Abstract
As one common malignant bone cancer, osteosarcoma is mainly occurred in young people with increasing incidences. Current treatment of osteosarcoma includes surgery and chemo-/radio-therapy. Due to the unclear pathogenesis mechanism, the overall treatment efficacy is still not satisfactory. As anti-apoptotic molecule Bcl-2 has been suggested to be related with osteosarcoma and is regulated by miR-143, we thus investigated the correlation between miR-143 and Bcl-2 in osteosarcoma patients, in an attempt to elucidate the role of miR-143 in cancer occurrence. Real-time fluorescent RT-PCR was used to quantify expression levels of miR-143 and Bcl-2 from a total of 5 osteosarcoma patients, along with protein contents determination by Western blotting. In vitro study was also performed to detect Bcl-2 expression and cell apoptosis via silencing or over-expressing miR-143 in cultured osteosarcoma cells. MiR-143 showed down-regulation in osteosarcoma tissues. Bcl-2, however, had elevated expression in cancer cells when compared to adjacent tissues (P<0.05). In cultured cells, Bcl-2 expression level was also potentiated after knock-down of miR-143, while those cells with miR-143 over-expression had depressed Bcl-2 levels. Those cells transfected with miR-143 mimics had higher percentage of apoptotic cells. MiR-143 can regulate the expression of Bcl-2 gene in osteosarcoma cells, and mediate the apoptotic process, thereby playing a critical role in the pathogenesis of osteosarcoma.
Keywords: MiR-143, Osteosarcoma, Bcl-2, Real-time fluorescent PCR, Western blotting
Introduction
As a common malignant bone tumor, osteosarcoma is mostly developed in children and younger populations [1]. Osteosarcoma is featured with higher incidence, malignancy and metastatic rates [2], thus causing its unfavorable prognosis and higher recurrence rate, leading to higher social and family costs [3]. Rapid progresses have been obtained regarding the treatment of osteosarcoma with the advancing technology, but still leaving the 5-year survival rate at relatively lower level [4]. In order to improve the diagnosis and treatment efficacy, further explorations are required targeting more specific biological markers. The emergence of micro RNA (miRNA) provides an alternative choice for specific diagnosis and individualized treatment of osteosarcoma. As one kind of small non-coding RNA , miRNA has highly conserved sequence containing about 18~24 nucleotides [5]. Mature miRNA with single-stranded form is transcribed from miRNA-coding gene under the direction of RNA polymerase II to synthesize pre-miRNA, which is undergone double splicing and editing by Drosha and Dicer enzymes [6]. Via binding onto 3’-untranslated region (UTR) of the target gene-coding mRNA, miRNA exert its biological function via degradation or inhibition of translation, thus regulating the expression of the target gene and related cellular functions [6]. Studies have shown the involvement of miRNA in multiple steps of tumor occurrence and activity via gene expression modulation [7,8]. Therefore, the quantification of specific miRNA level may own significant implication for the early diagnosis of tumors. MiR-143 has been found to be down-regulated in osteosarcoma tissues [9], suggesting the possible relationship between miR-143 and osteosarcoma formation. This study thus detected the expression of miR-143 and its target genes in osteosarcoma tissues and adjacent tissues, in addition to in vitro cell assay for the mediating effect of miR-143, in an attempt to illustrate the potential role of miR-143 in the occurrence and progression of osteosarcoma.
Materials and methods
Patient information
A total of five osteosarcoma patients who have received the surgical resections in Qilu Hospital, Shandong University between May 2013 and May 2014 were recruited as the patient group. Both tumor tissue and adjacent non-carcinoma tissue samples were collected during the surgery and stored in liquid nitrogen. The tumor-adjacent tissue was employed as the control group. This study has been preapproved by Qilu Hospital, Shandong University. Written consents have been obtained from patients and their guardians.
Cell line
MG63 osteosarcoma cell line was purchased from Chinese Academy of Science, Shanghai and was incubated in RPMI 1640 medium (Gibco, US) containing 10% fetal bovine serum (FBS, Gibco, US).
Real-time fluorescent PCR
Total tissue RNA was extracted by Trizol reagents (Invitrogen, US) following manual instruction, and was used as the template to synthesize cDNA. Fluorescent quantitative PCR (qPCR) was performed using cDNA, PCR primers (as Table 1) and SYBR qPCR Mix (Toyobo, Japan). Amplification conditions were: 95°C per-denature for 15 sec, followed by 40 cycles each containing 95°C denature (15 sec), 60°C annealing (15 sec) and 72°C elongation (45 sec). The relative mRNA level was determined by 2-ΔΔCt method.
