Abstract
Purpose: Rhabdomyolysis is a threatening syndrome because it causes the breakdown of skeletal muscle. Muscle destruction leads to the release of myoglobin, intracellular proteins, and electrolytes into the circulation. The aim of this study was to investigate the differences in gene expression profiles and signaling pathways upon rhabdomyolysis-induced acute kidney injury (AKI). Methods: In this study, we used glycerol-induced renal injury as a model of rhabdomyolysis-induced AKI. We analyzed data and relevant information from the Gene Expression Omnibus database (No: GSE44925). The gene expression data for three untreated mice were compared to data for five mice with rhabdomyolysis-induced AKI. The expression profiling of the three untreated mice and the five rhabdomyolysis-induced AKI mice was performed using microarray analysis. We examined the levels of Cyp3a13, Rela, Aldh7a1, Jun, CD14. And Cdkn1a using RT-PCR to determine the accuracy of the microarray results. Results: The microarray analysis showed that there were 1050 downregulated and 659 upregulated genes in the rhabdomyolysis-induced AKI mice compared to the control group. The interactions of all differentially expressed genes in the Signal-Net were analyzed. Cyp3a13 and Rela had the most interactions with other genes. The data showed that Rela and Aldh7a1 were the key nodes and had important positions in the Signal-Net. The genes Jun, CD14, and Cdkn1a were also significantly upregulated. The pathway analysis classified the differentially expressed genes into 71 downregulated and 48 upregulated pathways including the PI3K/Akt, MAPK, and NF-κB signaling pathways. Conclusion: The results of this study indicate that the NF-κB, MAPK, PI3K/Akt, and apoptotic pathways are regulated in rhabdomyolysis-induced AKI.
Keywords: Rhabdomyolysis, acute kidney injury, gene expression profile, signal pathway
Introduction
Rhabdomyolysis occurs when compression, exercise, fever, medication, or inflammation cause skeletal muscle destruction and disintegration, which releases intracellular contents such as creatine kinase and myoglobin from myocytes into the circulation [1]. This destruction can cause coagulopathies, metabolic acidosis, hypovolemia, electrolyte disturbances and myoglobinuric renal failure [2]. Acute kidney injury (AKI) is a severe syndrome caused by rhabdomyolysis. The prognosis of this syndrome is poor after renal failure occurs. Approximately 10-40% of rhabdomyolysis cases result in AKI, and these cases account for 2-15% of all AKI cases [3].
The decreased delivery of blood to the glomerulus [4], reduced glomerular filtration rate [5], and the leakage of filtrate across a damaged tubular epithelium or tubular obstruction by myoglobin casts [6] may explain the pathophysiology of rhabdomyolysis-induced renal failure. The aggregation of renal tubular myoglobin contributes to the combined effects of hypovolemia, aciduria, and direct cytotoxicity [6]. Myoglobin proteins are freely filtered by the glomerulus, absorbed by tubular epithelial cells, and then degraded into heme and globin. These pigment molecules are directly toxic to tubular cells via the generation of oxygen free radicals and cast precipitation, which results in tubular obstruction. AKI is associated with rhabdomyolysis (RML) in one-third of patients with myoglobinuria due to the oxidative stress and myoglobin cast nephropathy [7,8]. Although renal failure can be explained by the deposition of myoglobin in the kidney, the mechanism of how the deposition occurs has not been elucidated [9].
The advent of gene expression microarrays allows us to develop a more comprehensive understanding of the variations in some diseases. A limited number of studies have utilized genome-wide technology to investigate changes in gene expression and signal pathways in rhabdomyolysis-induced AKI. Therefore, we examined the differences in gene expression between normal and rhabdomyolysis-induced AKI using gene expression microarray analyses.
Materials and methods
Animal group data selection
Our data and related information can be accessed at the Gene Expression Omnibus database. The accession number is GSE44925. The following two groups of animals were used for microarray analyses: (1) controls, n = 3 untreated mice; the GEO data set accession numbers are GSM1093973, GSM1093974, GSM1093975 and (2) AKI, n = 5 with rhabdomyolysis; the GEO data set accession numbers are GSM1093979, GSM10939780, GSM10939781, GSM10939782, and GSM10939783. To induce AKI, we injected 50% glycerol (0.05 mL per 10 g body weight) intramuscularly into the left hind limb under inhalation anesthesia [10]. The control mice (n = 3) were compared with the AKI mice (n = 5) at day 1.
Differential gene expression data in rhabdomyolysis-induced AKI
The genes differentially expressed between normal and rhabdomyolysis-induced AKI tissues were identified using the TwoClassDif method and Gene Ontology (GO)-Analysis. According to the Random Variance Model (RVM) t-test, we filtered the differentially expressed genes between the normal tissue and AKI groups. The RVM t-test was applied to filter the differentially expressed genes for the control and experimental groups because the RVM t-test can increase the degrees of freedom in small sample data sets. We selected the differentially expressed genes according to the P-value threshold after the significance analysis and FDR analysis were performed. A P-value < 0.05 was considered significant [11-13].
GO analysis was applied to analyze the main functions of the differentially expressed genes and is the key functional classification of NCBI. The GO analysis program can organize genes into hierarchical categories and identify gene regulatory networks based on the biological process and molecular function [14].
Differential pathway analysis based on microarray data
Pathway analysis was used to determine the pathways associated with the differentially expressed genes according to KEGG, Biocarta, and Reatome. We used the Fisher’s exact test and x2 test to select the significant pathways. The significance threshold was defined by the P-value and FDR. The enrichment value was calculated as previously described [15-17].
