 71 The 8-mer 3UP8i has fewer off-target effects than the longer ASOs, significantly increases inclusion of SMN2 exon 7, and ameliorates neuromuscular junction (NMJ) pathology and increases survival in a mild SMA mouse model.72 ICV injection of an E1 intronic-repressor-binding ASO, E1MO-ASO, in the SMNΔ7 mouse model was sufficient to induce SMN expression in the spinal cord and brain, leading to increased weight, longer lifespan, and improved motor behaviors.73 A screen for orally-available, small-molecule SMN splicing modifiers in SMA patient fibroblasts led to the discovery of three small molecules: SMN-C1, SMN-C2, and SMN-C3,74 which are associated with a dose-dependent increase in SMN protein following oral treatment in the SMNC/C and SMNΔ7 mouse models. Orally-available molecules are now being tested in a Phase I clinical trial for safety and tolerability of RO6885247 (NCT02240355). Due to the invasive nature of other therapeutic strategies that rely on intravenous and intrathecal injections, an effective orally-available molecule could be beneficial for patient safety and dosing.
Gene therapy
Viral vectors for exogenous expression of gene targets
One main approach to mitigating the effects of SMN loss is exogenous expression of full-length SMN protein by introducing genetic material directly into motor neurons. The gene of interest, SMN1 in the case of SMA, is packaged into a carrier vector that is usually an attenuated or modified viral vector. For CNS targeting, lentiviral (LV) or adeno-associated viral (AAV) vectors are most common as they are able to transduce non-dividing cells like neurons (reviewed in Refs 75 and 76, respectively). Additionally, non-viral techniques use direct injection of genetic material or rely on cationic lipids or polymers to facilitate entry into the cells.77 A number of different therapeutic strategies have been investigated for development of in vivo gene therapy vectors for expression of full-length SMN.
The earliest promising SMA gene therapy efforts used intramuscular administration of LV vectors that were retrogradely transferred to the spinal cord and spinal motor neurons to demonstrate phenotypic improvement in SMNΔ7 mice.78 However, more recently AAV gene therapy vectors have been developed for the treatment of a number of monogenic CNS disorders. Because of their long-term gene expression, efficient transduction of non-dividing target cells without integrating into the host genome, and limited immunogenic potential, AAV are advantageous.79 Specifically, the AAV9 serotype has emerged as a frontrunner in CNS gene therapy trials because it can cross the BBB,80 and thus it can be administered intravenously.
Gene therapy development in SMA
SMA model mouse studies from several different labs have shown that systemic administration of scAAV9-SMN not only leads to widespread motor neuron transduction and SMN expression,81 but significantly rescues the SMNΔ7 phenotype and increases life span.82–84 Direct administration of scAAV9-SMN to the CNS via ICV or intrathecal injection also rescues the SMNΔ7 model and significantly increases lifespan.85, 86 Injection of scAAV9-SMN into the CSF is more invasive and carries additional risks, but requires a significantly lower dose of viral vector to achieve high levels of motor neuron transduction in both pig and primate models.87, 88 In addition to AAV9, several studies have investigated other viral vectors. Administration of AAV8-SMN directly into the CNS by intraventricular injection also improved NMJ morphology and muscle function of SMNΔ7 mice and increased life span.85 New lentiviral vectors that are integration-deficient are being developed, which are safer and more efficient in in vitro assays; however, they require extensive testing.89
Begun in 2014, the first Phase I clinical trial for gene therapy in SMA (NCT02122952) will evaluate the safety and efficacy of escalating doses of scAAV9.CB.SMN, by intravenous infusion, of three cohorts of SMA type I infants. The study is currently recruiting participants and is expected to finish data collection in mid-2017. A number of other AAV-SMN vectors are in pre-clinical development and, hopefully, will be advanced for clinical trials in the coming years.
Modulatory therapeutic strategies
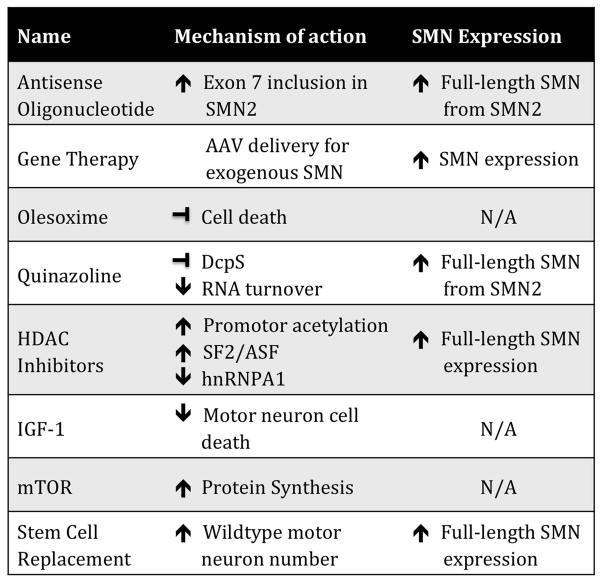
While directly increasing full-length SMN expression has a significant impact on ameliorating the SMA phenotype, many therapeutic approaches do not directly target SMN expression (Fig. 2). Instead, these therapies are aimed at modulating molecules and cellular pathways involved, such as neuroprotective mechanisms, cell survival, gene expression, and axon outgrowth. A number of drugs have been investigated (reviewed in Ref. 90); the following provides an overview of the current modulatory therapies in SMA trials.
Figure 2.
