Abstract
Purpose
To evaluate the performance of an edge-based registration technique in correcting for respiratory motion artifacts in MR renographic data and to examine the efficiency of a semi-automatic software package in processing renographic data from a cohort of clinical patients.
Materials and Methods
The developed software incorporates an image-registration algorithm based on the generalized Hough transform of edge maps. It was used to estimate GFR, RPF, and MTT from 36 patients who underwent free-breathing MR renography at 3T using saturation-recovery turbo-FLASH. Processing time required for each patient was recorded. Renal parameter estimates and model-fitting residues from the software were compared to those from a previously reported technique. Inter-reader variability in the software was quantified by the standard deviation of parameter estimates among three readers. GFR estimates from our software were also compared to a reference standard from nuclear medicine.
Results
The time taken to process one patient’s data with the software averaged 12 ± 4 minutes. The applied image registration effectively reduced motion artifacts in dynamic images by providing renal tracer-retention curves with significantly smaller fitting residues (P < 0.01) than unregistered data or data registered by the previously reported technique. Inter-reader variability was less than 10% for all parameters. GFR estimates from the proposed method showed greater concordance with reference values (P < 0.05).
Conclusion
These results suggest that the proposed software can process MR renography data efficiently and accurately. Its incorporated registration technique based on the generalized Hough transform effectively reduces respiratory motion artifacts in free-breathing renographic acquisitions.
Keywords: Magnetic resonance renography, Glomerular filtration rate, Image registration, Hough transform
INTRODUCTION
MR renography (MRR) is emerging as a reliable technique for evaluating single-kidney glomerular filtration and perfusion (1-6). Following intravenous injection of a bolus of contrast agent, MR images are repeatedly acquired to record the passage of tracer through the kidneys. Tracer enhancement and excretion from a kidney over time reflect its glomerular and tubular function (4,6). Using a tracer-kinetic model to analyze the tracer retention vs. time curves, we can estimate functional parameters such as glomerular filtration rate (GFR), renal plasma flow (RPF), and mean transit time (MTT) (4,6-8). Over the past decade, technical improvements have been made on multiple aspects of the technique, including data acquisition and analysis (9), making MRR a promising tool for the assessment of renal function.
One major obstacle to the widespread application of MRR is the lack of an efficient software package that performs rapid data post-processing (9), particularly one with robust image registration. In human subjects, respiratory motion may displace the kidneys by 5-10 mm (10), and this relative motion between images acquired at different time-points may introduce substantial error into the estimation of functional parameters. Techniques for the correction of respiratory motion include acquisition during suspended respiration (6,11,12), which can be challenging for elderly or infirm patients, or post-acquisition image registration (3,13-15).
Registration of dynamic kidney images faces multiple challenges. First, signal intensity is different between dynamic frames due to tracer enhancement, and different regions enhance differently. Therefore, correlation-based registration techniques may fail. Second, surrounding organs such as the liver and spleen might have similar signal-intensities as the kidneys and thereby interfere with registration. Third, poorly-functioning kidneys usually display little contrast enhancement, making their separation from background difficult. Because of these challenges, a robust registration scheme requires a combination of automatic registration and manual guidance. Involvement of manual guidance, however, introduces inter-observer variability.
Another strategy for respiratory motion correction makes use of the direction of respiratory motion. Since breathing primarily moves the kidneys along the head-to-toe direction, renal cortical signals sampled from an axial slice are less affected by motion (3) and therefore avoid the need for registration. Medullary signals, on the other hand, are better sampled from a coronal slice. We term this method “composite sampling” (3) because cortical and medullary signals are sampled from different slices. This method works well, but requires the acquisition and analysis of dynamic images from two slices.
In this study, we evaluated the performance of an edge-based registration technique in correcting respiratory motion artifacts in free-breathing MRR and examined the efficiency of a software package developed to accelerate the post-processing of MR renographic data with tracer-kinetic models.
MATERIALS AND METHODS
MRR Data Acquisition
This study was approved by the institutional review board (IRB) at the University of Utah. Thirty-six patients (24 male, 12 female; ages 28-81 years) with suspected liver diseases were recruited from the Liver Center at the University of Utah between February 2012 and May 2014. Subjects were instructed to fast overnight before their examination. Following written informed consent, MRI scans were performed on a 3T scanner (TimTrio; Siemens Medical Solutions, Erlangen, Germany) following a previously reported protocol (3).