The Path-Net is the interaction net containing the pathways associated with the differentially expressed genes and was built according to the interactions among pathways of the KEGG database. The objective of building this net was to directly and systemically identify the interactions between the pathways. This strategy summarizes the pathway interactions associated with the differentially expressed genes in diseases and can be used to evaluate why certain pathways are activated [16].
Signal-Net analysis
The gene-gene interaction network was constructed based on the differentially expressed gene data. The program allows users to build and analyze molecular networks and to construct network maps. For example, if there is confirmed evidence that two genes interact, then an interaction edge is assigned between the two genes. The evidence source is compared to the KEGG interaction database. The networks are stored and presented as graphs, and the nodes can be genes, proteins, compounds, etc. The edges represent relationships types between the nodes and may signify activation or phosphorylation. The graph nature of networks allowed us to investigate the data using powerful tools implemented in R. We were also able to search for differentially expressed genes. Two nodes were connected when their corresponding encoded gene products were either directly connected or indirectly connected by a linker gene in the interaction network. The network of each gene was measured by counting the numbers of upstream and downstream genes or binding genes, which were expressed as in-degree and out-degree or degree, respectively. A higher degree indicates that the gene is regulating or being regulated by a greater number of genes. Thus, a high degree implies that the gene has a more important role in the signaling network. A P-value < 0.05 was considered significant. Gene interactions were drawn based on the data [18-22].
Experimental animals and procedures
Eight- to twelve-week-old C57BL/6 male mice were provided by the Experimental Animal Center of the Academy of Military Medical Sciences. The mice were housed at a stable temperature and a 12 hour light/dark cycle. We provided adequate standard rodent chow and water. The animals were acclimated for seven days before initiating the experiment. All animal protocols were approved by the Animal Ethics Committee of the Chinese PLA General Hospital and Military Medical College. C57BL/6 mice were deprived of water for 24 hours and then administered half the dose of glycerol (50% v/v in sterile saline) in each hindlimb muscle following mild sedation with pentobarbital. Our dose-dependent studies defined the optimal dose of glycerol as 8 mL/kg body weight. Twenty male mice were randomly assigned to either the control group (control, n = 10), or glycerol group (Gly, n = 10). The blood and kidney tissue were harvested for further processing 24 h after rhabdomyolysis.
Real-time PCR
The total cellular RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA). A TaqMan Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and a Gene Amp® PCR System 9700 platform (Applied Biosystems) were used to generate cDNA. The gene expression analysis was conducted by quantitative real-time PCR using SYBR Green Mastermix and a 7500 Real-time PCR System (Applied Biosystems). The results were analyzed using the 2 -ΔΔCT method and were normalized (n = 10) to Gly-induced renal injury (n = 10). The primers are shown in Table 1.
Table 1.
Gene names and the primers in Real-time PCR
| Gene Name | Fw (5’→3’) | Rev (5’→3’) | Products length |
|---|---|---|---|
| Cyp3a13 | CGCTTTGGTCCAGTGGGTAT | CAGGGCTCGGATTCTCTTCC | 78 bp |
| Rela | GGATTCCGGGCAGTGACG | CACGGCGCGCTAAAGTAAAG | 81 bp |
| Aldh7a1 | GAGCTCAGTATGTGGCGTGT | CAGAGTAGACATGTGGGCGG | 105 bp |
| Jun | TGGGCACATCACCACTACAC | TCTGGCTATGCAGTTCAGCC | 90 bp |
| CD14 | CCCTTGCCACCTTAGACCTG | GGAACTTGCTCGGACACAGA | 81 bp |
| Cdkn1a | AGGCCACCATTTGAGGATGG | CGCCTATGGAATGGCTTGGT | 73 bp |
Statistical analysis
All data are presented as the mean ± standard deviation. The statistical analyses were performed using SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA). The significance of experimental differences between groups was evaluated using Student’s t-test. A P-value < 0.05 indicated a significant difference.
Results
The differences in gene expression profiles and signaling pathways upon rhabdomyolysis-induced AKI
The DNA microarray data are shown in Table 2. There were 1709 genes upregulated or downregulated more than 1.5-fold compared with normal tissue. The genes were related to 129 signal pathways in rhabdomyolysis-induced AKI tissues. There were 734 differential pathways and crosslinked genes in the Signal-Net analysis of rhabdomyolysis-induced AKI tissues compared with that of normal tissue.
Table 2.
Overall differential gene expression profiles and signaling pathways in rhabdomyolysis-induced AKI
| Number of significant differential genes | Number of significant differential pathways | Number of genes crosslink in Signal-Net | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Group | Upper | Down | Total | Upper | Down | Total | |
| AKI/normal | 659 | 1050 | 1709 | 48 | 71 | 129 | 734 |
Differential gene expression profiles in rhabdomyolysis-induced AKI
The differences between the experimental group and control group were used to develop a clustering map by cluster analysis (Figure 1). The microarray analysis revealed changes in the expression of 1709 genes upon rhabdomyolysis-induced AKI, and there were 1050 downregulated genes and 659 upregulated genes. There are 10 significantly upregulated and downregulated genes, as shown in Tables 3 and 4, respectively. A gene interaction network was constructed based on the genes expressed upon rhabdomyolysis-induced AKI (Figure 2). Cyp3a13 (upregulated), Rela (upregulated), and Aldh7a1 (downregulated) were the three main genes identified by the Signal-Net analysis. We found that Cyp3a13 and Rela had the most interactions with other genes in the gene profiles. Additionally, Rela and Aldh7a1 were the key nodes and had connections with many genes in the Signal-Net.
Figure 1.