Mechanisms of action of SMA therapeutics. There are a number of different therapeutic strategies. SMA therapies not only directly increase the full-length SMN expression, but also indirectly modulate pathways that effect SMN expression, and act as neuroprotective compounds to inhibit motor neuron cell death and provide trophic support for surviving cells.
Olesoxime
Olesoxime (TRO19622) was originally identified in a small molecule screen for potential modifiers of motor neuron degeneration in ALS.91 It is a cholesterol-like molecule that acts on mitochondria to inhibit cell death, rescue neurite outgrowth,91 and prevent astrogliosis.92 Additionally, olesoxime treatment has been shown to accelerate myelination in response to injury.93 Olesoxime entered a two-year Phase II/III clinical trial in SMA type II/III patients that ended in October 2013 (NCT01302600). Two year oral dosing prevented loss of motor function on the Motor Function Measure Scale (MFMS) and disease-related adverse events, compared to the placebo control group.94 If approved by the FDA, olesoxime will be the first neuroprotective treatment for SMA.
Quinazolines
A small-molecule screen for SMN2 transcriptional activators first identified the quinazoline family of compounds that increase SMN mRNA and protein as well as nuclear Cajal bodies in patient fibroblasts.95 The compound was manipulated to improve half-life, blood-brain barrier permeability, and SMN2 promoter activation for development in clinical trials.96 Oral treatment of neonatal SMNΔ7 mice with quinazoline compounds increased Smn promoter activity, ameliorated the neuromuscular phenotype, and increased lifespan by up to 30 percent.97 Quinazoline increases SMN levels by binding and inactivating DcpS, a decapping enzyme, to stabilize the mRNA.98 One quinazoline compound, RG3039, had good CNS distribution, was a potent inhibitor of DcpS, modestly increased Smn levels, and improved survival and motor function in multiple severe mouse models.99, 100 Additionally, RG3039 was shown to have a long half-life and increased not only the number of cells with nuclear gems, but also the number of gems per cell.100 RG3039 was well tolerated in a Phase 1 clinical trial for safety in healthy controls and clinical studies are ongoing.
Histone deacetylase inhibitors
Histone deacetylase (HDAC) inhibitors are a class of compounds that are epigenetic regulators of gene expression. In vitro studies of HDAC inhibitor treatment with valproic acid (VPA) or phenylbuterate (PB) in SMA patient-derived fibroblasts show a significant increase in full-length SMN levels.101–103 HDAC inhibitors traditionally increase transcriptional activation via hyperacetylation of a gene’s promoter. However, conflicting studies of SMN promoter acetylation state have shown that the mechanism could be more complex. RNAi studies of individual HDAC enzymes in vitro have shown that different enzymes affect SMN promoter activity differently.104 VPA treatment has also been shown to modulate SMN pre-RNA splicing by increasing expression of splicing factors SF2/ASF and decreasing hnRNPA1.105 Interestingly, SMN was recently reported to have a chromatin-binding ability, and may interact with methylated histone H3K79.106 Whether this function has implications for disease pathogenesis is not yet clear.
In a number of Phase I and Phase II clinical trials, VPA and PB have been tested for safety and efficacy in SMA type II/III patients. Increase in blood levels of full-length SMN, and patient motor benefits were moderate and inconsistent between studies (reviewed in Ref. 107). Reports of weight gain and loss of carnitine led to combined therapy of VPA with L-carnitine, but in multiple human trials there was no improvement with the combined treatment.108–110 Interestingly, heterogeneity in a cohort of 16 SMA patients correlated with increased expression of fatty acid translocase CD36, which repressed the response to VPA treatment in patient-derived IPS cells.111 In the future, CD36 may be a marker to identify patient populations that might receive the most benefit from VPA treatment.
Trichostatin A (TSA), LBH589 and suberoylanilide hydroxamic acid (SAHA) are potent inhibitors of a broader spectrum of HDACs112 and have been shown to significantly increase full-length SMN expression.112–115 Furthermore, TSA or SAHA treatment has been demonstrated to ameliorate motor deficits and increase lifespan in both the SMNΔ7114, 116 and the Taiwanese mouse models.117 Interestingly in SMN2b model mice, which lack the SMN2 transgene, TSA treatment is able to rescue the phenotype without directly modulating SMN expression.118 Additionally, two polyphenol compounds, curcumin and resveratrol, have HDAC inhibitor and neurotrophic properties that increase SMN2 gene expression119–121 and interact with SMN to affect neurite outgrowth.122 While the exact mechanism of action of these compounds is unclear, they are potential candidates for further exploration in mouse models and SMA clinical studies.
Additional pathways for drug targets and clinical intervention
Several studies have implicated the PTEN/mTOR/Akt pathway in SMA. Loss of PTEN expression by shRNA injection in SMNΔ7 mouse muscle rescues NMJ morphology, while systemic administration of an AAV9 PTEN knockdown construct increases life span and rescues motor function in the same mouse model.123 In the CNS, insulin-like growth factor 1 (IGF-1) plays an important role in normal brain development, axon outgrowth, and cell survival,124 making it an interesting target for SMA therapy. Previous studies using recombinant IGF-1 treatment (IPLEX)125 or AAV-IGF-1 administration to the CNS126 rescued motor neuron cell death but did not lead to significant increases in motor function or life span. Increases in mouse life span were only reported when IGF-1 was administered in combination with an ASO specific to SMN.127 Further, transgenic expression of mouse IGF-1 under a skeletal muscle–specific promoter in the SMNΔ7 mouse background led to an increase in life span and myofiber size, but did not ameliorate motor behavior, indicating muscle-specific rescue is not comprehensive.128 Interestingly, systemic delivery by intravenous injection of AAV1 expressing human IGF-1 at postnatal day 0 led to hepatic induction and increased serum levels of IGF-1; this reduced motor neuron death and increased motor function and lifespan into a severe mouse model of SMA.129 These results highlight not only the efficacy of IGF-1 in alleviating motor neuron pathology, but also the need for systemic IGF-1 expression for potential therapeutic value.
