Abstract
We examined the microbial composition in the diseased lung and early-phase microbial cultures from the blood of a patient with a rapidly progressing fatal pulmonary illness. Although no microbes could be isolated from such cultures during the initial study, the HTS-microbiome study revealed the presence of a unique mixture of alphaproteobacteria, composed mainly of different families of Rhizobiales microbes. Microbial 16S rDNA sequences matching closely to Afipia cberi were identified mainly in the patient’s diseased lung tissue, but only rarely in the early-phase blood cultures. Conversely, the high abundance of sequences found in early-phase blood cultures of different broth media matched closely with those of the families Methylobacteriaceae, Phyllobacteriaceae and Sphingomonadaceae. The two species that successfully adapted to grow in a laboratory culture system were A. cberi and Mesorhizobium hominis, which eventually were isolated from a previously cryopreserved blood culture of SP4 broth. Many other species, including members of the Bradyrhizobiaceae and Phyllobacteriaceae families, and all members of the Methylobacteriaceae and Sphingomonadaceae families identified by HTS remained non-cultivated. We developed specific PCR primers and FISH probes, which detected the target Rhizobiales microbes in former blood cultures and autopsy lung tissues. It is unclear what role these Rhizobiales microbes might have played in the patient’s complex disease process. However, the above mentioned assays should help in rapidly detecting and identifying these previously unrecognized Rhizobiales microbes in patients.
Keywords: Microbiome, MPS, HTS, Rhizobiales, alphaproteobacteria, Bradyrhizobiaceae, Phyllobacteriaceae, Methylobacteriaceae, Sphingomonadaceae, Afipia, Mesorhizobium
Introduction
Non-biased massively parallel sequencing (MPS) studies using high-throughput sequencing (HTS) technology have provided powerful means for the sequence-based discovery and characterization of microbes. The new technique has been particularly useful for studying difficult-to-culture microbes [1,2] and whole microbial communities in examined samples without the need for laboratory cultivation or isolation of individual species [3,4]. The capabilities of HTS technology led us to re-examine some of our cryopreserved broths of blood cultures that were suspected of having low numbers of inactive unknown microbes, despite having negative microbial isolation results. We recently reported the discovery and isolation of previously unknown Rhizobiales microbes from formerly cryopreserved blood sample cultures from three patients with chronic forms of poorly defined illnesses, as well as a patient who had an acute fatal pulmonary illness [5,6]. In the process, genomic sequencing proved to be highly effective in the characterization of these novel microbes.
A blood sample was obtained shortly before a patient dying of an acute on-set of pulmonary illness and sent to the Armed Forces Institute of Pathology (AFIP) for diagnostic consultation in 1999. The AFIP laboratory set up cultures of the received blood sample in various broth media at the beginning of routine laboratory microbial work-ups to isolate and identify possible infectious microbial agents and could not isolate continuously growing microbial agents from any of these blood cultures in almost three months of careful follow-up [6]. As part of the infectious disease work-up, we obtained DNA from the early-phase blood cultures containing RPMI-1640, SP4 and BHI broth media supplemented with 5% irradiated fetal bovine serum (FBS) designated R5, SP4_F and BCF media. These DNA samples obtained from the first 1-3 weeks of blood cultures and DNA extracted from a small piece of this patient’s frozen lung tissue obtained at autopsy were previously used for polymerase chain reaction (PCR) studies to investigate possible mycoplasmal and/or bacterial infections. However, PCR assays targeting M. fermantans insertion-like element sequences [7,8] did not produce any positive signals. PCR amplification of 16S rRNA genes using universal bacterial primers [9,10] and DNA sequencing of the amplicons using the Sanger method yielded mixed, uninterpretable sequencing results. The DNA specimens prepared from diseased lung tissue obtained at autopsy and early-phase blood cultures were archived and stored frozen.
Two different species of novel Rhizobiales microbes, Afipia cberi of the Bradyrhizobiaceae family and Mesorhizobium hominis of the Phyllobacteriaceae family, were recently isolated and characterized in our laboratory by re-cultivating a formerly cryopreserved SP4 broth culture of a blood sample from this same patient with acute fatal pulmonary disease using modified SP4 broth and culture condition [6]. To better understand the microbial compositions of these samples, we conducted microbiomic study using the stored DNA specimens prepared previously from the diseased lung tissue and the early-phase blood cultures. Microbial 16S rRNA gene sequences of ~440 bps covering the species-specific hyper-variable regions (V6, V7 and V8) were amplified from the DNA samples using broad-range PCR primers corresponding to the highly conserved eubacteria sequences [9,10]. We examined the sequences of amplicons by both Sanger method and HTS technology [11,12]. Polymorphisms of 16S DNA sequences across the three hyper-variable regions were used for taxonomic identification and classification of various microbial populations present in the examined samples. In this study, we designed Bradyrhizobiaceae-specific and Mesorhizobium hominis-specific primers for PCR-based studies of the target Rhizobiales microbes in the diseased lung tissues and early-phase blood sample-derived cultures. In addition, we prepared a Cy3 labeled probe specific to the Bradyrhizobiaceae 16S rRNA for Fluorescence in Situ Hybridization (FISH) studies to directly identify Bradyrhizobiaceae microbes in the patient’s formalin-fixed paraffin-embedded (FFPE) autopsy lung samples.
Materials and methods
Ethics
Studying previously frozen DNA samples from cultures of the blood samples were conducted under the FDA Research Involving Human Subjects Committee (RIHSC) protocol #10-008B entitled “Detection of Infectious Agents in Previously Frozen Blood Samples from Patients with Various Illnesses and Healthy Blood Donors”. For this study, Dr. Zucker provided the patient’s original clinical information and Dr. Olesnicky provided the preserved lung sections from an autopsy performed in 1999. The IRB of Newark Beth Israel Medical Center (NBIMC) and Bamabas Health Corporate legal representatives reviewed the activity and determined that it did not meet the definition of human subject research that requires written informed consent from the patient. The clinical case was described in the study without revealing the patient’s identity.
DNA preparation from the original microbial cultures of blood samples in diagnostic studies
The laboratory received 600 to 700 μL of partially hemolyzed whole blood (WB) for the diagnostic study in 1999. The details of the original microbial cultures used for diagnostic studies were previously described [6]. In the original microbial study, 100 µL aliquots of WB were inoculated into each of the four tube cultures. Each culture tube contained 5 mL of 1) SP4 broth medium supplemented with 5% fetal bovine serum (FBS) (SP4_F), 2) RPMI-1640 broth medium supplemented with 5% FBS (R5), 3) BHI broth medium (BD Diagnostics, Circle Sparks, MD) and 4) BHI broth medium supplemented with 5% FBS (BCF). The cultures were kept at 32°C under aerobic conditions. When none of the microbial cultures (cultures kept at different temperatures under aerobic conditions and in GasPack) in the study showed evidence of microbial growth after one week of incubation, DNA was extracted from 1 mL of the cultures of SP4_F and R5 broth media using the DNeasy Blood & Tissue Kit (Qiagen). Similarly, DNA was extracted from 1 mL of the cultures of BHI and BCF broth media at week three, when no microbial growth was identified in the study. These DNA specimens were used for PCR to detect target mycoplasma DNA [7,8] and amplification of microbial 16S rRNA sequences [9,10] for Sanger sequencing before being stored at a temperature of -80°C. In the study, DNA was prepared from an equal amount of each control culture medium and water using the same Qiagen kit. As previously described [6], we continuously followed the microbial cultures for three months and routinely streaked their broth on SP4 agar plates and BHI agar plates without successful isolation of any microbes.
DNA preparation from autopsy lung tissue
DNA was extracted from a small piece of lung tissue obtained at autopsy using the DNeasy Blood & Tissue Kit (Qiagen) and stored at -80°C.