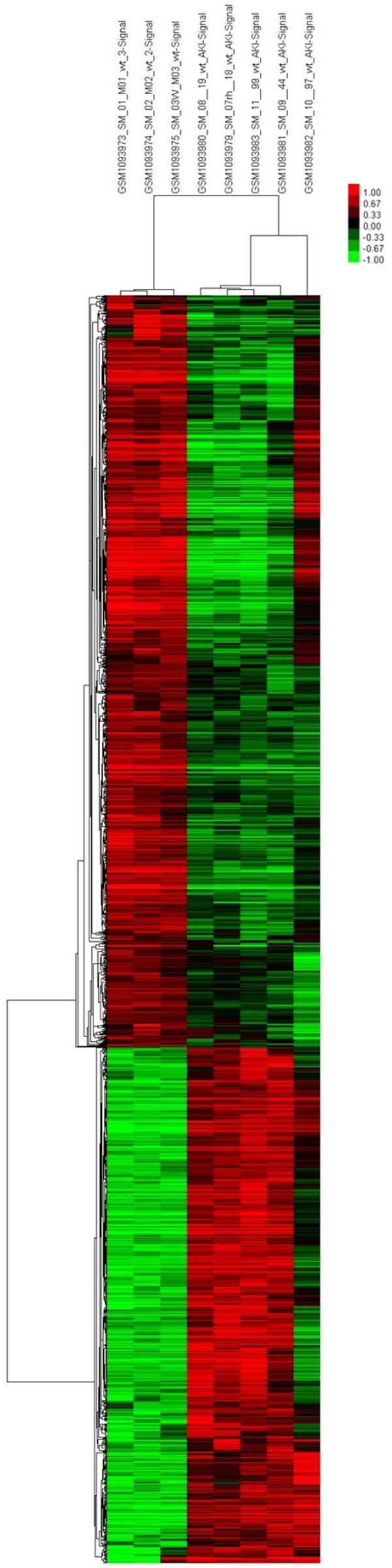
Cluster map of differentially expressed genes in rhabdomyolysis-induced AKI. X axis, the name of sample groups; Y axis, differentially expressed genes (red shows upregulated, green shows downregulated).
Table 3.
The top 10 differential upregulated genes expression in rhabdomyolysis-induced AKI
| Gene Symbol | P-value | Mean of intensities: exp | Mean of intensities: con | Fold-change |
|---|---|---|---|---|
| Mt2 | < 1.0×10-7 | 13688.24 | 5088.42 | 2.69 |
| Ptp4a1 | 4.0×10-7 | 2612.51 | 942.77 | 2.77 |
| Cast | 5.0×10-7 | 974.95 | 508.12 | 1.92 |
| Cdkn1a | 6.0×10-7 | 4171.6 | 316.51 | 13.18 |
| Slc38a2 | 9.0×10-7 | 3913.48 | 1443.53 | 2.71 |
| Lcn2 | 1.1×10-7 | 11277.44 | 381.88 | 29.53 |
| Cd14 | 1.4×10-7 | 369.94 | 98.72 | 3.75 |
| Serpina3n | 1.9×10-6 | 2312.12 | 135.76 | 17.03 |
| Mt1 | 2.9×10-6 | 5088.42 | 3128.59 | 2.85 |
| Fosl1 | 3.0×10-6 | 1565.68 | 105.57 | 14.83 |
Table 4.
The top 10 differential downregulated genes expression in rhabdomyolysis-induced AKI
| Gene Symbol | P-value | Mean of intensities: exp | Mean of intensities: con | Fold-change |
|---|---|---|---|---|
| Veph1 | 1.0×10-7 | 200.32 | 483.66 | 0.41 |
| Mir681 | 8.0×10-7 | 114.69 | 320.21 | 0.36 |
| Shank2 | 1.1×10-6 | 358.58 | 976.46 | 0.37 |
| Ipcef1 | 1.1×10-6 | 52.94 | 181.24 | 0.29 |
| Bbs9 | 1.4×10-6 | 326.17 | 532.6 | 0.61 |
| Pcmtd2 | 1.6×10-6 | 273.71 | 569.91 | 0.48 |
| Stard8 | 1.7×10-6 | 290.87 | 812.47 | 0.36 |
| Mbtps1 | 1.8×10-6 | 416.48 | 727.95 | 0.57 |
| 6720456H20Rik | 1.9×10-6 | 297.07 | 626.38 | 0.47 |
| 2310081J21Rik | 2.1×10-6 | 83.45 | 173.2 | 0.48 |
Figure 2.
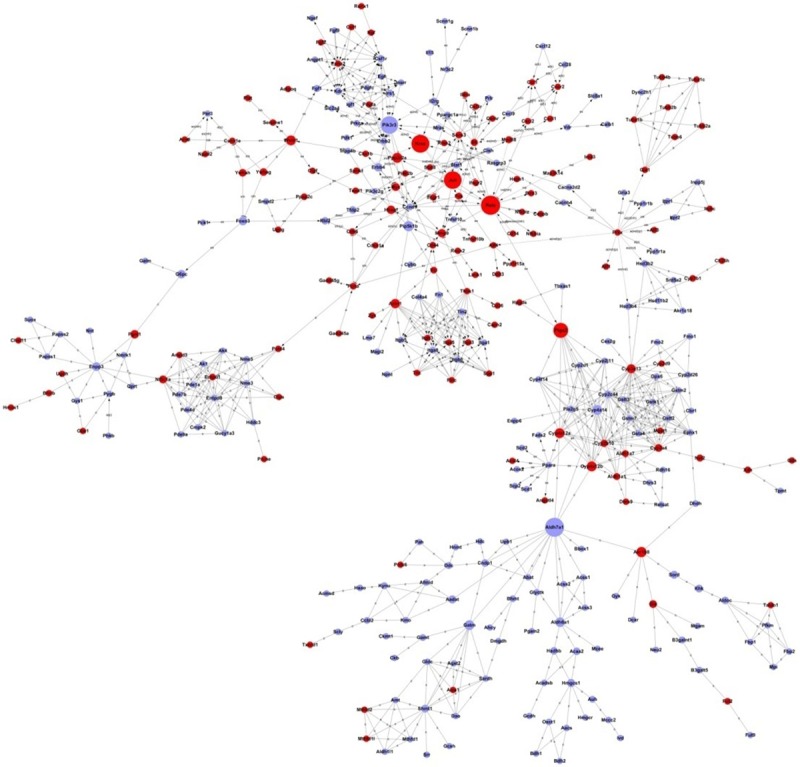
Signal-Net analysis of differentially expressed genes in rhabdomyolysis-induced AKI. Circles represent genes (red shows upregulated, blue shows downregulated). The size of the circle represents the number of other genes that interact with this gene. Lines indicate interactions between genes Pathways in rhabdomyolysis-induced AKI.