Our lab recently demonstrated that microRNA miR-183 is increased in SMA models and directly leads to altered expression of mTOR pathway components, as well as aberrant axonal local translation.23 Repressed mTOR activity and protein synthesis in SMN-deficient cells suggest that increasing full-length SMN transcript alone may not be sufficient because protein synthesis machinery may still be suppressed in SMA. If so, it may be necessary to “prime” the protein synthesis machinery by activating mTOR within critical therapeutic windows. These findings may have implications for therapeutic trial design in the future.
Other molecules that have shown promise in in vitro studies have been unsuccessful in clinical trials, including lithium130 and somatotropin.131 Such results highlight the differences between rodent and human cellular models, and the importance of clinical trials for confirming efficacy in human patients.
Therapeutic challenges
Therapeutic targeting: CNS or systemic?
Although SMN is ubiquitously expressed, spinal motor neurons are most susceptible to loss of full-length SMN protein; the specificity of this motor neuron degeneration remains an important, unanswered question. Notably, disruption of astrocytes,132 Schwann cells,133 and microglial activation in the spinal cord during motor neuron loss have also been reported.134 Ideally, therapeutic strategies would also target these other cell types.
A number of studies have described additional SMA pathology outside the CNS in SMA animal models and have highlighted the need for systemic or combination therapies in clinical trials. Targeted loss of SMN only in motor neurons recapitulates the SMA neuromuscular phenotype, however, the mice have lower mortality, and even improvement in some phenotypes with age.135 Conversely, motor neuron specific rescue of SMN expression in a severe SMA mouse model attenuates the motor phenotype and NMJ innervation, but does not improve lifespan due to disrupted autonomic innervation of the heart.136 Further, inducible SMN expression under the Emx1 promoter that expresses in cortical neurons also failed to extend lifespan.137 In fact, a number of cardiac abnormalities due to altered autonomic innervation have been reported in SMA mouse models, including heart failure and arrhythmia.138–140 In humans, several small cohort studies have noted arrhythmia and abnormal vascular perfusion in SMA infants, but further investigation is required to understand how this might affect SMA patient outcomes and treatment response.141, 142
Studies on ASO- and gene therapy vector–dosing to the CNS, versus systemic administration, in SMA mouse models have shown varying amounts of efficacy for each strategy. CNS targeting of SMA by ASO therapy is accomplished by intrathecal injection into the spinal cord. The scAAV9 vectors used in gene therapy cross the BBB and can be administered either intrathecally or systemically and still reach critical cell populations. Intrathecal injections are invasive and costly, and if repeated doses are necessary, treatments must be timed for maximum efficacy. Longer lasting, less invasive therapies that still have extensive motor neuron targeting would be ideal when designing new therapeutics and clinical trials for SMA.
Critical windows for SMA treatment
Due to the irreversible cell death and degeneration of the spinal motor neurons, determining an appropriate treatment window for SMA is crucial for the ultimate success of these therapies on SMA patients. To that end, studies on the exact timing of motor neuron loss and therapy efficacy are being completed in mouse and primate models.76 In an inducible hypomorphic mouse model, low SMN expression and early embryonic lethality can be rescued by induction of full-length SMN early in development (E7.5) but not later (E13.5).143 In an effort to diagnose SMA patients early for early intervention, a non-invasive blood-based postnatal screen for homozygous loss of SMN1 is also being developed (NCT02123186).
Clinically relevant outcome measures
Current clinical trial designs for SMA therapeutics rely on a number of different outcome measures to determine success. Among the most commonly used are compound muscle activated potential (CMAP), motor unit number estimation (MUNE), and, more recently, electrical impedance myography (EIM); but because of the age range of the patients and the heterogeneity of the rate of decline, these measures can be difficult to interpret. The discovery of new biomarkers for SMA is critical for identifying factors that could be measured as additional benchmarks of positive outcomes in these trials. To this end, several groups have looked for biomarkers in SMA patient samples144, 145 and in mouse models.146 SMN transcript levels do not correlate well with protein levels or disease severity, possibly due to variation in patient age, expression, and even sample storage and handling.147–149 A new two year longitudinal natural history study (NCT01736553) is in progress and will measure both SMN mRNA and protein in addition to total plasma protein levels with secondary motor function outcomes to address these issues and identify biomarkers.