Amplification of 16S rRNA gene sequences and BAMQ5 and MSMQ8 target sequences
Microbial 16S rRNA genes were amplified from tested DNA samples using broad-range (pan-bacteria) primers p91F (5’-CAAATGAATTGACGGGGGC-3’) and p13B (5’-AGGCCCGGGAACGTATTCAC-3’) which were adapted with minor modifications from previous studies [9,10]. The region of the microbial 16S rRNA gene sequences (corresponding to positions 911 to 1390 of the E. coli sequence) that were amplified covers three species-specific hyper-variable regions (V6, V7 and V8). To amplify the Bradyrhizobiaceae-specific BAMQ#8 primers-targeting gene, the BAMQ#8F (5’-GGMCCGGTGATCTACACCTT-3’) and the BAMQ#8R (5’-AGCGTGGTCCAGGTCTTCTG-3’) primer set was used. The M. hominis-specific MSMQ#5 primers-targeting gene was amplified by the MSMQ8F (5’-CAATGCCGCCAGATCTGAAT-3’) and the MSMQ8R (5’-ACTCCGCAATGCCTCTCTTA-3’) primer set. PCR was conducted by an initial denaturation at 95°C for 5 min, 40 cycles at 95°C for 30 sec; 55°C for 30 sec; 72°C for 1 min and a final extension at 72°C for 7 min using the Hotstar Taq-Plus DNA polymerase (Qiagen).
Amplification of 16S rRNA gene and BAMQ#8 target gene amplicons with Nextera XT indexes
The amplicons of the 16S rRNA gene and BAMQ#8 primers-targeting region were amplified for 10 PCR cycles using the primer set p91F-XT (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCAAATGAATTGACGGGGGC-3’) and p13B-XT (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGAGGCCCGGGAACGTATTCAC-3’), and the primer set BAMQ#8F-XT (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGMCCGGTGATCTACACCTT-3’) and BAMQ#8R-XT (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGAGCGTGGTCCAGGTCTTCTG-3’), respectively. The second-step PCR for adding XT tags to the tails of the respective amplicons was conducted using HotStart-It FideliTaq master mix (Affymetrix). Amplicons of the second-step PCR were purified using the Agencourt AMPure XP system (Beckman Coulter) and indexed by PCR using indexing primers. The amplification system included in the Nextera XT kit (Illumina) was used to construct amplicon libraries. These libraries were purified using the Agencourt AMPure XP system before being applied to the Miseq sequencer (Illumina, San Diego, CA).
High-throughput sequencing
We previously described conducting 2×250 bp pair-end sequencing using the MiSeq sequencer [5,6]. To optimize the sequencing quality of the amplicons, 3-4 multiplexed amplicon DNA libraries with proper indexing were sequenced together with DNA samples from whole bacterial genomes to increase the overall complexity of sequencing reads in each run. The FASTQ file for each amplicon library was de-multiplexed according to the unique index present in the library construct.
Bioinformatic and metagenomic analyses
Total raw reads obtained from amplicons of each examined sample by MiSeq sequencing were first qualified by removing poor quality reads and trimming out the sequences of PCR primers using CLC Genomics Workbench (version 7.0.3) (www.clcbio.com). The trimmed pair-reads were then merged using the CLC “Merge Overlapping Pairs” function with the default parameters. The qualified merged reads (~440 bp) from each examined sample were then aligned with the reference sequences of A. cberi or M. hominis. The numbers of reads with 100%, 95% and 90% sequence similarity to the reference sequences were identified. Sequences of all of the qualified merged-reads from each sample were then blast searched against the 16S ribosomal RNA sequences in the NCBI database using Easyblast on NIH Biowulf (http://biowulf.nih.gov). The BlastN results were then imported into the MEGAN program (version 4.70.4) [11]. The distribution of the taxonomic classifications for all sequence reads was set at the family level, sorted by abundance and displayed as a bar graph. The qualified merged reads with identical sequences from each sample were calculated and consolidated as one read using the Fastx Collapser program available on EMBOSS [13]. A phylogenetic tree for the top number reads that match with Rhizobiales and their neighboring species was constructed using MEGA4 [14].
Retrieval of DNA from FFPE tissue sections
DNAs were retrieved from a de-waxed and rehydrated FFPE autopsy tissue section using the on-slide non-destructive molecule extraction method (NDME) as previously described [15].
Infection of A549 lung cell culture with A. cberi and M. hominis
The human lung carcinoma A549 cell line (ATCC CCL-185) was cultured in 75 cm2 flasks using RPMI-1640 broth medium supplemented with 10% of FBS. Cultures at 80-90% confluence were infected using ~108 A. cberi or M. hominis at 32°C for 30 min. The infected A549 cell cultures and the non-infected control cultures were then incubated in a CO2 incubator at 35°C for 36 hrs. The cells were harvested from culture flasks by scraping and pelleted by centrifugation. The cell pellets were fixed with 4% paraformaldehyde in PBS for 24 hrs and then processed in centrifuge tubes and embedded in paraffin blocks. The FFPE A549 cell pellets were sectioned and processed for FISH studies as were the FFPE autopsy lung tissues.
Fluorescence in situ hybridization
The 16S sequences of all of the different species of Afipia bacteria and B. japonicum were aligned using MEGA4 [14] against those of M. hominis, E. coli, P. aeruginosa, Klebsiella pneumonia, S. pyogenes and S. aureus. Regions that displayed high conservation among Bradyrhizobiaceae microbes and maximum sequence heterogeneity from non-Afipia bacteria including M. hominis were subjected to computational evaluation using an RNA-targeted FISH probe design using mathFISH [16]. The P1-2 FISH probe (5’-Cy3/ATCTCTGGTACCGGTCAT-3’) was selected based on its overall predicted lowest free energy in hybridization against the target 16S rRNA sequence. The P1-1 FISH probe (5’-Cy3/ATGACCGGTACCAGAGAT-3’) with reverse polarity to P1-2 was prepared and used as a negative control. Cy-3 labelling of the P1-2 probe and in situ hybridization were conducted as described [17]. Briefly, FFPE autopsy lung tissues and culture cell pellet sections were deparaffinized using five changes of xylene, followed by three changes of 100% ethanol and two changes of 95% ethanol. The de-waxed sections were incubated with 1 mg/mL lysozyme for 15 minutes at 37°C. Probes were pre-diluted in ddH2O to a concentration of 50 ng/mL. Half microliter (0.5 μL) of the probe was added to 50 μL of hybridization buffer (0.88 M NaCl, 20 mM Tris HCl, pH 7.4, 0.05% SDS) containing 1% formamide and applied onto each slide. The samples were incubated with the probe solution for two hours in a 50°C water bath. Then, the slides were washed in a pre-warmed wash buffer (900 mM NaCl, 100 mM Tris HCl, pH 7.4, 0.03% SDS) for six minutes at 50°C in a shaking water bath. The slides were washed with ddH2O and mounted using Prolong Gold antifade with DAPI (Invitrogen, Carlsbad, CA) before microscopic examination.
Results
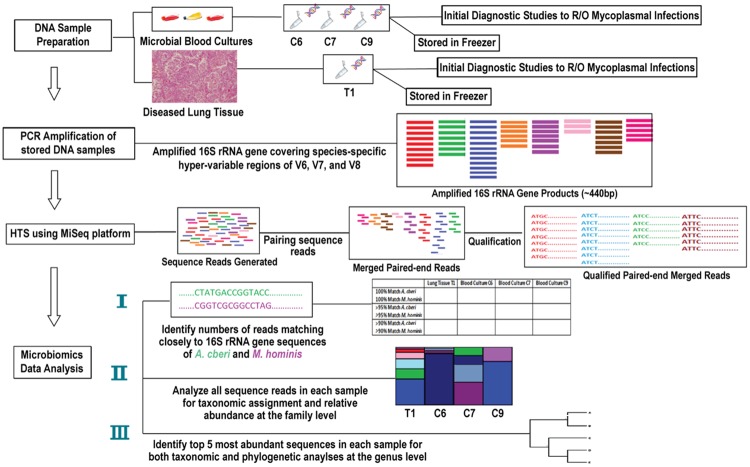
A schematic of the workflow to study microbial populations by HTS and taxonomical analysis of 16S rRNA gene sequence reads PCR-amplified from formerly stored frozen DNA samples of diseased lung tissue obtained at autopsy and early-phase (the first 1-3 weeks) cultures of the patient’s blood sample involving different broth media is shown in Figure 1.