Differentially represented pathways in rhabdomyolysis-induced AKI
The signaling pathways implicated in rhabdomyolysis-induced AKI were analyzed based on the functions and interactions of the differentially expressed genes. Figure 3 shows the interaction net for the pathways associated with differentially expressed genes in the Path-Net analysis. These interactions represent the pathways that are directly and systemically involved in rhabdomyolysis-induced AKI.
Figure 3.
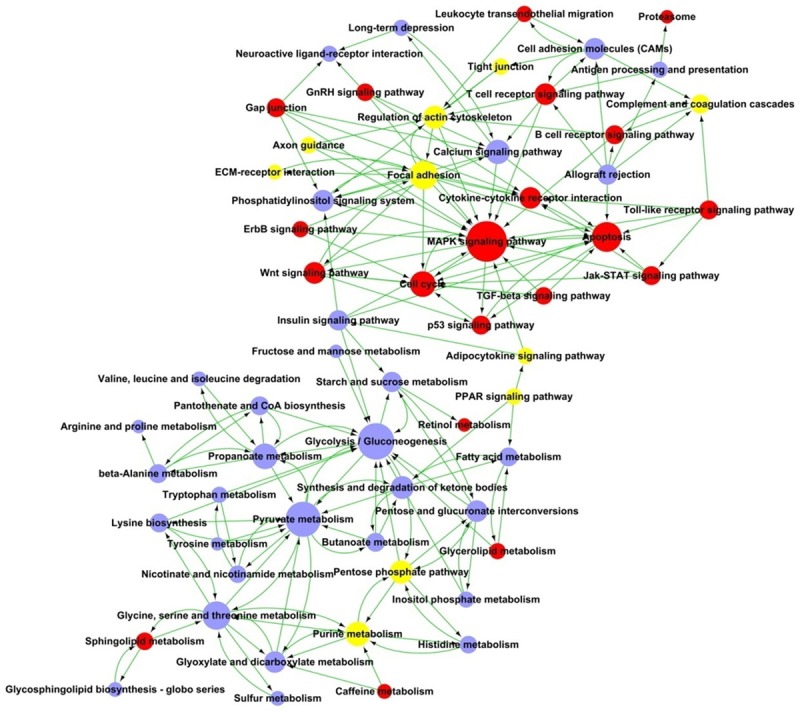
Path-Net analysis of differentially signaling pathways in rhabdomyolysis-induced AKI. Circles represent pathways (red shows upregulated, blue shows downregulated, yellow represents for both up- and down-regulated pathways). The size of the circle represents the number of other genes that interact with this gene. Lines indicate interactions between pathways in rhabdomyolysis-induced AKI.
As shown in Figure 3, the MAPK signaling pathway is the most significantly altered pathway in rhabdomyolysis-induced AKI, and it has an important position within the Path-Net. Furthermore, glycolysis, gluconeogenesis, pyruvate metabolism, and apoptosis pathways are additional pathways involved in rhabdomyolysis-induced AKI.
We also performed GO and KEGG pathway analyses to classify the genes that are differentially expressed in rhabdomyolysis-induced AKI. The results indicated that 71 pathways were downregulated, and 48 pathways were upregulated. Figure 4 shows the signaling pathways involved in rhabdomyolysis-induced AKI. The pathways were then divided into upregulated and downregulated pathways.
Figure 4.
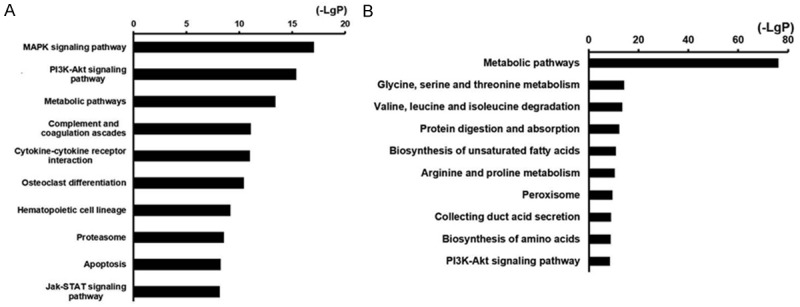
Histogram of top10 signal pathways that were significantly different in rhabdomyolysis-induced AKI showing (A) upregulated pathways; (B) Downregulated pathways. X axis, negative logarithm of the P value (-LgP); Y axis, the name of the pathway. The larger the value, the smaller the P value.
As shown in Figure 4, metabolic pathways are both upregulated and downregulated. However, the downregulated metabolic pathways are more important. The MAPK signaling pathway is the most strongly upregulated pathway in rhabdomyolysis-induced AKI. The PI3K/Akt signaling pathway is the second most strongly upregulated pathway.