Table 1.
Primer sequences for qPCR
| Target gene | Forward primer | Reverse primer |
|---|---|---|
| β-actin | 5’- GAGGG AAATC GTGCG TGAC-3’ | 5’- CTGGA AGGTG GACA GTGAG-3’ |
| Bcl-2 | 5’- CCTCT CGAGA AGGAT GGCGC ACGC TGG-3’ | 5’-CCGGA ATTCT TGGGC AGGCA TCTTG ACT-3’ |
| miR-143 | 5’-AGTCA GTGAG ATGAA GCACT G-3’ | 5’-GTGCA GGGTC CGAGG T-3’ |
| U6 | 5’-GCTTC GGCAG CACAT ATACT AAAAT-3’ | 5’-CGCTT CACGA ATTTG CGTGT CAT-3’ |
Western blotting
Tissues were homogenized and lysed in RIPA buffer (0.5 mL) with proteinase inhibitor cocktail. After complete tissue lysis, proteins were collected from the supernatant after iced vortex, sonic rupture and cold centrifugation (10000 g, 10 min). Protein contents were determined by BCA method using absorbance value. After boiled denature, protein samples were loaded onto SDS gel (Sigma, US) for electrophoresis separation. The protein band was then transferred to NC membrane in transferring buffer. The membrane was then blocked, washed and incubated with primary antibody (1:1000, Abcam, US) overnight. Secondary antibody (1:10000) was applied on the next day for 1-hour incubation at room temperature. After development and exposure, a gel imaging analysis system GIS-2020D was used to calculate the optical density (OD) value of all protein bands for illustrating relative protein level.
Flow cytometry
Cultured cells were collected (5 × 106) in tubes for 1000 g centrifugation for 5 min. After discarding the supernatant, 0.1 mL blocking buffer was added for 10-min incubation at room temperature. Buffer containing Ca2+ was then added to discard the supernatant. Labelling solution (0.1 mL) was then added with Annexin V substrate for another 15 min incubation. After centrifugation and discarding supernatants, 0.3 mL labelling solution was mixed with PI dye. The sample was then loaded on the flow cytometry for analysis.
Statistical analysis
All experiments were performed at least in triplicates. Data were presented as mean ± standard error of means (SEM). SPSS 19.0 software was used to analyze all collected data. Student t-test or analysis of variance (ANOVA) was employed for comparisons of means. A statistical significance was defined when P<0.05.
Results
MiR-143 expression in osteosarcoma tissues
Previous study has revealed the down-regulation of miR-143 in osteosarcoma cells. To further explore the role of miR-143 in osteosarcoma formation, we quantified the expression level of miR-143 in both osteosarcoma and adjacent non-tumor tissue samples. Results showed significantly depressed miR-143 expression in tumor tissues, which was about 56% of that in tumor adjacent tissues (P<0.05, Figure 1).
Figure 1.
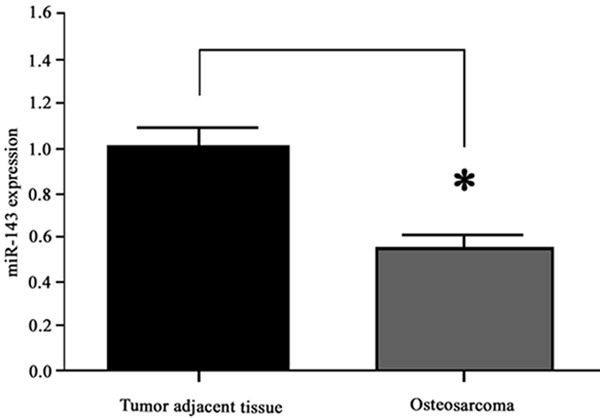
MiR-143 expression. *, P<0.05 compared to tumor adjacent tissue.
Bcl-2 expression level
To further elucidate the biological role of miR-143 in osteosarcoma occurrence, it is necessary to find its target gene. Based on software estimation, Bcl-2 may be the candidate. As one anti-apoptotic molecule, Bcl-2 played a critical role in the cell apoptosis. We thus examined the expressional prolife of Bcl-2 and found about 2.1-fold increased mRNA in osteosarcoma tissues compared to tumor adjacent tissue (P<0.05, Figure 2A). The protein level of Bcl-2 in tumor was also higher than control ones (P<0.05, Figure 2B).
Figure 2.