Briefly, MRR data was acquired using a two-dimensional (2D), T1-weighted turbo fast low-angle shot (FLASH) sequence prepared with saturation recovery, with the following sequence parameters: slice thickness 7 mm, TR 526 ms, TE 1.21 ms, saturation-recovery TI 300 ms, flip angle 16°, FOV 382×420 mm, matrix 154×176. In each acquisition, three 2D slices were covered: coronal and axial slices through the middle of the kidneys, and a coronal or sagittal slice through the abdominal aorta. Temporal resolution was ~1.5 sec per frame. After 5 baseline acquisitions, 4 ml gadoteridol (ProHance; Bracco Diagnostics, Milan, Italy) was administered intravenously at a rate of 2 ml/sec, followed by a 20 ml saline flush at the same rate. Data acquisition continued for 5 minutes. To quantify tracer concentration from MR signals, proton-density-weighted images (M0) were acquired from the same three slices and using the same sequence but with a longer TR of 4 sec. As part of the clinical protocol for these liver patients (16), high-resolution three-dimensional (3D) T1-weighted volumetric interpolated breath-hold examinations (VIBEs) were acquired to cover the liver and the kidneys before and after the injection of 0.1 mmol gadoteridol per kilogram of body weight: slice thickness 3 mm, TR 3.5 ms, TE 1.22 ms, flip angle 10°, FOV 450×340×240 mm, matrix 320×240×80. High corticomedullary contrast in the post-contrast-injection VIBE allowed the volume of the renal cortex to be measured. Medullary volume was measured by subtraction of the cortical volume from the whole kidney volume (without the collecting system) measured from the pre-contrast-injection VIBE. These volumes were then used in quantitative MRR analysis (3).
Reference GFR Measurement
As a reference standard, GFR values for the subjects were also measured from urinary clearance of 99mTc-DTPA on the same day as their MR scans. Following intravenous injection of a bolus of 5 mCi 99mTc-DTPA, two urine samples were collected at 150 and 240 minutes after contrast injection and peripheral venous blood samples were collected at 60, 150, and 240 minutes after contrast injection. The MRI scan was performed between the 60-minute and 150-minute blood samples. From each urine sample, GFR was estimated with the formula: UV/P, where U is the urine concentration of DTPA (in counts/min/ml), V is the urine flow rate (in ml/min), and P is the estimated concentration of contrast in the blood samples before and after urine collection (3,17,18). The GFR values estimated from the two urine samples were averaged and used as the reference GFR (termed “nuc-GFR” in this study).
Edge-based Image Registration
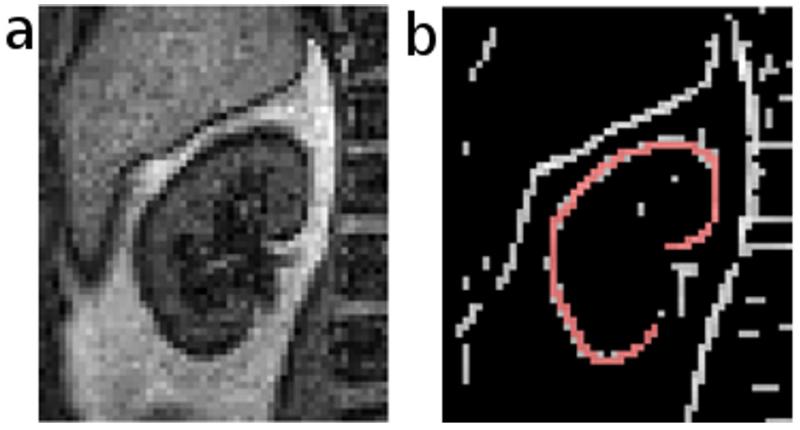
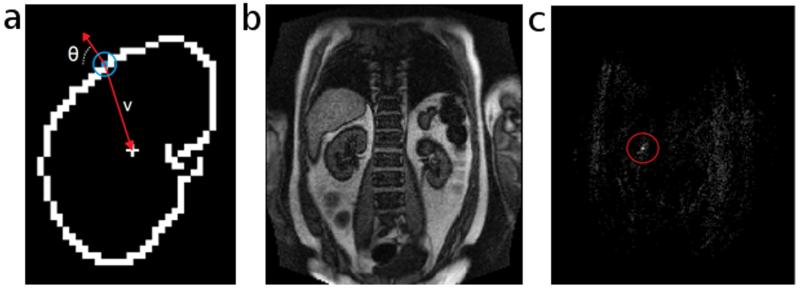
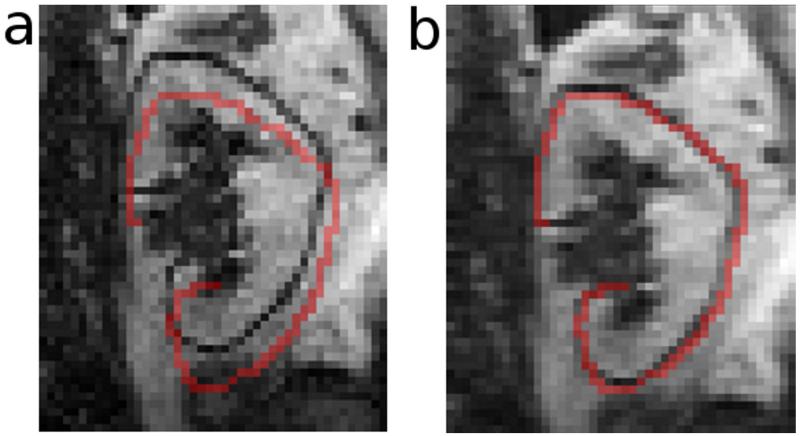