Use of an established glycerol-induced AKI animal model for verification
We established a glycerol-induced AKI animal model to investigate the accuracy of the microarray results. Glycerol administration resulted in significant increases in the serum creatinine (Scr) and blood urea nitrogen (BUN) levels in the Gly group compared with the control group (Table 5). We also examined the levels of Cyp3a13, Rela, Aldh7a1, Jun, CD14, and Cdkn1a using RT-PCR. Our results showed that compared with the control group, the mRNA expression levels of Aldh7a1 were significantly decreased. The mRNA expression levels of Cyp3a13, Rela, Jun, CD14, and Cdkn1a were significantly increased (Figure 5).
Table 5.
Serum creatinine (Scr) and blood urea nitrogen (BUN) concentrations
| Control (x̅ ± SEM) | Gly (x̅ ± SEM) | |
|---|---|---|
| Scr (mg/dl) | 0.19±0.05 | 1.17±0.21 |
| BUN (mg/dl) | 25.6±3.94 | 119.8±10.87 |
Figure 5.
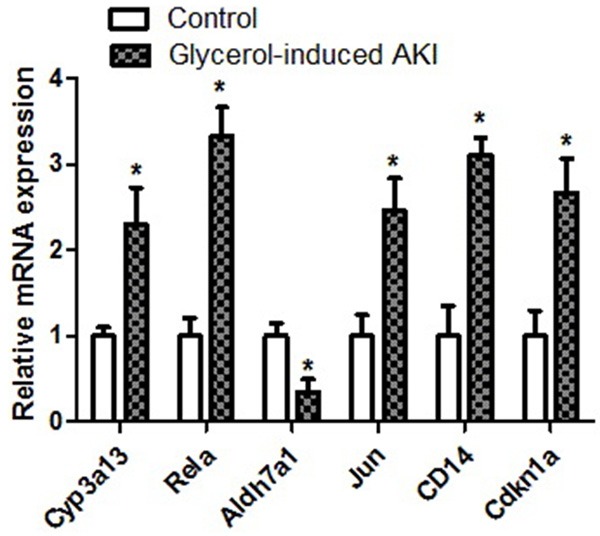
The results showed that compared with the control group, the mRNA expression levels of Aldh7a1 was significantly decreased and the mRNA expression levels of Cyp3a13, Rela, Jun, CD14 and Cdkn1a were significantly increased.
Discussion
Rhabdomyolysis causes high mortality and is accompanied by a series of clinical and laboratory findings resulting from exposure to the lysis of striated muscle cells [23]. While technological advances enable treatments for many diseases, we will continue to encounter traumatic rhabdomyolysis as a result of wars, terror attacks, earthquakes, and other natural disasters [24]. Thus, studies examining the prevention of rhabdomyolysis-related organ damage and AKI will continue. Hypertonic glycerol injection in mice is an established method used to study the pathogenesis of rhabdomyolysis-induced AKI [25]. Therefore, we also elected to use this method. Pronounced necrosis and cast formation in the tubules of the rhabdomyolysis group supported the development of rhabdomyolysis-induced AKI.
The pathogenesis of rhabdomyolysis-induced AKI needs to be further studied. Myoglobulin released from the lysed striated muscle cells reaches the tubules and produces casts by combining with Tamm-Harsfall protein. This process causes cast-dependent myoglobulin to increase nitric oxide (NO) consumption and vasoconstriction. Additionally, there are increases in the formation of reactive oxygen species (ROS) and a series of mechanisms involved in apoptosis have been implicated [26]. Studies have shown that apoptosis is activated by the activation of both the intrinsic and extrinsic apoptotic pathways.
We carefully studied the variations in gene expression profiles and signal pathways in rhabdomyolysis-induced AKI using microarray techniques. The Signal-Net analysis included many genes involved in rhabdomyolysis-induced AKI because of the relative numbers of differentially expressed genes. This result suggests the mechanisms of rhabdomyolysis-induced AKI are complex. The Signal-Net analysis showed that Cyp3a13 and Rela interacted most with other genes. Rela and Aldh7a1 were the key nodes and had strong connections throughout the Signal-Net. CYP3a13 is widely distributed in liver tissue. In addition to liver expression, CYP3a13 is also present in kidney, lung, and brain. CYP3a13 plays a role in altering drug metabolism in humans and many of the novel synthesized ligands/drugs targeting nuclear receptors influence CYP3A gene expression [27]. However, there are no data describing the role in rhabdomyolysis-induced AKI. Mammalian Aldh7a1 is an enzyme that protects against the effects of salinity, dehydration, and osmotic stress. The Aldh7a1 protein is expressed in the liver, kidney, brain, and pancreas based on mouse histological distribution studies. Aldh7a1 protein was found in the cytosol, mitochondria, and nucleus. Thus, Aldh7a1 is a unique aldehyde dehydrogenase enzyme [28]. Additional Aldh7a1 studies in rhabdomyolysis-induced AKI are ongoing.
The mechanisms of rhabdomyolysis-induced AKI are complex and many signaling pathways are altered as a consequence of gene expression changes. Our results indicate that Rela not only had the most relationships with other genes but also had one of the most important positions in the entire Signal-Net. Our results also suggest the NF-κB signaling pathway played an important role in rhabdomyolysis-induced AKI. Rela/p65 is a REL-associated protein involved in NF-κB heterodimer formation, nuclear translocation, and activation. Rela is a member of the NF-κB family, which is one of the most essential transcription factors. Rela is the prototypical heterodimer complex partner with NF-κB. Together with p50, Rela/p65 interacts with various proteins in both the cytoplasm and in the nucleus during the process of classical NF-κB activation and nuclear translocation [29]. Free radicals released by damaged tissues and inflammatory cells induce NF-κB activation [30]. Thus, it has been suggested that activation of the NF-κB pathway and Rela might play a role in the progression of rhabdomyolysis-induced AKI.