Figure 1.
Schematic workflow to study microbial populations of the stored frozen DNA samples from diseased lung tissue and early-phase (the first 1-3 weeks) cultures of the patient’s blood sample involving different broth media by HTS.
Identification of microbial 16S rRNA gene sequences in diseased lung tissue and early-phase blood cultures matching closely to those of A. cberi and M. hominis
Table 1 reveals the sequencing data sets generated using the MiSeq sequencer for the 16S rRNA gene sequences amplified using pan-bacteria PCR primers [10] from the DNA samples prepared from the diseased lung tissue (T1 sample) and the early-phase blood cultures involving RPMI-1640 with 5% FBS (R5, C6 sample), SP4 with 5% FBS (SP4_F, C7 sample) and BHI with 5% FBS (BCF, C9 sample) broth media. The sequence reads matching closely to those of A. cberi and M. hominis, the two novel Rhizobiales microbes isolated from our re-initiated SP4 broth culture of the blood sample [6], were first sought in each of these examined samples using CLC Genomics Workbench (Table 1). Of the nearly two million total qualified 440 bp sequence reads identified in the T1 sample, ~4.8% of the sequences had a 100% match and ~18% of the sequences had a high similarity (>95% matching) to the A. cberi sequence. On the other hand, only a very small fraction (less than 0.03% of total reads) of sequences with close similarity (>95% match) to that of A. cberi were found in the early phase of blood cultures in the C6 or C7 samples (Table 1). There were few sequences with close similarity (>95%) to that of M. hominis in the T1 sample. The numbers of reads with sequences closely matching (>95%) to those of M. hominis were also low in blood cultures of either the C6 or C7 samples. However, nearly 27% of total qualified reads in the early-phase blood culture of the C9 sample had sequences with high similarity (>95% matching) to the 16S rRNA sequence of M. hominis; 0.45% had the exact (100% match) sequence of M. hominis.
Table 1.
Datasets generated by MPS for microbiomics analyses
| DNA Samples | T1 | C6 | C7 | C9 | Culture Broth Control | Culture Broth Control | Culture Broth Control | Blank/Reagent Control |
|---|---|---|---|---|---|---|---|---|
| Prepared from | Diseased lung tissue | Culture of R5 medium | Culture of SP4_F medium | Culture of BCF medium | R5 medium | SP4_F medium | BCF medium | H2O |
| Time of Culture | NA | Week 1 | Week 1 | Week 3 | NA | NA | NA | NA |
| Qualified raw reads | 5,573,804 | 4,692,312 | 6,988,910 | 3,581,211 | 626 | 2,453,560 | 771,844 | 538,744 |
| No. of reads with paired-ends | 3,933,870 (70.61%*) | 3,558,936 (75.86%*) | 3,933,342 (56.3%*) | 981,934 (27.4%*) | 12 (1.9%) | 704 (0.03%) | 968 (0.13%) | 182 (0.03%) |
| No. of paired-end reads with 440 bp joint sequences | 1,965,819 | 1,778,015 | 1,965,500 | 490,967 | 0 | 1 | 0 | 0 |
| Numbers of reads matching: | ||||||||
| 1) 100% to sequences of | ||||||||
| A. cberi | 95,247 (4.84%**) | 15 | 7 | 0 | 0 | 0 | 0 | 0 |
| M. hominis | 0 | 0 | 0 | 2.164 (0.45%**) | 0 | 0 | 0 | 0 |
| 2) >95% to sequences of | ||||||||
| A. cberi | 356,393 (18.13%**) | 243 | 599 | 0 | 0 | 0 | 0 | 0 |
| M. hominis | 0 | 21 | 12 | 129,667 (26.41%**) | 0 | 0 | 0 | 0 |
| 3) >90% to sequence of | ||||||||
| A. cberi | 392,413 (19.96%**) | 22,902 (1.29%**) | 207,281 (10.55%**) | 30 | 0 | 0 | 0 | 0 |
| M. hominis | 7 | 12,469 (0.70%**) | 31,506 (1.60%**) | 142,745 (29.08%**) | 0 | 0 | 0 | 0 |
Percentage of the number of reads with qualified paired-ends out of the numbers of qualified raw reads.
Percentage of the number of reads out of the numbers of 440 by merged sequences derived from paired-ends reads.
The percentage of <0.1% is not marked.
The concurrent 16S rRNA PCR products amplified in parallel from DNA preparations of R5, SP4_F and BCF culture broths as well as water control using primers p91F/p13B were minimal, barely detectable in gel electrophoresis. In the study, sequencing libraries were similarly prepared for the retrieved amplicon DNA of these control samples. Sequencing of these samples by MiSeq yielded very low numbers of paired reads (Table 1). Blast analysis revealed the low numbers of sequence reads with paired-ends as well as the unpaired raw sequence reads obtained from the culture broth and water controls were best-matched to some gamma-proteobacteria and Bacteroides. No sequence reads matching with those of Rhizobiales microbes were found.
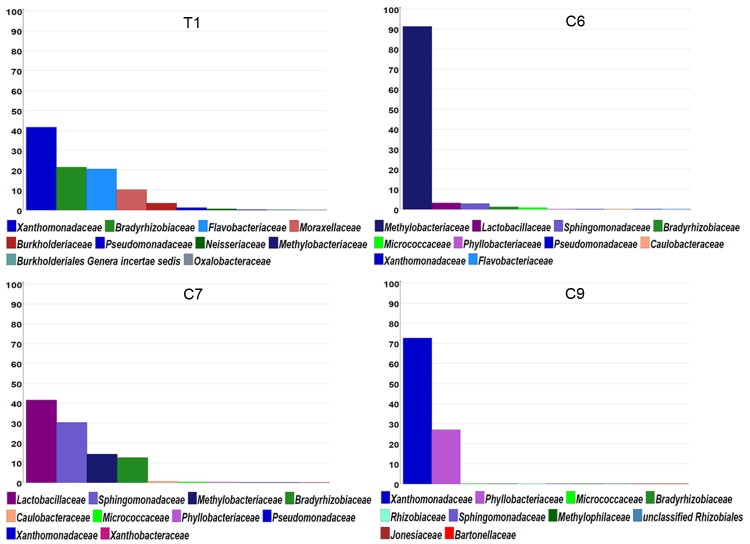
Taxonomic analyses of all the sequence reads obtained in each examined sample for relative abundance at the family level
Figure 2 reveals the MEGAN display for relative abundance of 16S rRNA gene sequences with taxonomic classification at the level of microbial families based on BlastN results of all the qualified sequence reads from each of the samples. In the T1 sample, ~40% of the total 16S rRNA gene sequence reads were found to correspond to those of Pseudomonadaceae/Xanthomonadaceae microbes. There was no significant difference of rRNA gene sequences in the amplified region for the two families of bacteria. However, more than 20% of the total qualified sequence reads (near two million) were classified as those of Bradyrhizobiaceae microbes (Figure 2), with ~4.8% of the reads with the sequence matching 100% with that of A. cberi (Table 1). In the C6 sample, ~90% of the total 16S rRNA gene sequence reads obtained were those from Methylobacteriaceae microbes. The second and third highest abundant sequence reads found in the C6 sample were those from Lactobacillaceae (~3%) and Sphingomonadaceae (~2%) microbes. In the C7 sample, most of the sequence reads found were those of Lactobacillaceae (~40%), Sphingomonadaceae (~30%), Methylobacteriaceae (~13%) and Bradyrhizobiaceae (~12%) microbes. In the C9 sample, similar to the finding with the T1 sample, the sequences of the most abundant reads (nearly 70% of total reads) belonged to those of Pseudomonadaceae/Xanthomonadaceae microbes. However, there were ~30% of reads with sequences belonging to Phyllobacteriaceae microbes (Figure 1) with a large majority of these reads (~27%) having sequences with very close similarity (>95%) to that of M. hominis (Table 1).