Conclusions
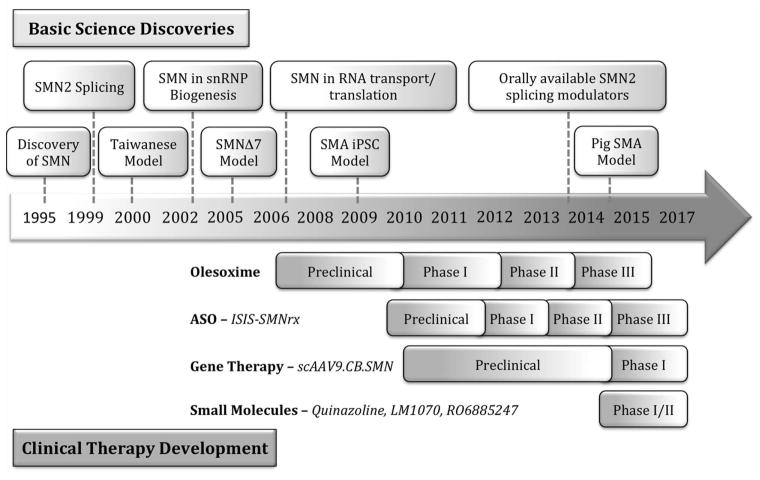
In the two decades since the discovery of the SMN gene many important discoveries have contributed to our basic understanding of SMA pathogenesis and the development of treatment paradigms (Fig. 3). Due to the well-characterized discrete genetic basis of the disease, SMA is a good candidate for SMN2 splice-site modulating ASOs, as well as exogenous SMN1 replacement by gene therapy vectors. In vitro and in vivo animal studies have provided strong evidence that the SMA neuromuscular phenotype can be significantly ameliorated by expression of full-length SMN protein or by treatment with neuroprotective compounds. There are still many questions as to the relative importance and timing of SMN expression in motor neurons and its subcellular functions in cell bodies versus axons and dendrites that are important for designing novel therapeutics. However, recent advances in pre-clinical drug development and clinical trials have shown great promise towards viable therapies for SMA patients.
Figure 3.
Important milestones in SMA basic science and clinical development. Timeline of important basic science discoveries in SMA: SMN basic biology and SMA modeling from the discovery of SMN to today (top), emerging and ongoing pre-clinical development and clinical trials of SMA therapeutics (bottom).
Acknowledgments
We thank Basil Darras and members of the Sahin lab for critically reviewing the manuscript. Due to limited space, we have not quoted all literature in the field, and we apologize to those whose articles are not referenced. Research in the Sahin lab has been supported by the NIH (P30 HD018655), Slaney Family Fund, Whitehall Foundation, SMA foundation, CureSMA, Boston Children’s Hospital Translational Research Program, Novartis, Roche, and Shire.
References
- 1.Sugarman EA, Nagan N, Zhu H, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet. 2012;20:27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Russman BS. Spinal muscular atrophy: clinical classification and disease heterogeneity. J Child Neurol. 2007;22:946–951. doi: 10.1177/0883073807305673. [DOI] [PubMed] [Google Scholar]
- 4.Dubowitz V. Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype. Eur J Paediatr Neurol. 1999;3:49–51. doi: 10.1053/ejpn.1999.0181. [DOI] [PubMed] [Google Scholar]
- 5.Monani UR, Lorson CL, Parsons DW, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 6.Lorson CL, Hahnen E, Androphy EJ, et al. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailman MD, Heinz JW, Papp AC, et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 11.Pellizzoni L, Baccon J, Rappsilber J, et al. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J Biol Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 12.Tisdale S, Lotti F, Saieva L, et al. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3′-end formation of histone mRNAs. Cell Rep. 2013;5:1187–1195. doi: 10.1016/j.celrep.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillai RS, Grimmler M, Meister G, et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Workman E, Veith A, Battle DJ. U1A regulates 3′ processing of the survival motor neuron mRNA. J Biol Chem. 2014;289:3703–3712. doi: 10.1074/jbc.M113.538264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Xing L, Rossoll W, et al. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J Neurosci. 2006;26:8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piazzon N, Rage F, Schlotter F, et al. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 17.Tadesse H, Deschenes-Furry J, Boisvenue S, et al. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 18.Akten B, Kye MJ, Hao le T, et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci U S A. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fallini C, Zhang H, Su Y, et al. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubers L, Valderrama-Carvajal H, Laframboise J, et al. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum Mol Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- 21.Fallini C, Rouanet JP, Donlin-Asp PG, et al. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev Neurobiol. 2014;74:319–332. doi: 10.1002/dneu.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saal L, Briese M, Kneitz S, et al. Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. RNA. 2014;20:1789–1802. doi: 10.1261/rna.047373.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kye MJ, Niederst ED, Wertz MH, et al. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet. 2014;23:6318–6331. doi: 10.1093/hmg/ddu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rathod R, Havlicek S, Frank N, et al. Laminin induced local axonal translation of beta-actin mRNA is impaired in SMN-deficient motoneurons. Histochem Cell Biol. 2012;138:737–748. doi: 10.1007/s00418-012-0989-1. [DOI] [PubMed] [Google Scholar]
- 25.Peter CJ, Evans M, Thayanithy V, et al. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum Mol Genet. 2011;20:1701–1711. doi: 10.1093/hmg/ddr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Custer SK, Todd AG, Singh NN, et al. Dilysine motifs in exon 2b of SMN protein mediate binding to the COPI vesicle protein alpha-COP and neurite outgrowth in a cell culture model of spinal muscular atrophy. Hum Mol Genet. 2013;22:4043–4052. doi: 10.1093/hmg/ddt254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ting CH, Wen HL, Liu HC, et al. The spinal muscular atrophy disease protein SMN is linked to the Golgi network. PLoS One. 2012;7:e51826. doi: 10.1371/journal.pone.0051826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez G, Dury AY, Murray LM, et al. A novel function for the survival motoneuron protein as a translational regulator. Hum Mol Genet. 2013;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki T, Chen S, Yu Y, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerbino V, Carri MT, Cozzolino M, et al. Mislocalised FUS mutants stall spliceosomal snRNPs in the cytoplasm. Neurobiol Dis. 2013;55:120–128. doi: 10.1016/j.nbd.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Groen EJ, Fumoto K, Blokhuis AM, et al. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum Mol Genet. 2013;22:3690–3704. doi: 10.1093/hmg/ddt222. [DOI] [PubMed] [Google Scholar]