The most upregulated pathway in rhabdomyolysis-induced AKI was the MAPK signaling pathway. From the Path-Net analysis, we concluded that the MAPK signaling pathway had the most relationships with other signaling pathways. Moreover, the JUN gene was significantly upregulated in the Signal-Net analysis. CD14 is one of the top 10 differentially upregulated genes in rhabdomyolysis-induced AKI. Mitogen-activated protein kinases (MAPKs) are a one member of serine/threonine protein kinases. The function of MAPKs is to respond to extracellular stimuli and regulate a variety of cellular activities such as differentiation, proliferation, cell survival, and apoptosis [31]. The MAPK family includes the extracellular-signal-regulated kinases (ERK), c-Jun N-terminal kinases (JNK), and p38. MAPKs play an important role in the regulation of cell death and survival. It has been reported that several anti-cancer drugs kill Renal-cell carcinoma (RCC) cells by altering the activities of ERK and JNK [32,33]. CD14 is a GPI-anchored protein expressed by macrophages (MΦ) and neutrophils. In response to danger-associated molecular patterns (DAMPs), CD14 facilitates pro- and anti-inflammatory cytokine production [34]. Lin et al. indicated that p38 MAPK-specific inhibitor could block the expression of TLR2/4 or CD14 and the activity of PU.1 in human monocytes. This finding suggests p38 MAPK and ERK1/2 may be the upstream signals inhibited by E. chaffeensis [35]. c-Jun protein is encoded by the JUN gene in humans. The ERK pathway can regulate the activity of c-jun protein. Constitutively active ERK increases c-jun transcription. This result indicates that c-jun was activated and that its downstream targets could increase the activity of the JNK pathway. The activation of JNK signaling can play a role in the regulation of c-jun activity [36]. After renal injury, MAPKs play an important role in determining the fate of renal tubular cells [37]. Overwhelming evidence has unequivocally defined the role of the JNKs, especially JNK1, in cell apoptosis or tumor suppression [38-40]. Recent studies have indicated that JNK activates the intrinsic apoptotic pathway in response to cellular stress [41-44]. Our results indicate that MAPKs and apoptosis activation were upregulated in rhabdomyolysis-induced AKI and that MAPKs contributed to the activation of the apoptotic pathway.
The data suggest that the PI3K/Akt signaling pathway plays an important role in rhabdomyolysis-induced AKI. The Path-Net analysis showed that PI3K/Akt signaling was the second most upregulated and the tenth most downregulated pathway in rhabdomyolysis-induced AKI. The top 10 differentially upregulated genes in rhabdomyolysis-induced AKI showed that Cdkn1a (p21) was the fourth most upregulated gene. The Cdkn1a (p21) protein plays a complex role in cell-cycle control, DNA repair, and blocks apoptosis [45]. In terms of cell proliferation, Akt-dependent phosphorylation of p21WAF/Cip1 reduces the inhibitory effect of p21WAF/Cip1 [46]. Heliez C et al. reported that Akt participated in p21WAF/Cip1 regulation by phosphorylation [47]. The PI3K/Akt signaling pathway is one of the most important survival pathways within cells [48]. Various types of cellular stimulation can activate the PI3K/Akt signaling pathway. The PI3K/Akt pathway also regulates cell growth, proliferation, and the cell cycle [49]. Although the pathogenesis of rhabdomyolysis-induced AKI is unclear, myoglobin released from muscle cells plays major roles in renal injury. It is known that myoglobin has direct tubular toxicity, causes vasoconstriction, and increases the formation of ROS, NO consumption, and various mechanisms involved in apoptosis [50,51]. The PI3K/Akt signaling pathway plays an important role in preventing the apoptosis induced by oxidative stress [52,53]. In addition, members of the MAPK family function in the response to oxidative stress [54]. MAPKs are major mediators of signal transduction and play an important role in the regulation of cell growth, proliferation, differentiation, and apoptosis. We speculate that the upregulation of apoptosis-related genes stimulated the PI3K/Akt signaling pathway. The PI3K/Akt pathway is an important signaling pathway for cell survival [55], and phosphorylated Akt can decrease apoptosis [56]. However, our analysis concluded that the PI3K/Akt pathway cannot prevent the effects of apoptosis.
The pathway analysis showed that metabolic pathways were the most strongly downregulated and third most strongly upregulated pathways in rhabdomyolysis-induced AKI. Our results suggested that metabolic pathways play an important role in rhabdomyolysis-induced AKI. Metabolic pathways involved in rhabdomyolysis-induced AKI are associated with metabolic disorders.
In conclusion, rhabdomyolysis-induced AKI will continue to be a major health problem as long as natural disasters and injury persist. The exact mechanisms of rhabdomyolysis-induced AKI need to be further elucidated. Our results show that differential gene expression occurs in rhabdomyolysis-induced AKI. These genes encode proteins involved in different signaling pathways. The disruption of these pathways plays an important role in rhabdomyolysis-induced AKI. Our findings provide a platform for the further characterization of the genes implicated in rhabdomyolysis-induced AKI and may provide new insights into the mechanisms of rhabdomyolysis-induced AKI.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (No. 2011AA020115), Major State Basic Research Development Program of China (No. 2014CBA02005), Chinese National Natural Sciences Foundation (No. 81470949; No. 31170810; No. 81370785), the Medical Technology Youth Training Project of PLA (No. 13QNP180) and Beijing NOVA Program (No. Z121107002512078). Microarray data were analyzed by Genminix Informatics (Shanghai, China).