Figure 2.
The distribution of taxonomic classification at the level of Families for all qualified merged paired reads of 16S rRNA sequences amplified from the diseased lung tissue and early-stage blood cultures set up with R5 medium (C6), SP4_F medium (C7) and BCF medium (C9). Only the top 10 families for each sample are shown in the MEGAN display.
Taxonomic classification of the five most abundant sequences identified in each examined sample at the genus level for phylogenetic analysis
The individual sequences of the five top-number reads in each examined sample were specifically identified and taxonomically classified at the level of microbial genus through BlastN against the NCBI 16S bacteria ribosomal sequence database (Table 2). The sequence with the most abundant reads in the T1 sample (T1_1, ~12% of total sequence reads) was that of Pseudomonas genus microbes of the Pseudomonadaceae/Xanthomonadaceae family. The sequence with the second highest abundance of reads (T1_2, 4.86% of total reads) was that of Afipia genus microbes of the Bradyrhizobiaceae family, matching 100% to that of A. cberi (Table 1). The sequences with the most abundant reads in the early-phase blood cultures involving various media were those of genera in the Methylobacteriaceae (C6_1 and C6_2, Methylobacterium), Sphingomonadaceae (C7_1, Sphingomonas) and Phyllobacteriaceae (C9_2, Mesorhizobium) families. However, no microbes of the Methylobacteriaceae (C6_1 or C6_2) and Sphingomonadaceae families (C7_1) were isolated by culture. The high abundant read sequence of C9_2 Mesorhizobium microbes with 12.1% of the total reads amplified from the culture of BCF media had only one nucleotide mismatch with that of M. hominis isolated subsequently from the re-initiated cryopreserved SP4 broth culture. Figure 3 shows the phylogenetic relationship among Rhizobiales-related 16S rRNA gene sequences identified as the top-number reads in the diseased lung tissue and the early-phase cultures of blood samples. The phylogenetic tree also includes the related Rhizobiales microbes with their 16S rRNA genes sequences available in the NCBI database as reference species.
Table 2.
Identification of the 5 most abundant sequences reads in each examined sample for taxonomic classification
| DNA samples studied by MPS | Top 5 most abundant sequences | Percentage reads of total qualified 440 bp joint sequences | Taxonomic classification of the 16S rRNA gene sequences |
|---|---|---|---|
| Diseased Lung Tissue (T1 Sample) | TI_1 | 8.46% | Pseudomonas genus of Pseudomonadaceae |
| TI_2 | 4.84% | Afipia genus of Bradyrhizobiaceae | |
| TI_3 | 3.74% | Pseudomonas genus of Pseudomonadaceae | |
| TI_4 | 2.57% | Flavobacteriaceae | |
| TI_5 | 2.48% | Flavobacteriaceae | |
| Blood Culture of R5 Medium (C6 Sample) | C6_1 | 28.47% | Methylobacterium genus of Methylobacteriaceae |
| C6_2 | 2.19% | Methylobacterium genus of Methylobacteriaceae | |
| C6_3 | 0.47% | Sphingomonas genus of Sphingomonadaceae | |
| C6_4 | 0.31% | Lactobacillaceae | |
| C6_5 | 0.18% | Methylobacterium genus of Methylobacteriaceae | |
| Blood Culture of SP4_F Medium (C7 sample) | C7_1 | 10.30% | Sphingomonas genus of Sphingomonadaceae |
| C7_2 | 5.58% | Lactobacillaceae | |
| C7_3 | 2.77% | Lactobacillaceae | |
| C7_4 | 2.77% | Methylobacterium genus of Methylobacteriaceae | |
| C7_5 | 2.32% | Bradyrhizobium genus of Bradyrhizobiaceae | |
| Blood Culture of BCF Medium (C9 Sample) | C9_1 | 27.48% | Pseudomonas genus of Pseudomonadaceae |
| C9_2 | 12.09% | Mesorhizobium genus of Phyllobacteriaceae | |
| C9_3 | 3.73% | Pseudomonas genus of Pseudomonadaceae | |
| C9_4 | 0.72% | Pseudomonas genus of Pseudomonadaceae | |
| C9_5 | 0.52% | Pseudomonas genus of Pseudomonadaceae |
Figure 3.
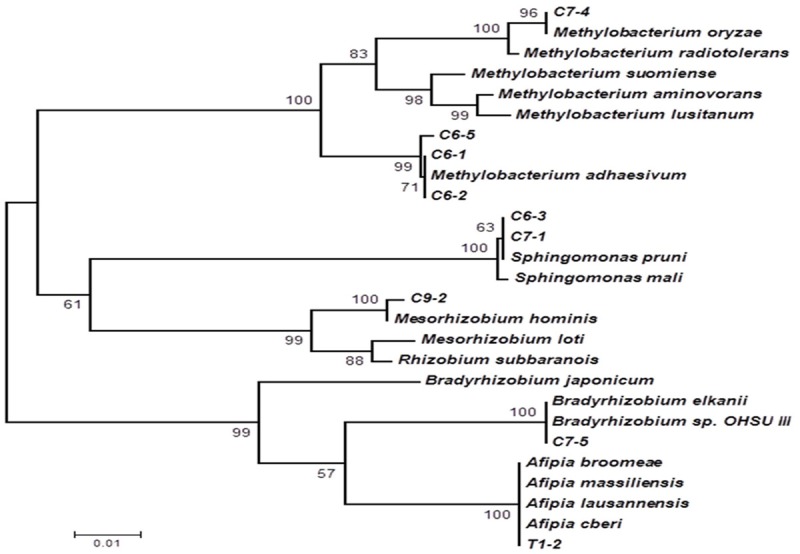
Phylogenetic analysis based on 16S rRNA gene sequences of the top-number reads amplified from the diseased lung tissue and early-stage blood cultures using the neighboring-joining method. Some of the neighboring Rhizobiales-related species: Methylobacterium oryzae (NR_043104.1), Methylobacterium radiotolerans (NR_112235.1), Methylobacterium suomiense (NR_041030.1), Methylobacterium aminovorans (NR_041025.1), Methylobacterium lusitanum (NR_112233.1), Methylobacterium adhaesivum (AB698708), Sphingomonas pruni (NR_113760.1), Sphingomonas mali (NR_113762.1), Mesorhizobium hominis (KF040403), Mesorhizobium loti (NC_002678.2), Rhizobium subbaraonis (NR_108508.1), Bradyrhizobium japonicum (NC_004463.1), Bradyrhizobium elkanii (AB931148), Bradyrhizobium sp. OHSU_III (KC677617), Afipia broomeae (NR_029200.1), Afipia massiliensis (NR_025646.1), Afipia lausannensis (DQ123622.1) and Afipia cberi (KF040404) are included as references. The scale bar unit represents the number of substitutions per 100 sites. Numerals indicate bootstrap percentages over 50 after 500 replications.