- 32.Ishihara T, Ariizumi Y, Shiga A, et al. Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22:4136–4147. doi: 10.1093/hmg/ddt262. [DOI] [PubMed] [Google Scholar]
- 33.Onodera O, Ishihara T, Shiga A, et al. Minor splicing pathway is not minor any more: implications for the pathogenesis of motor neuron diseases. Neuropathology. 2014;34:99–107. doi: 10.1111/neup.12070. [DOI] [PubMed] [Google Scholar]
- 34.Turner BJ, Alfazema N, Sheean RK, et al. Overexpression of survival motor neuron improves neuromuscular function and motor neuron survival in mutant SOD1 mice. Neurobiol Aging. 2014;35:906–915. doi: 10.1016/j.neurobiolaging.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrank B, Gotz R, Gunnersen JM, et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bebee TW, Dominguez CE, Chandler DS. Mouse models of SMA: tools for disease characterization and therapeutic development. Hum Genet. 2012;131:1277–1293. doi: 10.1007/s00439-012-1171-5. [DOI] [PubMed] [Google Scholar]
- 37.Hsieh-Li HM, Chang JG, Jong YJ, et al. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 38.Le TT, Pham LT, Butchbach ME, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 39.Dimitriadi M, Sleigh JN, Walker A, et al. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praveen K, Wen Y, Gray KM, et al. SMA-causing missense mutations in survival motor neuron (Smn) display a wide range of phenotypes when modeled in Drosophila. PLoS Genet. 2014;10:e1004489. doi: 10.1371/journal.pgen.1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timmerman C, Sanyal S. Behavioral and electrophysiological outcomes of tissue-specific Smn knockdown in Drosophila melanogaster. Brain Res. 2012;1489:66–80. doi: 10.1016/j.brainres.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McWhorter ML, Monani UR, Burghes AH, et al. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gassman A, Hao le T, Bhoite L, et al. Small molecule suppressors of Drosophila kinesin deficiency rescue motor axon development in a zebrafish model of spinal muscular atrophy. PLoS One. 2013;8:e74325. doi: 10.1371/journal.pone.0074325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.See K, Yadav P, Giegerich M, et al. SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum Mol Genet. 2014;23:1754–1770. doi: 10.1093/hmg/ddt567. [DOI] [PubMed] [Google Scholar]
- 45.Wishart TM, Mutsaers CA, Riessland M, et al. Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J Clin Invest. 2014;124:1821–1834. doi: 10.1172/JCI71318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao le T, Burghes AH, Beattie CE. Generation and Characterization of a genetic zebrafish model of SMA carrying the human SMN2 gene. Mol Neurodegener. 2011;6:24. doi: 10.1186/1750-1326-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hao le T, Duy PQ, Jontes JD, et al. Temporal requirement for SMN in motoneuron development. Hum Mol Genet. 2013;22:2612–2625. doi: 10.1093/hmg/ddt110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doktor TK, Schroder LD, Andersen HS, et al. Absence of an intron splicing silencer in porcine Smn1 intron 7 confers immunity to the exon skipping mutation in human SMN2. PLoS One. 2014;9:e98841. doi: 10.1371/journal.pone.0098841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duque SI, Arnold WD, Odermatt P, et al. A large animal model of Spinal Muscular Atrophy and correction of phenotype. Ann Neurol. 2014 doi: 10.1002/ana.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Z, Gao F. Stem cell challenges in the treatment of neurodegenerative disease. CNS Neurosci Ther. 2012;18:142–148. doi: 10.1111/j.1755-5949.2011.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ebert AD, Yu J, Rose FF, Jr, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ebert AD, Shelley BC, Hurley AM, et al. EZ spheres: a stable and expandable culture system for the generation of pre-rosette multipotent stem cells from human ESCs and iPSCs. Stem Cell Res. 2013;10:417–427. doi: 10.1016/j.scr.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corti S, Nizzardo M, Nardini M, et al. Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J Clin Invest. 2008;118:3316–3330. doi: 10.1172/JCI35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyatt TJ, Rossi SL, Siegenthaler MM, et al. Human motor neuron progenitor transplantation leads to endogenous neuronal sparing in 3 models of motor neuron loss. Stem Cells Int. 2011;2011:207230. doi: 10.4061/2011/207230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corti S, Nizzardo M, Simone C, et al. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4:165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veltrop M, Aartsma-Rus A. Antisense-mediated exon skipping: taking advantage of a trick from Mother Nature to treat rare genetic diseases. Exp Cell Res. 2014;325:50–55. doi: 10.1016/j.yexcr.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 57.Touznik A, Lee JJ, Yokota T. New developments in exon skipping and splice modulation therapies for neuromuscular diseases. Expert Opin Biol Ther. 2014;14:809–819. doi: 10.1517/14712598.2014.896335. [DOI] [PubMed] [Google Scholar]