The image registration implemented in our developed software is based on the generalized Hough transform (19-22) of edge maps of the dynamic MR images. First, a template for registration is generated using the following procedure: We choose an image frame with a visually detectable kidney boundary, crop the image, and generate an edge map (Fig. 1a) using Canny edge-detection (23); our software provides slider widgets that enable the user to control multiple edge-detection parameters, such as the smoothing factor and upper and lower thresholds for edge-pixels to ensure the generation of well-defined kidney contours. With a built-in paintbrush tool in the software, the user then highlights the kidney contour in the edge map (Fig. 1b), and this user-defined contour will be used as a template for aligning all other frames. Second, using the generalized Hough transform, we characterize each pixel on the kidney contour in the template by two parameters: an angle (θ) that defines the orientation of the contour at this location and a vector (v) pointing from the pixel to the center of the contour (Fig. 2a). A lookup table is generated containing angles θ and vectors v for all pixels on the contour. Third, to align an image frame (example in Fig. 2b) to the template, an edge map of the image is computed using Canny edge-detection. Without the user’s manual highlight, this edge map contains spurious edges due to either noise or non-kidney related boundaries in addition to the true kidney boundary. Fourth, for each edge pixel in the edge map to be registered, the orientation angle θ is computed and compared to those in the lookup table for the template. If the angle matches any angle in the template, the corresponding vector v in the lookup table is used to point to a candidate location for the center of the kidney contour. This process is repeated for all edge pixels. The location with the most vectors pointing to it is the true center of the kidney in the current image frame (Fig. 2c). Once the center of the kidney is determined, the frame is aligned to the template based on the relative displacement of the kidney-centers using an affine transformation (Fig. 3). Upsampling of the images occurs prior to alignment to account for sub-voxel displacements. This process is repeated for all dynamic frames.
Figure 1.
Template generation after edge detection. a) Cropped image of a kidney. b) Edge map of the image overlaid with a user-defined contour template (red).
Figure 2.
Identifying the center of the kidney with the generalized Hough transform. a) Template of a kidney contour. Each edge pixel (colored blue in the blue circle) along the contour is indexed by its orientation angle (θ) and a vector (v) that points towards the contour’s center. b) An MRR image frame that we want to register to the template. c) A map with the same matrix size as the image frame. The value of each element represents the number of edge pixels that regard the current element as the center of the kidney contour in the current frame. The brightest point in the red circle is the true center, and is used for registering the current frame to the template.
Figure 3.
Registration of a dynamic frame to the template. This figure shows an MRR frame before (a) and after (b) upsampling and affine alignment to the template shown by the red overlay. Upsampling allows for correction of sub-voxel displacements.