PCR detection of Bradyrhizobiaceae microbes and M. hominis in the diseased lung tissues from autopsy samples and early-phase blood cultures
Specific PCR primers were developed for the detection of Bradyrhizobiaceae microbes in the diseased lung tissues and in early-phase blood cultures. The Bradyrhizobiaceae-specific primer set (BAMQ#8) corresponds to highly conserved sequences in the genomes of microbes in the Bradyrhizobiaceae family, including all Afipia sp., B. japonicum and B. elkanii. However, the intervening sequences of the amplicons amplified using the BAMQ#8 primer set varied among different microbes of the Bradyrhizobiaceae family. As predicted from the genomic sequence datasets, BAMQ#8 amplified DNA sequences from Bradyrhizobiaceae, A. felis and B. japonicum with high sensitivity, but not DNAs from M. loti of the family of Phyllobacteriaceae, or non-alphaproteobacteria species. Figure 4A shows that primer set BAMQ#8 amplified specific sequences using DNA of A. cberi, with high sensitivity using 10 fg per PCR reaction, but not using genomic DNA from non-target M. hominis, E. coli, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pyogenes or human PBMC at more than 10 million-fold higher concentration of 500 ng per reaction. As shown in part (Figure 4A), primer set BAMQ#8 would not amplify genomic DNAs from 20 different non-target species of commonly-encountered Gram positive and Gram negative bacteria tested at the concentration 10 million-fold higher than target DNA of A. cberi. The PCR primers BAMQ#8 positively amplified the T1 sample and DNA specimens freshly retrieved from FFPE autopsy lung tissues of all four different lobes (RUL, RML, LUL, LLL) (Figure 4A). The C6 sample also tested positive by PCR. However, the size of the C6 amplicon appeared to be slightly larger than expected. DNA specimens prepared from the early-phase blood cultures involving SP4, BCF or BHI broth medium all tested negative by PCR using BAMQ#8 primers.
Figure 4.
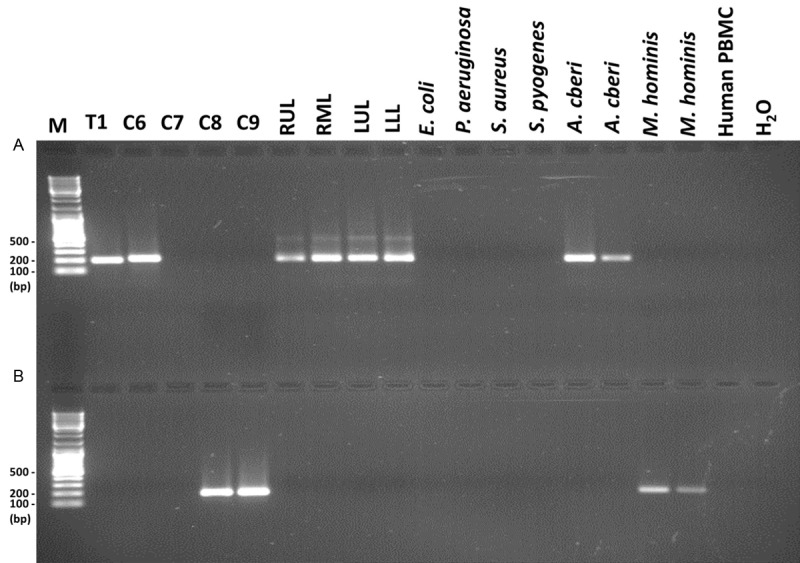
PCR studies of Bradyrhizobiaceae microbes and M. hominis in the patient’s autopsy lung tissues and early-phase cultures of the blood sample. The DNA samples from diseased lung tissue obtained at autopsy (T1) and early-phase blood cultures involving broth media R5 (C6), SP4 (C7), BHI (C8) and BCF (C9) as well as FFPE autopsy lung tissues right upper lobe, right middle lobe, left upper lob and left low lobe (RUL, RML, LUL and LLL) were PCR amplified using the BAMQ#8 primer set (upper panel, A) and the MSMQ#5 primer set (lower panel, B). Genomic DNAs of E. coli, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pyogenes and human PBMC (500 ng per reaction) were included in the negative control for both panels. In the upper panel, M. hominis DNAs (500 ng, duplicated) were used as negative controls and A. cberi DNAs (50 fg and 10 fg, respectively) as positive controls. In the lower panel, A. cberi DNAs (500 ng, duplicated) were used as negative controls and M. hominis DNAs (50 fg and 10 fg, respectively) as positive controls.
Specific primers were also developed for PCR detection of M. hominis of the Phyllobacteriaceae family in these samples. Figure 4B reveals that the M. hominis-specific primers MSMQ#5 amplified DNA sequences of M. hominis with high sensitivity using ~10 fg per reaction, but produced no positive products when using genomic DNA of A. cberi of the Bradyrhizobiaceae family at 10 million-fold higher concentration. The primer set MSMQ#5 also did not amplify genomic DNA from non-target E. coli, P. aeruginosa, S. aureus, S. pyogenes and human PBMC using 500 ng per reaction. As shown in part in Figure 4B, primer set BAMQ#8 would not amplify genomic DNAs from 20 different non-target species of commonly encountered Gram positive and Gram negative bacteria tested at the concentration 10 million-fold higher than target DNA of M. hominis. Consistent with the earlier results of the HTS-microbiome analyses, DNA previously prepared from the diseased lung tissue obtained at autopsy and DNA retrieved from the FFPE autopsy lung tissues tested negative by PCR using the M. hominis-specific primer set MSMQ#5. The early-phase blood cultures involving R5 and SP4 broth media also tested negative. On the other hand, the early-phase blood cultures involving BHI medium with or without FBS supplement (C9 and C8 samples) PCR-tested positive (Figure 4B).
Alignments of sequences amplified from the diseased lung tissues and the early-phase blood cultures using the PCR primer sets BAMQ#8 and MSMQ#5
All PCR amplicons obtained with the primer set BAMQ#8 in Figure 4A were retrieved from the agarose gel and initially subjected to Sanger sequencing. However, the amplicons obtained using DNAs extracted from different lobes of FFPE autopsied lung tissues had mixed populations of sequences and yielded uninterpretable signals by Sanger sequencing. We then conducted MPS to study the sequence composition of amplicons derived from 3 different lobes of the FFPE autopsy lung tissues. The top three reads of the BAMQ#8 amplicon sequences obtained with FFPE autopsy lung samples from each lobe of were aligned against A. cberi and amplicon sequences amplified from the T1 sample using the BAMQ#8 primer set. The sequence amplified by the BAMQ#8 primer set using the T1 sample appeared to be closely related to that of A. cberi, although some sequence differences were noticed. Various degrees of sequence variations were also found among the most highly abundant reads amplified from different lobes of the patient’s FFPE autopsy lung tissues and that of A. cberi (Figure 5). The amplicon obtained using the blood culture C6 sample was found to be 15 nucleotides longer and had many more sequence differences in comparison with A. cberi (Figure 5). Since an earlier analysis showed that more than 90% of microbial population in the early-phase blood culture of the C6 sample consistent of Methylobacteriaceae microbes (Figure 2), the BAMQ#8 PCR amplicon sequence of the C6 sample was likely derived from those of the Methylobacteriaceae family and not the Bradyrhizobiaceae family.
Figure 5.
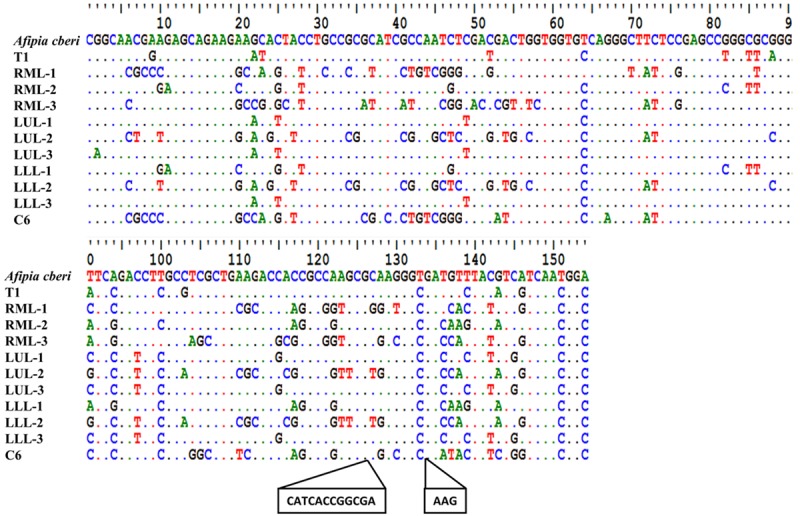
Alignment of amplicon sequences obtained using the BAMQ#8 primer set from different lobes of the patient’s diseased lung tissues and from early-phase R5 culture of a blood sample with that of A. cberi. Sequences of amplicons from T1 and C6 samples were obtained using the Sanger Method. Sequencing of amplicons from FFPE autopsy lung tissues, right middle lobe, left upper lobe and left lower lobe (RML, LUL and LLL), that failed using the Sanger method were sequenced through MPS using MiSeq. Sequences of the three most-abundant reads from each amplicon sequenced by MPS were used for the alignment. The sequence variations seen between the high-abundance reads from the autopsy lung tissues and those of A. cberi are marked. The amplicon of the C6 sample was found to be distinct with 15 nts longer in size compared to the expected amplicons from Bradyrhizobiaceae microbes. Since phylogenetic analysis showed that more than 90% of microbial populations in the C6 sample were Methylobacteriaceae microbes (Figure 1), the C6 amplicon sequence was likely from microbes of Methylobacteriaceae, not Bradyrhizobiaceae.