- 58.Baughan T, Shababi M, Coady TH, et al. Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol Ther. 2006;14:54–62. doi: 10.1016/j.ymthe.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Baughan TD, Dickson A, Osman EY, et al. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dickson A, Osman E, Lorson CL. A negatively acting bifunctional RNA increases survival motor neuron both in vitro and in vivo. Hum Gene Ther. 2008;19:1307–1315. doi: 10.1089/hum.2008.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monia BP, Lesnik EA, Gonzalez C, et al. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 62.McKay RA, Miraglia LJ, Cummins LL, et al. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J Biol Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 63.Hua Y, Vickers TA, Baker BF, et al. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hua Y, Vickers TA, Okunola HL, et al. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh RN. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- 66.Singh NN, Shishimorova M, Cao LC, et al. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hua Y, Sahashi K, Rigo F, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hua Y, Sahashi K, Hung G, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Passini MA, Bu J, Richards AM, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rigo F, Chun SJ, Norris DA, et al. Pharmacology of a central nervous system delivered 2′-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther. 2014;350:46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh NN, Seo J, Rahn SJ, et al. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS One. 2012;7:e49595. doi: 10.1371/journal.pone.0049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keil JM, Seo J, Howell MD, et al. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol Ther Nucleic Acids. 2014;3:e174. doi: 10.1038/mtna.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Osman EY, Miller MR, Robbins KL, et al. Morpholino antisense oligonucleotides targeting intronic repressor Element1 improve phenotype in SMA mouse models. Hum Mol Genet. 2014;23:4832–4845. doi: 10.1093/hmg/ddu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naryshkin NA, Weetall M, Dakka A, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 75.Edry E, Lamprecht R, Wagner S, et al. Virally mediated gene manipulation in the adult CNS. Front Mol Neurosci. 2011;4:57. doi: 10.3389/fnmol.2011.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karda R, Buckley SM, Mattar CN, et al. Perinatal systemic gene delivery using adeno-associated viral vectors. Front Mol Neurosci. 2014;7:89. doi: 10.3389/fnmol.2014.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang W, Li W, Ma N, et al. Non-viral gene delivery methods. Curr Pharm Biotechnol. 2013;14:46–60. [PubMed] [Google Scholar]
- 78.Azzouz M, Le T, Ralph GS, et al. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J Clin Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weinberg MS, Samulski RJ, McCown TJ. Adeno-associated virus (AAV) gene therapy for neurological disease. Neuropharmacology. 2013;69:82–88. doi: 10.1016/j.neuropharm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H, Yang B, Mu X, et al. Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol Ther. 2011;19:1440–1448. doi: 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duque S, Joussemet B, Riviere C, et al. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Foust KD, Wang X, McGovern VL, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Valori CF, Ning K, Wyles M, et al. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 84.Dominguez E, Marais T, Chatauret N, et al. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum Mol Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- 85.Passini MA, Bu J, Roskelley EM, et al. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Glascock JJ, Shababi M, Wetz MJ, et al. Direct central nervous system delivery provides enhanced protection following vector mediated gene replacement in a severe model of spinal muscular atrophy. Biochem Biophys Res Commun. 2012;417:376–381. doi: 10.1016/j.bbrc.2011.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Passini MA, Bu J, Richards AM, et al. Translational fidelity of intrathecal delivery of self-complementary AAV9-survival motor neuron 1 for spinal muscular atrophy. Hum Gene Ther. 2014;25:619–630. doi: 10.1089/hum.2014.011. [DOI] [PubMed] [Google Scholar]
- 88.Meyer K, Ferraiuolo L, Schmelzer L, et al. Improving Single Injection CSF Delivery of AAV9-mediated Gene Therapy for SMA: A Dose-response Study in Mice and Nonhuman Primates. Mol Ther. 2014 doi: 10.1038/mt.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peluffo H, Foster E, Ahmed SG, et al. Efficient gene expression from integration-deficient lentiviral vectors in the spinal cord. Gene Ther. 2013;20:645–657. doi: 10.1038/gt.2012.78. [DOI] [PubMed] [Google Scholar]
- 90.Singh P, Liew WK, Darras BT. Current advances in drug development in spinal muscular atrophy. Curr Opin Pediatr. 2013;25:682–688. doi: 10.1097/MOP.0b013e32836565ac. [DOI] [PubMed] [Google Scholar]
- 91.Bordet T, Buisson B, Michaud M, et al. Identification and characterization of cholest-4-en-3-one, oxime (TRO19622), a novel drug candidate for amyotrophic lateral sclerosis. J Pharmacol Exp Ther. 2007;322:709–720. doi: 10.1124/jpet.107.123000. [DOI] [PubMed] [Google Scholar]
- 92.Sunyach C, Michaud M, Arnoux T, et al. Olesoxime delays muscle denervation, astrogliosis, microglial activation and motoneuron death in an ALS mouse model. Neuropharmacology. 2012;62:2346–2352. doi: 10.1016/j.neuropharm.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 93.Magalon K, Zimmer C, Cayre M, et al. Olesoxime accelerates myelination and promotes repair in models of demyelination. Ann Neurol. 2012;71:213–226. doi: 10.1002/ana.22593. [DOI] [PubMed] [Google Scholar]
- 94.Dessaud ECA, Scherrer B, Berna P, Pruss R, Cuvier V, Hauke W, 4, Bruno C, Chabrol B, Comi G, Cuisset JM, Deconinck N, Goemans N, Estournet B, Fontaine-Carbonel S, Gorni K, Kirschner J, Lusakowska A, Lochmuller H, Mayer M, Mercuri E, Müller-Felber W, Muntoni F, Rivier F, Roper H, Schara U, Van den Berg L, Vita G, Walter M, Bertini E. Results of a phase II study to assess safety and efficacy of olesoxime (TRO19622) in 3–25 years old spinal muscular atrophy patients. Neuromuscul Disord. 2014;24:920–921. [Google Scholar]