Disclosure of conflict of interest
None.
References
- 1.Zager RA. Rhabdomyolysis and myohemoglobinuric acute renal failure. Kidney Int. 1996;49:314–326. doi: 10.1038/ki.1996.48. [DOI] [PubMed] [Google Scholar]
- 2.Dayer-Berenson L. Rhabdomyolysis: a comprehensive guide. ANNA J. 1994;21:15–18. quiz 19-20. [PubMed] [Google Scholar]
- 3.Beetham R. Biochemical investigation of suspected rhabdomyolysis. Ann Clin Biochem. 2000;37:581–587. doi: 10.1258/0004563001899870. [DOI] [PubMed] [Google Scholar]
- 4.Chedru MF, Baethke R, Oken DE. Renal cortical blood flow and glomerular filtration in myohemoglobinuric acute renal failure. Kidney Int. 1972;1:232–239. doi: 10.1038/ki.1972.33. [DOI] [PubMed] [Google Scholar]
- 5.Cox JW, Baehler RW, Sharma H, O’Dorisio T, Osgood RW, Stein JH, Ferris TF. Studies of the mechanism of oliguria in a model of unilateral acute renal failure. J Clin Invest. 1974;53:1546–1558. doi: 10.1172/JCI107705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan FY. Rhabdomyolysis: a review of the literature. Neth J Med. 2009;67:272–283. [PubMed] [Google Scholar]
- 7.Twardowski ZJ. Treatment time and ultrafiltration rate are more important in dialysis prescription than small molecule clearance. Blood Purif. 2007;25:90–98. doi: 10.1159/000096403. [DOI] [PubMed] [Google Scholar]
- 8.Katzarski KS, Charra B, Luik AJ, Nisell J, Divino Filho JC, Leypoldt JK, Leunissen KM, Laurent G, Bergstrom J. Fluid state and blood pressure control in patients treated with long and short haemodialysis. Nephrol Dial Transplant. 1999;14:369–375. doi: 10.1093/ndt/14.2.369. [DOI] [PubMed] [Google Scholar]
- 9.Boutaud O, Roberts LJ 2nd. Mechanism-based therapeutic approaches to rhabdomyolysis-induced renal failure. Free Radic Biol Med. 2011;51:1062–1067. doi: 10.1016/j.freeradbiomed.2010.10.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahling M, Mathia S, Paliege A, Koesters R, Mrowka R, Peters H, Persson PB, Neumayer HH, Bachmann S, Rosenberger C. Tubular von Hippel-Lindau knockout protects against rhabdomyolysis-induced AKI. J Am Soc Nephrol. 2013;24:1806–1819. doi: 10.1681/ASN.2013030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Crawford N, Lukes L, Finney R, Lancaster M, Hunter KW. Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis. 2005;22:593–603. doi: 10.1007/s10585-005-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke R, Ressom HW, Wang A, Xuan J, Liu MC, Gehan EA, Wang Y. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8:37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. Whole Pathway Scope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24:1175–1182. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- 20.Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23:1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JD, Wiemann S. KEGG graph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25:1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parekh R, Care DA, Tainter CR. Rhabdomyolysis: advances in diagnosis and treatment. Emerg Med Pract. 2012;14:1–15. quiz 15. [PubMed] [Google Scholar]
- 24.Kim JH, Lee SS, Jung MH, Yeo HD, Kim HJ, Yang JI, Roh GS, Chang SH, Park DJ. N-acetylcysteine attenuates glycerol-induced acute kidney injury by regulating MAPKs and Bcl-2 family proteins. Nephrol Dial Transplant. 2010;25:1435–1443. doi: 10.1093/ndt/gfp659. [DOI] [PubMed] [Google Scholar]
- 25.Heyman SN, Lieberthal W, Rogiers P, Bonventre JV. Animal models of acute tubular necrosis. Curr Opin Crit Care. 2002;8:526–534. doi: 10.1097/00075198-200212000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Bosch X, Poch E, Grau JM. Rhabdomyolysis and acute kidney injury. N Engl J Med. 2009;361:62–72. doi: 10.1056/NEJMra0801327. [DOI] [PubMed] [Google Scholar]
- 27.Anakk S, Kalsotra A, Shen Q, Vu MT, Staudinger JL, Davies PJ, Strobel HW. Genomic characterization and regulation of CYP3a13: role of xenobiotics and nuclear receptors. FASEB J. 2003;17:1736–1738. doi: 10.1096/fj.02-1004fje. [DOI] [PubMed] [Google Scholar]
- 28.Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 30.Guijarro C, Egido J. Transcription factor-kappa B (NF-kappa B) and renal disease. Kidney Int. 2001;59:415–424. doi: 10.1046/j.1523-1755.2001.059002415.x. [DOI] [PubMed] [Google Scholar]
- 31.Weston CR, Lambright DG, Davis RJ. Signal transduction. MAP kinase signaling specificity. Science. 2002;296:2345–2347. doi: 10.1126/science.1073344. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Gao Y, Zhang L, Zeng J, He D, Sun Y. Silibinin inhibits cell growth and induces apoptosis by caspase activation, down-regulating survivin and blocking EGFR-ERK activation in renal cell carcinoma. Cancer Lett. 2008;272:61–69. doi: 10.1016/j.canlet.2008.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Takano Y, Iwata H, Yano Y, Miyazawa M, Virgona N, Sato H, Ueno K, Yano T. Up-regulation of connexin 32 gene by 5-aza-2’-deoxycytidine enhances vinblastine-induced cytotoxicity in human renal carcinoma cells via the activation of JNK signalling. Biochem Pharmacol. 2010;80:463–470. doi: 10.1016/j.bcp.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Wooten RM, Weis JJ. Host-pathogen interactions promoting inflammatory Lyme arthritis: use of mouse models for dissection of disease processes. Curr Opin Microbiol. 2001;4:274–279. doi: 10.1016/s1369-5274(00)00202-2. [DOI] [PubMed] [Google Scholar]