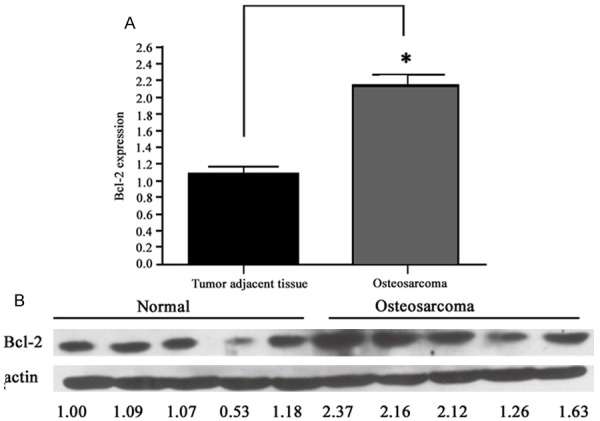
Bcl-2 expressions in tumor and adjacent tissues. A. mRNA level of Bcl-2; B. Western blotting bands. *, P<0.05 compared to tumor adjacent tissue.
Bcl-2 as the target gene of miR-143
To prove the regulatory relationship between miR-143 and Bcl-2, we used the dual luciferase reporter system to test its interaction. We constructed reporter vector of both wild type and mutant forms of 3’-UTR region of Bcl-2 gene, and co-transfected those vectors with pTK vector and miR-143 mimics into MG63 cells. Results found significantly inhibited luciferase activity after cotransfection of wild type 3’-UTR of Bcl-2 gene and miR-143 mimic, compared to those cells transfected with mutant Bcl-2 and miR-143 vectors (Figure 3). These results suggest that Bcl-2 works as one target gene of miR-143.
Figure 3.
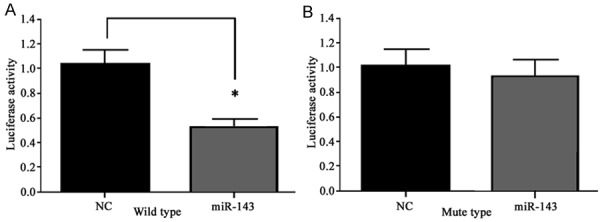
Dual-luciferase reporter assay. *, P<0.05 compared to negative control (NC).
MiR-143 regulates Bcl-2 gene expression
We further tested the effect of miR-143 on Bcl-2 gene expression via manipulating endogenous miR-143 level in MG63 cells. As shown in Figure 4, the transfection of miR-143 mimic effectively depressed Bcl-2 expression by more than 40% compared to control group (P<0.05). Meanwhile, the transfection of miR-143 inhibitor endowed cells with elevated Bcl-2 expression by more than 30% (P<0.05). These results suggest the role of miR-143 in mediating Bcl-2 gene expression.
Figure 4.
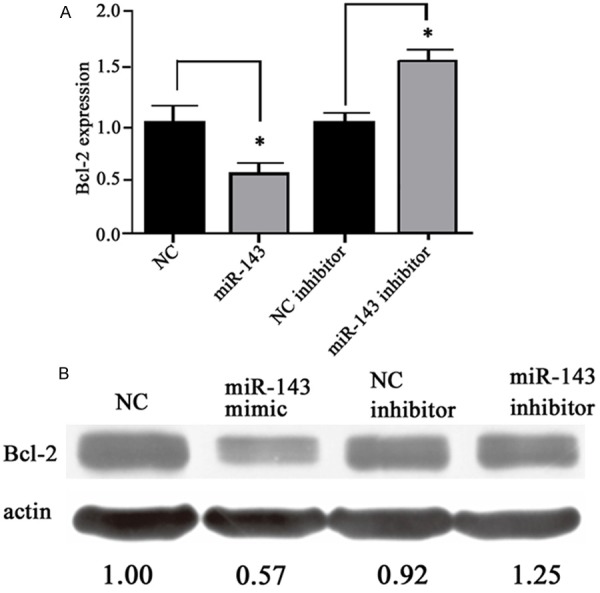
Bcl-2 expressions in MG63 cells. A. mRNA level of Bcl-2 gene; B. Protein bands reflecting Bcl-2 level in cell extract. *, P<0.05 compared to negative control.
Effects of miR-143 on apoptosis of osteosarcoma cells
As the regulatory role of miR-143 on endogenously expression of Bcl-2, we thus tested the effect of miR-143 in tumor cell apoptosis. In MG63 cells, we firstly transfected them with miR-143 mimics to elevated its expression level, followed by Annexin V/PI double labelling on flow cytometry to determine the apoptotic level. As shown in Figure 5, the introduction of miR-143 mimics remarkable increased the percentage of apoptotic cells, suggesting the involvement of miR-143 in tumor cell apoptosis.
Figure 5.