- 95.Jarecki J, Chen X, Bernardino A, et al. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 96.Thurmond J, Butchbach ME, Palomo M, et al. Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy. J Med Chem. 2008;51:449–469. doi: 10.1021/jm061475p. [DOI] [PubMed] [Google Scholar]
- 97.Butchbach ME, Singh J, Thorsteinsdottir M, et al. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet. 2010;19:454–467. doi: 10.1093/hmg/ddp510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singh J, Salcius M, Liu SW, et al. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem Biol. 2008;3:711–722. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Meerbeke JP, Gibbs RM, Plasterer HL, et al. The DcpS inhibitor RG3039 improves motor function in SMA mice. Hum Mol Genet. 2013;22:4074–4083. doi: 10.1093/hmg/ddt257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gogliotti RG, Cardona H, Singh J, et al. The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum Mol Genet. 2013;22:4084–4101. doi: 10.1093/hmg/ddt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brichta L, Hofmann Y, Hahnen E, et al. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 102.Sumner CJ, Huynh TN, Markowitz JA, et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 103.Andreassi C, Angelozzi C, Tiziano FD, et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 104.Evans MC, Cherry JJ, Androphy EJ. Differential regulation of the SMN2 gene by individual HDAC proteins. Biochem Biophys Res Commun. 2011;414:25–30. doi: 10.1016/j.bbrc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harahap IS, Saito T, San LP, et al. Valproic acid increases SMN2 expression and modulates SF2/ASF and hnRNPA1 expression in SMA fibroblast cell lines. Brain Dev. 2012;34:213–222. doi: 10.1016/j.braindev.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 106.Sabra M, Texier P, El Maalouf J, et al. The Tudor protein survival motor neuron (SMN) is a chromatin-binding protein that interacts with methylated lysine 79 of histone H3. J Cell Sci. 2013;126:3664–3677. doi: 10.1242/jcs.126003. [DOI] [PubMed] [Google Scholar]
- 107.Lunke S, El-Osta A. Applicability of histone deacetylase inhibition for the treatment of spinal muscular atrophy. Neurotherapeutics. 2013;10:677–687. doi: 10.1007/s13311-013-0209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kissel JT, Elsheikh B, King WM, et al. SMA valiant trial: a prospective, double-blind, placebo-controlled trial of valproic acid in ambulatory adults with spinal muscular atrophy. Muscle Nerve. 2014;49:187–192. doi: 10.1002/mus.23904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swoboda KJ, Scott CB, Crawford TO, et al. SMA CARNI-VAL trial part I: double-blind, randomized, placebo-controlled trial of L-carnitine and valproic acid in spinal muscular atrophy. PLoS One. 2010;5:e12140. doi: 10.1371/journal.pone.0012140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kissel JT, Scott CB, Reyna SP, et al. SMA CARNIVAL TRIAL PART II: a prospective, single-armed trial of L-carnitine and valproic acid in ambulatory children with spinal muscular atrophy. PLoS One. 2011;6:e21296. doi: 10.1371/journal.pone.0021296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garbes L, Heesen L, Holker I, et al. VPA response in SMA is suppressed by the fatty acid translocase CD36. Hum Mol Genet. 2013;22:398–407. doi: 10.1093/hmg/dds437. [DOI] [PubMed] [Google Scholar]
- 112.Hahnen E, Eyupoglu IY, Brichta L, et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 113.Riessland M, Brichta L, Hahnen E, et al. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 114.Avila AM, Burnett BG, Taye AA, et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garbes L, Riessland M, Holker I, et al. LBH589 induces up to 10-fold SMN protein levels by several independent mechanisms and is effective even in cells from SMA patients non-responsive to valproate. Hum Mol Genet. 2009;18:3645–3658. doi: 10.1093/hmg/ddp313. [DOI] [PubMed] [Google Scholar]
- 116.Narver HL, Kong L, Burnett BG, et al. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Riessland M, Ackermann B, Forster A, et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet. 2010;19:1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 118.Liu H, Yazdani A, Murray LM, et al. The Smn-independent beneficial effects of trichostatin A on an intermediate mouse model of spinal muscular atrophy. PLoS One. 2014;9:e101225. doi: 10.1371/journal.pone.0101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sakla MS, Lorson CL. Induction of full-length survival motor neuron by polyphenol botanical compounds. Hum Genet. 2008;122:635–643. doi: 10.1007/s00439-007-0441-0. [DOI] [PubMed] [Google Scholar]
- 120.Dayangac-Erden D, Bora G, Ayhan P, et al. Histone deacetylase inhibition activity and molecular docking of (e)-resveratrol: its therapeutic potential in spinal muscular atrophy. Chem Biol Drug Des. 2009;73:355–364. doi: 10.1111/j.1747-0285.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 121.Dayangac-Erden D, Bora-Tatar G, Dalkara S, et al. Carboxylic acid derivatives of histone deacetylase inhibitors induce full length SMN2 transcripts: a promising target for spinal muscular atrophy therapeutics. Arch Med Sci. 2011;7:230–234. doi: 10.5114/aoms.2011.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bora-Tatar G, Erdem-Yurter H. Investigations of curcumin and resveratrol on neurite outgrowth: perspectives on spinal muscular atrophy. Biomed Res Int. 2014;2014:709108. doi: 10.1155/2014/709108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Little D, Valori CF, Mutsaers CA, et al. PTEN Depletion Decreases Disease Severity and Modestly Prolongs Survival in a Mouse Model of Spinal Muscular Atrophy. Mol Ther. 2014 doi: 10.1038/mt.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ozdinler PH, Macklis JD. IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat Neurosci. 2006;9:1371–1381. doi: 10.1038/nn1789. [DOI] [PubMed] [Google Scholar]