We constructed a phylogenetic tree based on the heterogeneity among the sequences with the most highly abundant reads amplified from different lobes of the diseased lung (Figure 6). Some of the BAMQ#8 PCR amplicon sequences were clustered with those of microbes in Afipia genus, while others were clustered with those in the Bradyrhizobium genus. But, many sequences with apparently more variations were not clustered with those in either genus of the Bradyrhizobiaceae family. The PCR amplicons amplified by primers MSMQ#5 from the early-phase blood culture involving BHI medium (C8) and BHI medium with FBS (C9) (Figure 4B) were also retrieved and subjected to Sanger sequencing. The amplicon sequences were highly uniform, matching 100% to the sequence of M. hominis.
Figure 6.
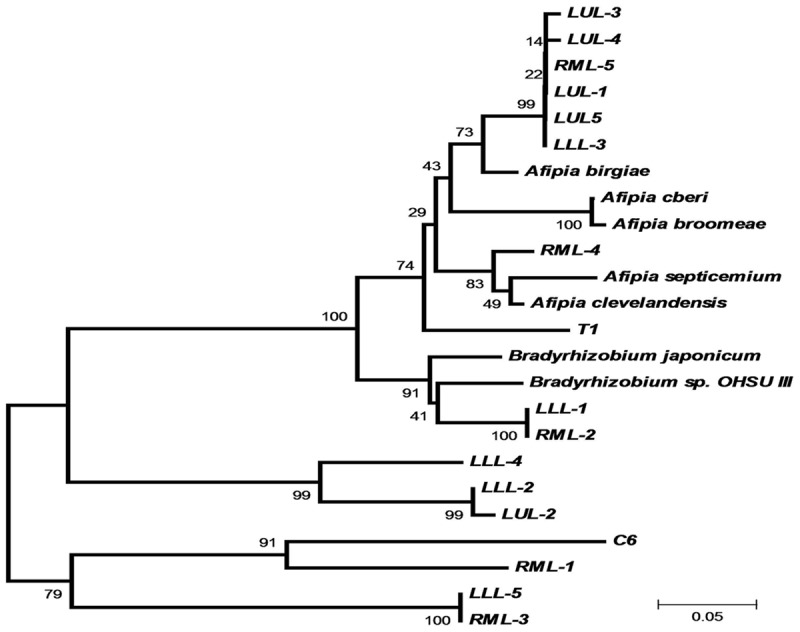
Phylogenetic analysis based on heterogeneity of sequences amplified from different lobes of the patient’s diseased lung tissues using the BAMQ#8 PCR primer set. Sequences of PCR amplicons from FFPE autopsy lung tissues using the BAMQ#8 primer set revealed a high heterogeneity. There were evidently wide variations among different strains or species members of Bradyrhizobiaceae microbes in the patient’s diseased lungs. None of the highly abundant or top-number sequence reads from different lobes of the patient’s diseased lung were the same as the sequence of A. cberi.
Direct identification of infections by Bradyrhizobiaceae microbes in the autopsy lung tissues by FISH
We designed a Bradyrhizobiaceae-specific DNA probe (P1-2) from a region of the 16S rRNA gene sequence that was highly conserved among microbes of the Bradyrhizobiaceae family and exhibited substantial differences from rRNA sequences of microbes of the Phyllobacteriaceae family for the FISH study. The selected Bradyrhizobiaceae-specific 16S rRNA gene sequence, as expected, also displayed major differences when compared with those of gamma-proteobacteria such as E. coli and P. aeruginosa. Figure 7 shows that the Cy3-labeled P1-2 probe hybridized specifically with A. cberi in FFPE cell pellets of cultured A549 human lung cells infected with A. cberi, but not with M. hominis in similarly processed FFPE cell pellets of A549 human lung cells infected by M. hominis. Nuclei of human cells in the sections of the FISH study were counterstained by DAPI (blue). Interestingly, A549 lung cells in cultures infected by A. cberi or M. hominis for 36 hours exhibited significant morphological differences (Figure 7A-D). A Cy3-labelled probe of P1-1 with the reversed polarity in the same 16S rRNA gene region of P1-2 was used as a negative control that produced no positive hybridization signal with A. cberi in infected A549 cells (Figure 7B). Figure 8 revealed that definite positive hybridization signals could be identified in all four different lobes of FFPE autopsy lung tissues studied by FISH using the Cy3-labelled P1-2 probe. The positive FISH signals found in areas of inflammation with or without apparent necrosis the autopsy lung tissues had characteristics highly similar to those elicited by A. cberi in A. cberi-infected A549 cells (Figure 8A-C). Occasional positive signals were found to be associated with inflammatory cells and degenerating pneumocytes in the intra-alveolar space (Figure 8D).
Figure 7.
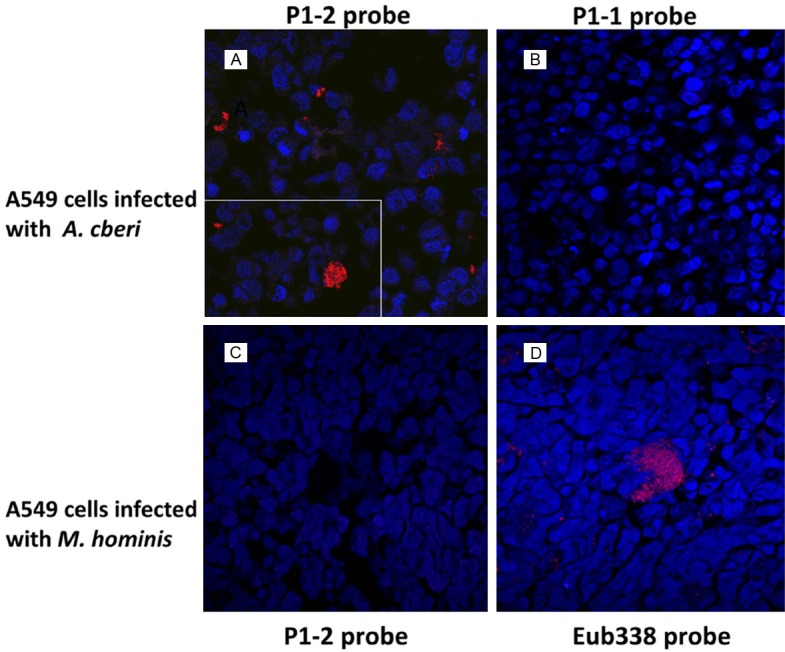
Detection and identification of Bradyrhizobiaceae microbes in infected cell cultures by FISH using the Bradyrhizobiaceae-specific P1-2 probe. The Cy-3 labelled P1-2 probe positively identified (red) A. cberi in sections of FFPE pellets of A549 lung cells experimentally infected by A. cberi (A). The inset shows positively hybridized clumping of A. cberi in the infected A549 cell culture. A FISH study using the Cy-3 labelled P1-1 probe with a sequence of reversed polarity compared to P1-2 as a negative control produced no positive signal in A. cberi-infected A549 lung cell cultures (B). The Cy-3 labelled P1-2 probe produced no positive signal in the A549 cell culture infected by M. hominis (C). In a parallel FISH study, single M. hominis cells and clumps of different sizes, in the A549 cell culture infected by M. hominis were positively identified (red) by the Cy-3 labelled universal eubacteria probe Eub338 [25] (D). Nuclei of A549 cells in all FISH study sections were counterstained (blue) with 4’, 6-diamidino-2-phenylindole (DAPI).