Rotational motion is not a significant problem with the kidneys, but the software still provides optional correction for in-plane rotation. Within a user-defined range of rotation angles, the software rotates the template in increments and compares the rotated template to each frame. The angle with the best match between the image frame and the template is subsequently used to rotate the image frame.
MRR Image Processing
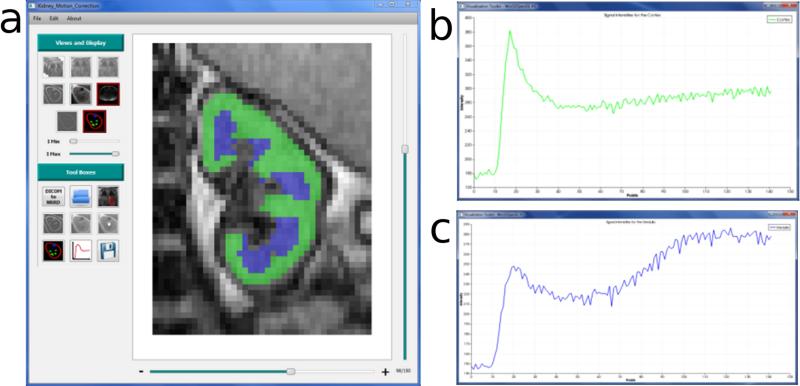
A software package was developed in C++ that incorporates robust image registration and all other post-processing steps to facilitate MRR data processing. With an intuitive graphical user interface (GUI) (Fig. 4), the software enables rapid visualization and cropping of the dynamic images to isolate the kidneys. After manually cropping the kidney, image registration as described above is performed. Following registration, regions of interest (ROIs) can be drawn over the cortex and medulla in a frame displaying high corticomedullary contrast (Fig. 4a). These ROIs are automatically applied to all the registered frames, and the average signal intensity within the ROIs is computed for each frame (Fig. 4b and 4c), as well as for the proton-density (M0) images. With our current acquisition protocol, MR images of the kidneys are acquired from axial and coronal slices as required for composite sampling, but now with our effective edge-based registration, we can sample both cortical and medullary signals from the coronal slice.
Figure 4.
Region-of-interest (ROI) analysis using our developed software. a) The user interface of the software displays control buttons that enable efficient post-processing of MRR data. In the stage of processing shown, ROIs have been drawn for the cortex (green) and medulla (blue). After drawing the ROIs, averaged signals within each ROI are automatically converted to tracer concentration based on algorithms embedded in the software. b) Tracer concentration vs. time curve for the cortical ROI. c) Tracer concentration vs. time curve for the medullary ROI.
Using the saturation-recovery formula and T1-shortening effect of gadolinium, the tracer concentration at each time point is calculated from the signal intensity and M0 value (3,4,12). Arterial signals are sampled from the abdominal aorta and are converted to arterial tracer concentration (otherwise known as the arterial input function (AIF)) using the same method as for the kidney tissue signals. Image registration is not needed for the aortic frames because the aorta is minimally affected by respiratory motion (24).
The volumes of the renal cortex and medulla, required for GFR estimation, can be obtained by segmenting the 3D VIBE data using ITK-SNAP (25), a widely used image-analysis tool conveniently linked to our software package. Once the volumes are available, our software then proceeds to automatically fit tissue retention vs. time curves to estimate renal functional parameters. While other tracer kinetic models can be easily incorporated, the current version of the software performs curve fitting with two models: a whole-kidney (WK) model that considers renal parenchyma as a single compartment (26), and a corticomedullary (CM) model that treats cortex and medulla separately (11). Curve fitting with both models provides estimates of GFR, RPF, and vascular and whole-kidney MTTs. By separating cortical and medullary signals, the CM model also gives an insight into tubular function (6,11,27).
Performance of Image Registration
We compared results from the 36 patient datasets with and without registration of the coronal kidney images. For the same datasets, we also applied composite sampling, using cortical signals from unregistered axial images and medullary signals from manually registered coronal images (3). All processing was done by the same user (CC). To estimate the motion artifacts in the raw and registered images, we computed the residues between the tissue retention curves from the images and their best curve-fit by the tracer-kinetic models, using the residues as an approximation of the amount of respiration-induced oscillation in the retention curves. Parameter estimates (GFR, RPF and MTT) obtained using the different strategies were compared using two-tailed, paired t-tests. P-values less than 0.05 were considered to be significant.