- 125.Murdocca M, Malgieri A, Luchetti A, et al. IPLEX administration improves motor neuron survival and ameliorates motor functions in a severe mouse model of spinal muscular atrophy. Mol Med. 2012;18:1076–1085. doi: 10.2119/molmed.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tsai LK, Chen YC, Cheng WC, et al. IGF-1 delivery to CNS attenuates motor neuron cell death but does not improve motor function in type III SMA mice. Neurobiol Dis. 2012;45:272–279. doi: 10.1016/j.nbd.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 127.Shababi M, Glascock J, Lorson CL. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum Gene Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- 128.Bosch-Marce M, Wee CD, Martinez TL, et al. Increased IGF-1 in muscle modulates the phenotype of severe SMA mice. Hum Mol Genet. 2011;20:1844–1853. doi: 10.1093/hmg/ddr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tsai LK, Chen CL, Ting CH, et al. Systemic administration of a recombinant AAV1 vector encoding IGF-1 improves disease manifestations in SMA mice. Mol Ther. 2014;22:1450–1459. doi: 10.1038/mt.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dachs E, Piedrafita L, Hereu M, et al. Chronic treatment with lithium does not improve neuromuscular phenotype in a mouse model of severe spinal muscular atrophy. Neuroscience. 2013;250:417–433. doi: 10.1016/j.neuroscience.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 131.Kirschner J, Schorling D, Hauschke D, et al. Somatropin treatment of spinal muscular atrophy: a placebo-controlled, double-blind crossover pilot study. Neuromuscul Disord. 2014;24:134–142. doi: 10.1016/j.nmd.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 132.McGivern JV, Patitucci TN, Nord JA, et al. Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia. 2013;61:1418–1428. doi: 10.1002/glia.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hunter G, Aghamaleky Sarvestany A, Roche SL, et al. SMN-dependent intrinsic defects in Schwann cells in mouse models of spinal muscular atrophy. Hum Mol Genet. 2014;23:2235–2250. doi: 10.1093/hmg/ddt612. [DOI] [PubMed] [Google Scholar]
- 134.Tarabal O, Caraballo-Miralles V, Cardona-Rossinyol A, et al. Mechanisms involved in spinal cord central synapse loss in a mouse model of spinal muscular atrophy. J Neuropathol Exp Neurol. 2014;73:519–535. doi: 10.1097/NEN.0000000000000074. [DOI] [PubMed] [Google Scholar]
- 135.Park GH, Maeno-Hikichi Y, Awano T, et al. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010;30:12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gogliotti RG, Quinlan KA, Barlow CB, et al. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J Neurosci. 2012;32:3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taylor AS, Glascock JJ, Rose FF, Jr, et al. Restoration of SMN to Emx-1 expressing cortical neurons is not sufficient to provide benefit to a severe mouse model of Spinal Muscular Atrophy. Transgenic Res. 2013;22:1029–1036. doi: 10.1007/s11248-013-9702-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bevan AK, Hutchinson KR, Foust KD, et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet. 2010;19:3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Heier CR, Satta R, Lutz C, et al. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shababi M, Habibi J, Yang HT, et al. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet. 2010;19:4059–4071. doi: 10.1093/hmg/ddq329. [DOI] [PubMed] [Google Scholar]
- 141.Hachiya Y, Arai H, Hayashi M, et al. Autonomic dysfunction in cases of spinal muscular atrophy type 1 with long survival. Brain Dev. 2005;27:574–578. doi: 10.1016/j.braindev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 142.Araujo A, Araujo M, Swoboda KJ. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J Pediatr. 2009;155:292–294. doi: 10.1016/j.jpeds.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hammond SM, Gogliotti RG, Rao V, et al. Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late. PLoS One. 2010;5:e15887. doi: 10.1371/journal.pone.0015887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Finkel RS, Crawford TO, Swoboda KJ, et al. Candidate proteins, metabolites and transcripts in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study. PLoS ONE. 2012;7:e35462. doi: 10.1371/journal.pone.0035462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kobayashi DT, Shi J, Stephen L, et al. SMA-MAP: a plasma protein panel for spinal muscular atrophy. PLoS ONE. 2013;8:e60113. doi: 10.1371/journal.pone.0060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mutsaers CA, Lamont DJ, Hunter G, et al. Label-free proteomics identifies Calreticulin and GRP75/Mortalin as peripherally accessible protein biomarkers for spinal muscular atrophy. Genome Med. 2013;5:95. doi: 10.1186/gm498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Crawford TO, Paushkin SV, Kobayashi DT, et al. Evaluation of SMN protein, transcript, and copy number in the biomarkers for spinal muscular atrophy (BforSMA) clinical study. PLoS ONE. 2012;7:e33572. doi: 10.1371/journal.pone.0033572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tiziano FD, Lomastro R, Di Pietro L, et al. Clinical and molecular cross-sectional study of a cohort of adult type III spinal muscular atrophy patients: clues from a biomarker study. Eur J Hum Genet. 2013;21:630–636. doi: 10.1038/ejhg.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hunter G, Roche SL, Somers E, et al. The influence of storage parameters on measurement of survival motor neuron (SMN) protein levels: Implications for pre-clinical studies and clinical trials for spinal muscular atrophy. Neuromuscul Disord. 2014;24:973–977. doi: 10.1016/j.nmd.2014.05.013. [DOI] [PubMed] [Google Scholar]