Figure 8.
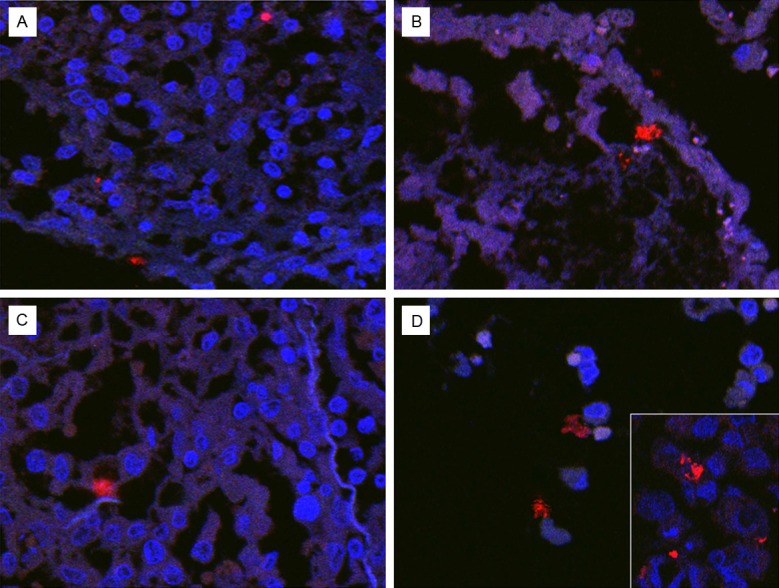
Detection and identification of Bradyrhizobiaceae microbes in FFPE autopsy lung tissues by FISH using the Cy-3 labelled Bradyrhizobiaceae-specific P1-2 probe. The Cy-3 positive signals (red) were identified in areas with many chronic inflammatory cells, their nuclei stained (blue) by DAPI (A & C). Positive signals could be found in areas with fulminant necrosis and alveolar degradation. Few viable cells with their nuclei positively stained by DAPI were present in this area (B). Positive signals were also seen associated with degenerating inflammatory cells and pneumocytes in the intra-alveolar space (D).
Discussion
The standard method used to identify microbial agent(s) in an infection or sepsis in a patient involves recovering microbe(s) from infected tissue(s) and/or a blood culture from the patient. However, this method can miss important insights into the possible diverse nature of microbes that exist in infected tissues and blood. The ability to grow in a laboratory culture system varies greatly among different microbes. MPS or HTS using various next generation sequencing (NGS) platforms has provided a critical means to assess in situ full microbial communities and to observe growing patterns of different microbial populations over time in the habitats, bypassing a cultivation-based bias [1-4]. A recent report summarized the tremendous growth in finding and understanding the presence of uncultivable microbial communities in the lungs of patients with various diseases and in healthy individuals using state-of-the-art culture-independent techniques [18]. In the present HTS-microbiome study using DNAs prepared from diseased lung tissue obtained at autopsy and from three early-phase cultures of blood samples involving different media, we did a comprehensive analysis of microbial populations present in a patient who had a rapidly-progressing pulmonary disease.
Although microbiome studies are highly effective for analyzing a complex microbial community, analysts need to be cautious. Reagent, water or other unknown system contamination in PCR can critically impact a sequence-based microbiome analysis. This contamination could be particularly challenging when studying low biomass samples without culture confirmation [19]. In this study, the concurrent 16S rRNA PCR products amplified in parallel from control culture broths of R5, SP4_F and BCF, as well as water samples using primers p91F/p13B, were minimal. Parallel sequencing of the PCR amplified products by MiSeq yielded only very low numbers of paired-ends reads (Table 1). The low numbers of sequence reads obtained with paired ends as well as the unpaired raw reads obtained from the culture broth and water controls best matched to some gamma-proteobacteria and Bacteroides. There were no sequence reads matching those of Rhizobiales microbes. In this particular case, the sequence-based finding has been confirmed by successful isolation of two new species of Rhizobiales microbes of the two specific families by culture from the blood sample [6]. Moreover, highly specific PCR primers and FISH probes developed from the genomic and 16S rRNA sequences of the Rhizobiales isolates clearly detected the target Rhizobiales microbes in former blood cultures and the diseased lung tissues (Figures 4A, 4B and 8).
Analysis of DNA specimens from both the autopsy lung tissue and the early-phase blood cultures involving BCF broth medium (C9) revealed that Pseudomonadaceae-related 16S rRNA gene sequences were the most highly abundant reads. Since the patient developed Pseudomonas infections at the terminal stage of illness after extracorporeal membrane oxygenation (ECMO) was initiated in the hospital, finding Pseudomonadaceae-related gene sequences by MPS was not unexpected. However, antibiotic treatments against infections of Pseudomonas-related microbes appeared to be effective. No Pseudomonas-related microbes were recovered in any of our previous microbial cultures, nor were they recovered in subsequent re-initiated culture using cryopreserved SP4 culture broth.
Importantly, the top sequence reads of Bradyrhizobiaceae found in the diseased lung (T1-2, 4.86% of total sequence reads) matched 100% to that of A. cberi, previously isolated from the re-initiated blood cultures using SP4 medium. However, the majority of sequences taxonomically classified as Bradyrhizobiaceae (~20% of total sequence reads) were those that had a >95% match with that of A. cberi (Table 1 and Figure 2). Microbes in the family of Bradyrhizobiaceae are known to have very high levels of homology in their 16S rRNA gene sequences, and thus are poor determinants of species or genera diversity [20,21]. Minor differences of 16S rRNA gene sequences in the Afipia genus or in the Bradyrhizobiaceae family could be significant. The differences of 16S rRNA gene sequences found in the diseased lung indicated that there were likely a mixed group of different Bradyrhizobiaceae microbes in addition to A. cberi. The marked sequence variations found in the PCR amplicons obtained with the diseased lung tissue (T1) and FFPE autopsied lung tissues (RML, LUL and LLL) using the Bradyrhizobiaceae-specific primer set BAMQ#8 (Figure 5) further provide strong evidence for a highly mixed group of different species of Bradyrhizobiaceae microbes to be present in the diseased lung.
One of the most abundant sequences (~30% of total reads) found in the early-phase blood culture set up with BCF medium (C9) belonged to microbes of Phyllobacteriaceae, another Rhizobiales family (Figure 3). These sequence reads matched closely (>95% of 440 bp, Table 1) with that of M. hominis isolated subsequently from the re-initiated SP4 broth culture of the patient’s blood sample. The sequence of the most abundant number reads identified in the C9 sample corresponding to those of Mesorhizobium in the family of Phyllobacteriaceae (C9_2 with 12.1% of total reads, Table 2) had only one nucleotide mismatch with that of M. hominis. However, the 16S rRNA sequence reads with a 100% match with that of M. hominis were identified as only 0.45% of total reads (Table 1). Therefore, there were also a mixed group of different species of Phyllobacteriaceae microbes, in addition to M. hominis, present in the patient’s blood. The M. hominis strain that successfully grew from the previously cryopreserved SP4 broth of blood culture [6] was one among several different strains/species of Phyllobacteriaceae microbes present in the patient’s blood. Differing from BAMQ#8 amplicons of Bradyrhizobiaceae microbes found in the diseased lung, MSMQ#5 amplicons of different strains/species of Phyllobacteriaceae microbes found in the patient’s blood samples appeared to be genetically more homogeneous. All of the amplicons obtained using the M. hominis-specific primer set MSMQ#5 have exactly the same sequence of M. hominis.