Performance of the Software in Processing MRR Patient Data
We evaluated inter-reader variability effects on functional-parameter estimates. Three readers with different levels of experience (CC with 3 years of experience processing MRR data, JZ with 12 years, and YZ with 3 months) processed the same 36 datasets using the developed software. For each parameter of each kidney, the standard deviation (SD) of the estimates by the three readers was computed and averaged across all kidneys. We also computed the coefficient of variation (CV = SD/mean) as a relative metric of variation. Paired t-tests were used to compare the inter-reader variability of estimates of the same parameter by the two tracer kinetic models. To evaluate the efficiency of the software, we recorded the time each reader spent processing each MRR dataset.
Using the developed software, we estimated GFR values from the 36 patient datasets, and the values, denoted as MR-GFR, were compared to the reference nuc-GFR values. To evaluate the overall accuracy of MR-GFR for the 36 cases, we computed the percentage of cases whose MR-GFR was within ±10%, ±20%, and ±30% of the nuc-GFR, in accordance with National Kidney Foundation guidelines (28). For comparison, we also processed the data using our in-house MATLAB (Mathworks; Natick, PA) program that uses composite sampling (manual registration of coronal images for sampling medullary signals). The program implements the same algorithms for signal-concentration conversion and tracer-kinetic model fitting as the new software, and is termed the manual processing method.
RESULTS
Performance of Image Registration
To quantify the motion artifacts in contrast-enhancement curves from kidney tissue, and thereby the performance of the applied image-registration, we computed the fitting residues between the curves and their model-fits. Without registration, the fitting residues for all the patient cases were 14% ± 6% of their respective curve magnitudes. Using composite sampling and the proposed registration technique, the residue was reduced to 10% ± 6% and 8% ± 4%, respectively, indicating a mitigation of motion artifacts. Residues for the separate cortical and medullary retention curves in the CM-model were significantly larger than the whole-kidney retention curve in the WK-model (P < 0.01): 18% ± 7% without registration, 13% ± 6% for composite sampling, and 10% ± 4% for our proposed technique. Compared to composite sampling, the reduction in curve-fitting residues achieved using our technique was significant (P < 0.01).
Table 1 lists the parameter estimates obtained using different registration techniques: no registration (using coronal images), composite sampling, and the edge-based registration in our software. For both the WK model and the CM model, estimated GFR values were significantly different between the edge-based registration and the no-registration estimation, with differences ranging from −9 to 14 ml/min for the WK model (P < 0.01) and −20 to 11 ml/min for the CM model (P < 0.01). No significant difference was observed between GFR values estimated using the edge-based registration and those by composite sampling (P = 0.06). RPF estimates obtained without registration and with composite sampling differed significantly from the edge-based registration (P < 0.01). RPF values from composite sampling were found to be significantly higher than those from the other two techniques (P < 0.01). No significant difference between the methods was observed for the MTT estimates (P = 0.34).
Table 1.
Comparison of different registration strategies in estimating GFR, RPF, and MTT.
| Registration | GFR (ml/min) | RPF (ml/min) | MTT (s) | |||
|---|---|---|---|---|---|---|
| WK | CM | WK | CM | WK | CM | |
| No-registration | 30 ± 10* | 32 ± 12* | 154 ± 48* | 158 ± 50* | 211 ± 49 | 219 ± 69 |
| Composite sampling | 32 ± 10 | 33 ± 12 | 191 ± 68* | 195 ± 72* | 220 ± 61 | 203 ± 54 |
| Edge-based method | 32 ± 11 | 33 ± 12 | 161 ± 51 | 165 ± 52 | 225 ± 61 | 215 ± 55 |
The values are mean ± standard deviation across all 72 kidneys. Asterisks indicate a significant difference (P < 0.05) as evaluated by a paired t-test between the value and the estimate from the edge-based method (bottom row). WK refers to the whole-kidney model, CM to the corticomedullary model.
Performance of the MRR-processing Software
All MRR patient data were successfully processed using the software. Processing time for each patient (two kidneys) averaged 12 ± 4 minutes, as compared to 40 ± 16 minutes with the manual processing method based on composite sampling.