- 35.Lin M, Rikihisa Y. Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU. 1 and inhibits lipopolysaccharide activation of NF-kappa B, ERK 1/2 and p38 MAPK in host monocytes. Cell Microbiol. 2004;6:175–186. doi: 10.1046/j.1462-5822.2003.00355.x. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Bergami P, Huang C, Goydos JS, Yip D, Bar-Eli M, Herlyn M, Smalley KS, Mahale A, Eroshkin A, Aaronson S, Ronai Z. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YK, Kim HJ, Kwon CH, Kim JH, Woo JS, Jung JS, Kim JM. Role of ERK activation in cisplatin-induced apoptosis in OK renal epithelial cells. J Appl Toxicol. 2005;25:374–382. doi: 10.1002/jat.1081. [DOI] [PubMed] [Google Scholar]
- 38.Tseng SH, Wang CH, Lin SM, Chen CK, Huang HY, Chen Y. Activation of c-Jun N-terminal kinase 1 and caspase 3 in the tamoxifen-induced apoptosis of rat glioma cells. J Cancer Res Clin Oncol. 2004;130:285–293. doi: 10.1007/s00432-004-0546-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Minemoto Y, Lin A. c-Jun N-terminal protein kinase 1 (JNK1), but not JNK2, is essential for tumor necrosis factor alpha-induced c-Jun kinase activation and apoptosis. Mol Cell Biol. 2004;24:10844–10856. doi: 10.1128/MCB.24.24.10844-10856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi H, Ogata H, Nishigaki R, Broide DH, Karin M. Tobacco smoke promotes lung tumorigenesis by triggering IKKbeta- and JNK1-dependent inflammation. Cancer Cell. 2010;17:89–97. doi: 10.1016/j.ccr.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donovan N, Becker EB, Konishi Y, Bonni A. JNK phosphorylation and activation of BAD couples the stress-activated signaling pathway to the cell death machinery. J Biol Chem. 2002;277:40944–40949. doi: 10.1074/jbc.M206113200. [DOI] [PubMed] [Google Scholar]
- 43.Tsuruta F, Sunayama J, Mori Y, Hattori S, Shimizu S, Tsujimoto Y, Yoshioka K, Masuyama N, Gotoh Y. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han JY, Jeong EY, Kim YS, Roh GS, Kim HJ, Kang SS, Cho GJ, Choi WS. C-jun N-terminal kinase regulates the interaction between 14-3-3 and Bad in ethanol-induced cell death. J Neurosci Res. 2008;86:3221–3229. doi: 10.1002/jnr.21759. [DOI] [PubMed] [Google Scholar]
- 45.Gorospe M, Wang X, Holbrook NJ. Functional role of p21 during the cellular response to stress. Gene Expr. 1999;7:377–385. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 47.Heliez C, Baricault L, Barboule N, Valette A. Paclitaxel increases p21 synthesis and accumulation of its AKT-phosphorylated form in the cytoplasm of cancer cells. Oncogene. 2003;22:3260–3268. doi: 10.1038/sj.onc.1206409. [DOI] [PubMed] [Google Scholar]
- 48.Maddika S, Ande SR, Panigrahi S, Paranjothy T, Weglarczyk K, Zuse A, Eshraghi M, Manda KD, Wiechec E, Los M. Cell survival, cell death and cell cycle pathways are interconnected: implications for cancer therapy. Drug Resist Updat. 2007;10:13–29. doi: 10.1016/j.drup.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Sussman M. “AKT”ing lessons for stem cells: regulation of cardiac myocyte and progenitor cell proliferation. Trends Cardiovasc Med. 2007;17:235–240. doi: 10.1016/j.tcm.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homsi E, Janino P, de Faria JB. Role of caspases on cell death, inflammation, and cell cycle in glycerol-induced acute renal failure. Kidney Int. 2006;69:1385–1392. doi: 10.1038/sj.ki.5000315. [DOI] [PubMed] [Google Scholar]
- 51.Wei Q, Hill WD, Su Y, Huang S, Dong Z. Heme oxygenase-1 induction contributes to renoprotection by G-CSF during rhabdomyolysis-associated acute kidney injury. Am J Physiol Renal Physiol. 2011;301:F162–170. doi: 10.1152/ajprenal.00438.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B, Shravah J, Luo H, Raedschelders K, Chen DD, Ansley DM. Propofol protects against hydrogen peroxide-induced injury in cardiac H9c2 cells via Akt activation and Bcl-2 up-regulation. Biochem Biophys Res Commun. 2009;389:105–111. doi: 10.1016/j.bbrc.2009.08.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura R, Okouchi M, Fujioka H, Ichiyanagi A, Ryuge F, Mizuno T, Imaeda K, Okayama N, Kamiya Y, Asai K, Joh T. Glucagon-like peptide-1 (GLP-1) protects against methylglyoxal-induced PC12 cell apoptosis through the PI3K/Akt/mTOR/GCLc/redox signaling pathway. Neuroscience. 2009;162:1212–1219. doi: 10.1016/j.neuroscience.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 54.Matsuzawa A, Ichijo H. Stress-responsive protein kinases in redox-regulated apoptosis signaling. Antioxid Redox Signal. 2005;7:472–481. doi: 10.1089/ars.2005.7.472. [DOI] [PubMed] [Google Scholar]
- 55.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