The present study shows that Rhizobiales microbes of different families clearly favor different culture media, even though they may still not be able to grow to the extent that can be recovered or isolated by culture. The Mesorhizobium microbes of the Phyllobacteriaceae family were found mainly in cultures involving BHI-based media (C8 and C9) (Figure 4B). In comparison, the large majority (more than 70%) of 16S rRNA gene sequence reads found in the blood culture using R5 medium (C6) were those of Methylobacteriaceae, another Rhizobiales family (Figure 1). The sequences with the two highest abundant reads corresponded to those of two different Methylobacterium genera of the Methylobacteriaceae family (C6_1 and C6_2, Table 2). On the other hand, the sequence with the most abundant reads in the early-phase blood culture using SP4 medium (C7_1) corresponded to those of the Sphingomonas genus of the Sphingomonadaceae family (Table 2). However, no species members of the Methylobacteriaceae or Sphingomonadaceae families were isolated from the re-initiated culture of cryopreserved SP4 culture broth. Instead, A. cberi of the Bradyrhizobiaceae family, and M. hominis of the Phyllobacteriaceae family, with their sequences found to be present only in very low numbers in the early-phase blood culture of SP4 medium, (15 or none after more than a million reads analyzed of the C7 sample), evidently continued to grow and were eventually isolated from the re-initiated culture after a long course of 2-3 months of incubation [6].
Highly sensitive Bradyrhizobiaceae-specific (BAMQ#8) and M. hominis-specific (MSMQ#5) PCR primers were developed to detect the target Rhizobiales microbes (at ≤ 10 fg/PCR reaction) in the patient’s diseased lung tissues and the early-stage blood cultures. In the study, as in part shown in Figure 4, the specificity of the primer sets designed from comparative bacteria genomic sequence alignments have been specifically examined experimentally against genomic DNAs from more than 20 different non-target species of commonly encountered bacteria at the concentration more than 10 million-fold higher than target DNA of A. cberi or M. hominis. In the future, these PCR primers may prove to be useful for molecular diagnostic studies and identification of Rhizobiales microbes in humans. Although the sequences obtained using the BAMQ#8 primer set are highly conserved among Bradyrhizobiaceae microbes, the intervening sequences of its amplicons were evidently heterogeneous among different species or strains (Figure 5). The heterogeneity of the amplicon sequences obtained using the BAMQ#8 primer set shows that they can be used to analyze the relatedness of different strains/species compositions of Bradyrhizobiaceae microbes (Figure 6). A Bradyrhizobiaceae-specific 16S rDNA probe was also developed (Figure 7) for direct identification of microbial infections in the diseased lung tissues by FISH (Figure 8). Definite positive hybridization signals could be identified in all four different lobes of FFPE autopsy lung tissues with or without apparent necrosis using the Cy3-labelled P1-2 probe. The RNA preservation in the FFPE autopsy tissue of more than 15 years could clearly result in the lower number of cells with positive hybridization signals identified.
The study revealed the presence of a mixed group of Rhizobiales microbes of different taxonomic families in the patient’s diseased lung and blood. Viral infections, particularly viremias involving RNA viruses, are often composed of swarms of viral variants [22,23]. The degrees of intra-host variability could be associated with the onset and/or severity of the viral disease. However, the presence of a mixed group of different families of Rhizobiales microbes has not been previously reported in humans. More HTS-microbiome studies in various diseased and/or non-diseased states in the future could provide insightful understanding of the nature of the mixed group of microbes [24]. Any role these Rhizobiales microbes might have played individually or together in any disease process will require further investigation. In this aspect, highly sensitive molecular assays and specific probes developed in the study are expected to be highly valuable in rapid detection and identification of these microbes in humans.
Acknowledgements
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). The authors thank Drs. Jakob Reiser and Syed R. Husain for critical review and editing of the manuscript. Mr. Philip Yang drew the schematic of workflow. The study is supported in part by an FDA Modernizing Science Grant.
Disclosure of conflict of interest
None.
References
- 1.Biesecker LG. Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Genet Med. 2012;14:393–398. doi: 10.1038/gim.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-R LM, Konstantinidis KT. Bypassing Cultivation To Identify Bacterial Species. Microbe. 2014;9:111–118. [Google Scholar]
- 3.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan XC, Huttenhower C. Chapter 12: Human microbiome analysis. PLoS Comput Biol. 2012;8:e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo SC, Hung GC, Li B, Lei H, Li T, Nagamine K, Zhang J, Tsai S, Bryant R. Isolation of novel Afipia septicemium and identification of previously unknown bacteria Bradyrhizobium sp. OHSU_III from blood of patients with poorly defined illnesses. PLoS One. 2013;8:e76142. doi: 10.1371/journal.pone.0076142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo SC, Li B, Hung GC, Lei H, Li T, Zhang J, Nagamine K, Tsai S, Zucker MJ, Olesnicky L. Isolation and characterization of two novel bacteria Afipia cberi and Mesorhizobium hominis from blood of a patient afflicted with fatal pulmonary illness. PLoS One. 2013;8:e82673. doi: 10.1371/journal.pone.0082673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson MS, Hayes MM, Wang RY, Armstrong D, Kundsin RB, Lo SC. Detection and isolation of Mycoplasma fermentans from urine of human immunodeficiency virus type 1-infected patients. Arch Pathol Lab Med. 1993;117:511–514. [PubMed] [Google Scholar]
- 8.Wang RY, Hu WS, Dawson MS, Shih JW, Lo SC. Selective detection of Mycoplasma fermentans by polymerase chain reaction and by using a nucleotide sequence within the insertion sequence-like element. J Clin Microbiol. 1992;30:245–248. doi: 10.1128/jcm.30.1.245-248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen K, Neimark H, Rumore P, Steinman CR. Broad range DNA probes for detecting and amplifying eubacterial nucleic acids. FEMS Microbiol Lett. 1989;48:19–24. doi: 10.1016/0378-1097(89)90139-0. [DOI] [PubMed] [Google Scholar]
- 10.Relman DA, Loutit JS, Schmidt TM, Falkow S, Tompkins LS. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 11.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James GS. PCR for Clinical Microbiology: An Australian and International Perspective. New York: Springer; 2010. Universal Bacterial Identification by PCR and DNA Sequencing of 16S rRNA Gene. [Google Scholar]
- 13.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 15.Chu WS, Liang Q, Liu J, Wei MQ, Winters M, Liotta L, Sandberg G, Gong M. A nondestructive molecule extraction method allowing morphological and molecular analyses using a single tissue section. Lab Invest. 2005;85:1416–1428. doi: 10.1038/labinvest.3700337. [DOI] [PubMed] [Google Scholar]
- 16.Yilmaz LS, Parnerkar S, Noguera DR. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl Environ Microbiol. 2011;77:1118–1122. doi: 10.1128/AEM.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiley JP, Caler EV. The lung microbiome. A new frontier in pulmonary medicine. Ann Am Thorac Soc. 2014;11(Suppl 1):S66–70. doi: 10.1513/AnnalsATS.201308-285MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox GE, Wisotzkey JD, Jurtshuk P Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 21.La Scola B, Mallet MN, Grimont PA, Raoult D. Description of Afipia birgiae sp. nov. and Afipia massiliensis sp. nov. and recognition of Afipia felis genospecies A. Int J Syst Evol Microbiol. 2002;52:1773–1782. doi: 10.1099/00207713-52-5-1773. [DOI] [PubMed] [Google Scholar]
- 22.Domingo E, Martin V, Perales C, Grande-Perez A, Garcia-Arriaza J, Arias A. Viruses as quasispecies: biological implications. Curr Top Microbiol Immunol. 2006;299:51–82. doi: 10.1007/3-540-26397-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito Y, Grivel JC, Margolis L. Real-time PCR assay of individual human immunodeficiency virus type 1 variants in coinfected human lymphoid tissues. J Clin Microbiol. 2003;41:2126–2131. doi: 10.1128/JCM.41.5.2126-2131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short FL, Murdoch SL, Ryan RP. Polybacterial human disease: the ills of social networking. Trends Microbiol. 2014;22:508–516. doi: 10.1016/j.tim.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]