Table 2 shows the SD and CV computed from the parameters estimated by the three readers using the software. All three parameters (GFR, RPF, and MTT) showed low inter-reader variability, with an average CV of less than 9.2%. In particular, inter-reader variability for GFR averaged 2-3 ml/min. The WK model provided GFR and RPF with slightly lower variability than the CM model, but the differences were not significant (P values of 0.40 and 0.14 for GFR and RPF, respectively). MR-GFR estimates from the new software were more likely to agree with nuc-GFR values than those from the manual processing method (Table 3).
Table 2.
Inter-reader variability of MRR parameters estimated with the proposed software.
| GFR (ml/min) | RPF (ml/min) | MTT (s) | ||||
|---|---|---|---|---|---|---|
| Model | WK | CM | WK | CM | WK | CM |
| SD (CV) |
2.0 ± 2.0 (7.4% ± 6.4%) |
3.0 ± 3.0 (8.0% ± 8.8%) |
13.1 ± 12.0 (8.6% ± 7.5%) |
14.7 ± 15.0 (9.2% ± 8.6%) |
21 ± 20 (8.6% ± 7.1%) |
20 ± 19 (8.8% ± 8.3%) |
Standard deviation (SD) values were computed from the parameter values estimated by the three readers and then were averaged across all kidneys. The coefficient of variation (CV) for a parameter is the ratio of the SD over the mean value between the three readers. WK refers to the whole-kidney model, CM to the corticomedullary model.
Table 3.
Percentage of MR GFR values within ±10, ±20, and ±30% of the nuc-GFR values obtained from 99mTc clearance on the same day as the MR examination.
| Accuracy interval | Percentage of cases within accuracy interval |
|
|---|---|---|
| Manual processing | Proposed software | |
| ±10% | 11% | 17% |
| ±20% | 27% | 33% |
| ±30% | 50% | 58% |
DISCUSSION
In this study, we demonstrated the efficiency and precision of a new software package for processing MRR data. With 36 patients, we showed that the robust image registration effectively corrected respiratory motion artifacts and greatly accelerated MRR data analysis, with low inter-reader variability.
Despite increasing acceptance of MRR as a means to assess single-kidney function, time-intensive post-processing has limited its adoption in clinical practice. Post-processing of renographic images commonly takes up to 45 minutes, largely because of labor-intensive image registration (1,3). In our software, we applied an edge-based registration algorithm that utilizes the generalized Hough transform to concisely characterize edge maps of dynamic image frames. With minimal effort, the user helps to localize the true kidney boundary in a template image, and the overall time to register both kidneys in a given patient averaged less than 10 minutes among three users with varying degrees of experience.
A previous protocol used composite sampling which requires the acquisition of at least two slices: an axial slice for cortical signals and a coronal slice for medullary signals. With effective registration of coronal images, however, both cortical and medullary signals can be recorded from a single coronal slice. Eliminating the need for axial images either enables increased spatial resolution, increased temporal resolution, or both. Higher temporal resolution would enable improved definition of the arterial input function and vascular peaks in renal tissue curves, improving measures of voxel-wise perfusion for detecting heterogeneous renal ischemia. In addition, sampling cortical and medullary signals from the same slice ensures that these signals do not vary artificially as a result of different coil-sensitivity profiles between the slices.
The proposed software provided GFR estimates with an inter-reader variability of 2-3 ml/min. GFR values agreed more often with reference methods than estimates from the manual processing did. While GFR is relatively robust to motion artifacts, other parameters of renal physiology are not. Using the vascular phase of the dynamic data, for example, voxel-wise perfusion can be estimated to study the spatial distribution of renal perfusion (29). Voxel-wise perfusion is highly sensitive to respiratory motion. Similarly, registration is also critical for analyzing MR renographic data of renal tumors (5,30), in which kidney and tumor tissue needs to be separated accurately and analyzed by different tracer-kinetic models. We expect that improved registration algorithms will benefit these clinical applications.
This study has multiple limitations. First, while most of our subjects had impaired renal function, none had diseases causing renal structural abnormality. The benefit of effective image registration for such diseases therefore warrants further study. Second, our software does not currently have an embedded component for measuring renal volumes, which are necessary for quantifying GFR in the selected tracer kinetic models. Third, due to the direction of respiratory motion and kidney anatomy, the proposed edge-based registration technique works best for dynamic images acquired in the coronal plane and not the axial plane. Fourth, we use a saturation recovery sequence for data acquisition and then measure proton density (M0) to convert the signals to Gd concentration. Other groups might choose to use other sequences, or may measure pre-contrast T1 for signal conversion. This would not impact other steps of post-processing such as image registration, signal sampling, or model fitting, but the formula for signal conversion may need to be changed accordingly.
In conclusion, the proposed software for analyzing MRR data utilizes a robust image registration algorithm and an intuitive graphical user interface for efficient post-processing. The software greatly accelerates the analysis of MRR data and provides parameter estimates with high precision. Its potential benefit in other kidney diseases will be explored in future studies. The proposed software has the potential to increase the adoption of MRR as a clinical tool for the assessment of renal function.
Acknowledgments
Grant support: NIH R01 DK088375
REFERENCES
- 1.Rusinek H, Boykov Y, Kaur M, et al. Performance of an automated segmentation algorithm for 3D MR renography. Magn Reson Med. 2007;57:1159–1167. doi: 10.1002/mrm.21240. [DOI] [PubMed] [Google Scholar]
- 2.Song T, Lee VS, Rusinek H, Wong S, Laine AF. Integrated Four Dimensional Registration and Segmentation of Dynamic Renal MR Images. Med Image Comput Comput Assist Interv – MICCAI. 2005;3750:205–213. doi: 10.1007/11866763_93. [DOI] [PubMed] [Google Scholar]
- 3.Vivier PH, Storey P, Rusinek H, et al. Kidney function: glomerular filtration rate measurement with MR renography in patients with cirrhosis. Radiology. 2011;259:462–470. doi: 10.1148/radiol.11101338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley DL, Shurrab AaE, Cheung CM, Jones AP, Mamtora H, Kalra PA. Measurement of single kidney function using dynamic contrast-enhanced MRI: Comparison of two models in human subjects. J Magn Reson Imaging. 2006;24:1117–1123. doi: 10.1002/jmri.20699. [DOI] [PubMed] [Google Scholar]
- 5.Sourbron SP, Buckley DL. Tracer kinetic modelling in MRI: estimating perfusion and capillary permeability. Phys Med Biol. 2012;57:R1–R33. doi: 10.1088/0031-9155/57/2/R1. [DOI] [PubMed] [Google Scholar]
- 6.Sourbron SP, Michaely HJ, Reiser MF, Schoenberg SO. MRI-Measurement of Perfusion and Glomerular Filtration in the Human Kidney With a Separable Compartment Model. Invest Radiol. 2008;43:40–48. doi: 10.1097/RLI.0b013e31815597c5. [DOI] [PubMed] [Google Scholar]
- 7.Tofts PS, Brix G, Buckley DL, et al. Estimating Kinetic Parameters From Dynamic Contrast-Enhanced T1-Weighted MRI of a Diffusable Tracer: Standardized Quantities and Symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Sourbron SP, Buckley DL. Classic models for dynamic contrast-enhanced MRI. NMR Biomed. 2013;26:1004–1027. doi: 10.1002/nbm.2940. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JL, Rusinek H, Chandarana H, Lee VS. Functional MRI of the kidneys. J Magn Reson Imaging. 2013;37:282–293. doi: 10.1002/jmri.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siva S, Pham D, Gill S, et al. An analysis of respiratory induced kidney motion on four-dimensional computed tomography and its implications for stereotactic kidney radiotherapy. Radiat Oncol Investig. 2013;8:248. doi: 10.1186/1748-717X-8-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JL, Rusinek H, Bokacheva L, et al. Functional assessment of the kidney from magnetic resonance and computed tomography renography: impulse retention approach to a multicompartment model. Magn Reson Med. 2008;59:278–288. doi: 10.1002/mrm.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokacheva L, Rusinek H, Zhang JL, Lee VS. Assessment of renal function with dynamic contrast-enhanced MR imaging. Magn Reson Imaging Clin N Am. 2008;16:597–611. doi: 10.1016/j.mric.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zollner FG, Sance R, Rogelj P, et al. Assessment of 3D DCE-MRI of the kidneys using non-rigid image registration and segmentation of voxel time courses. Comput Med Imaging Graph. 2007;33:171–181. doi: 10.1016/j.compmedimag.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Tofts PS. The Measurement Process: MR Data Collection and Image Analysis. In: Tofts PS, editor. Quantitative MRI of the Brain: Measuring Changes Caused by Disease. The Atrium, Southern Gate, Chichester, West Sussex PO19. John Wiley & Sons, Ltd; England: 2003. pp. 17–54. [Google Scholar]
- 15.Melbourne A, Atkinson D, White MJ, Collins D, Leach M, Hawkes D. Registration of dynamic contrast-enhanced MRI using a progressive principal component registration (PPCR) Phys Med Biol. 2007;52:5147–5156. doi: 10.1088/0031-9155/52/17/003. [DOI] [PubMed] [Google Scholar]
- 16.Rofsky NM, Lee VS, Laub G, et al. Abdominal MR Imaging with a Volumetric Interpolated Breath-hold Examination. Radiology. 1999:212. doi: 10.1148/radiology.212.3.r99se34876. [DOI] [PubMed] [Google Scholar]
- 17.Levey AS, Greene T, Schluchter MD, et al. Glomerular filtration rate measurements in clinical trials. Modification of Diet in Renal Disease Study Group and the Diabetes Control and Complications Trial Research Group. J Am Soc Nephrol. 1993;4:1159–1171. doi: 10.1681/asn.v451159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skluzacek PA, Szewc RG, Nolan CRI, Riley DJ, Lee S, Pergola PE. Prediction of GFR in liver transplant candidates. Am J Kidney Dis. 2003;42:1169–1176. doi: 10.1053/j.ajkd.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Illingworth J, Kittler J. A survey of the Hough transform. Computer Vision, Graphics, and Image Processing. 1988;44:87–116. [Google Scholar]
- 20.Ballard DH. Generalizing the Hough Transform to Detect Arbitrary Shapes. Pattern Recognition. 1981;13:111–122. [Google Scholar]
- 21.Gerig G, Kikinis R, Kuoni W, Schulthess GKv, Kubler O. Semiautomated ROI Analysis in Dynamic MRI-Studies: Part I: Image Analysis Tools for Automatic Correction of Organ Displacements. IEEE Trans Med Imaging. 1992;11:221–232. [Google Scholar]
- 22.Zana F, Klein JC. A multimodal registration algorithm of eye fundus images using vessels detection and Hough transform. IEEE Trans Med Imaging. 1999;18:419–428. doi: 10.1109/42.774169. [DOI] [PubMed] [Google Scholar]
- 23.Canny J. A Computational Approach to Edge Detection. IEEE Trans Pattern Anal Mach Intell. 1986;8:679–698. [PubMed] [Google Scholar]
- 24.Mendichovszky IA, Cutajar M, Gordon I. Reproducibility of the aortic input function (AIF) derived from dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) of the kidneys in a volunteer study. Eur J Radiol. 2009;71:576–581. doi: 10.1016/j.ejrad.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 25.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Koh TS, Zhang JL, Ong CK, Shuter B. A biphasic parameter estimation method for quantitative analysis of dynamic renal scintigraphic data. Phys Med Biol. 2006;51:2857–2870. doi: 10.1088/0031-9155/51/11/012. [DOI] [PubMed] [Google Scholar]
- 27.Lee VS, Rusinek H, Bokacheva L, et al. Renal function measurements from MR renography and a simplified multicompartmental model. Am J Physiol. 2007;292:F1548–1559. doi: 10.1152/ajprenal.00347.2006. [DOI] [PubMed] [Google Scholar]
- 28.Glomerular Filtration Rate (GFR) National Kidney Foundation; 2015. [Google Scholar]
- 29.Zhang JL, Rusinek H, Conlin C, Lee VS. Feasibility of regional renal blood flow and vascular volume fraction measurement with cardiac-output corrected MR renography; Proceedings of the 20th Annual Meeting of ISMRM; Melbourne, Australia. 2012. [Google Scholar]
- 30.Chandarana H, Amarosa A, Huang WC, et al. High temporal resolution 3D gadolinium-enhanced dynamic MR imaging of renal tumors with pharmacokinetic modeling: preliminary observations. J Magn Reson Imaging. 2013;38:802–808. doi: 10.1002/jmri.24035. [DOI] [PubMed] [Google Scholar]