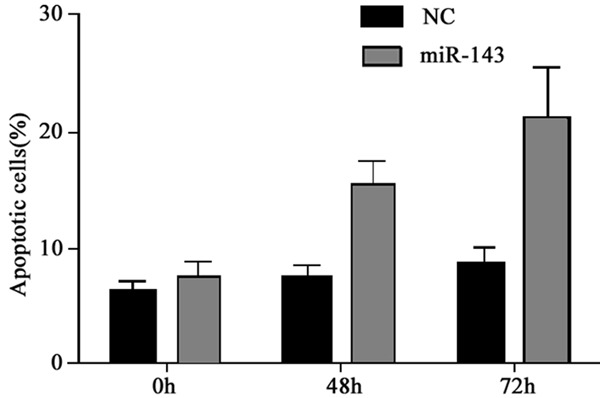
MG63 cell apoptosis after miR-143 mimics transfection. *, P<0.05 compared to negative control.
Discussion
As a common malignant bone tumor, osteosarcoma is mostly developed in children and younger populations [1]. Due to its high malignancy, metastatic level and unfavorable prognosis, classical treatment including surgery combined with chemo-/radio-therapy still cannot obtain satisfactory treatment efficacy. With the advancement of medical technology, the treatment of osteosarcoma has been remarkably improved, but still leaving the tumor metastasis and/or re-occurrence as major reasons causing tumor related mortality Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration [10]. Recently emerged genetic therapy against tumors has brought a revolution to cancer treatment [11-13]. Targeting the key molecule or gene in tumor pathogenesis, gene therapy can modulate proliferation and differentiation of tumor cells via specifically modifying biological functions [14]. Therefore, the identification of critical molecule/gene in the formation of osteosarcoma can bring revolutionary changes to diagnosis and treatment of tumors.
As one kind of small non-coding RNA, miRNA has highly conserved sequence and function, and may regulate the expression of target genes via binding onto 3’-UTR region of the gene [7] Studies have found that miRNAs could regulate nearly 30% of all genes in our body. MiR-143 was found to be down-regulated in various malignant tumors including cervical cancer, rectal cancer, osteosarcoma and nasopharyngeal carcinoma [15,16]. This study also revealed the decreased level of miR-143 in osteosarcoma tissues when compared to tumor adjacent tissues. To further illustrate the role of miR-143, we selected some candidates of miR-143 target genes including TGF-β, ERK-5 and Bcl-2 [17-20]. Early study has established Bcl-2 as one important anti-apoptotic factor for regulating cell proliferation [21].
Our study showed elevated Bcl-2 and depressed miR-143 in osteosarcoma tissues compared to tumor adjacent tissues. With the help of software screening, Bcl-2 may be one target gene of miR-143. To further confirm the relationship, we constructed wild type and mutant forms and 3’-UTR of Bcl-2 gene vectors, which were tested under a dual luciferase reporter system. This result further validated the specific binding between miR-143 and 3’-UTR of Bcl-2 gene. We further manipulated the intracellular miR-143 level to observe the effect on Bcl-2. Results showed that both mRNA and protein levels of Bcl-2 were depressed after over-expression of miR-143. Meanwhile, the inhibition of miR-143 potentiated intracellular Bcl-2 level. These results suggested that miR-143 can regulate Bcl-2 expression. In the last part of this study, it is found that elevated percentage of apoptotic cells occurred in osteosarcoma cells transfected with miR-143 mimics, suggesting the facilitation of miR-143 on tumor cell apoptosis. The occurrence of osteosarcoma is a complex process involving multiple factors [22]. Our study demonstrated the role of miR-143 in cancer pathogenesis via regulating Bcl-2 expression. This study provided further knowledge on the pathogenesis of osteosarcoma and gene therapy strategy, although detailed mechanism is worth further investigations.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Osborne TS, Khanna C. A review of the association between osteosarcoma metastasis and protein translation. J Comp Pathol. 2012;146:132–42. doi: 10.1016/j.jcpa.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 4.Tu B, Peng ZX, Fan QM, Du L, Yan W, Tang TT. Osteosarcoma cells promote the production of pro-tumor cytokines in mesenchymal stem cells by inhibiting their osteogenic differentiation through the TGF-beta/Smad2/3 pathway. Exp Cell Res. 2014;320:164–73. doi: 10.1016/j.yexcr.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Chen T. The role of MicroRNA in chemical carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2010;28:89–124. doi: 10.1080/10590501.2010.481477. [DOI] [PubMed] [Google Scholar]
- 6.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. OncomirsmicroRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi E, Satow R, Ono M, Masuda M, Honda K, Sakuma T, Kawai A, Morioka H, Toyama Y, Yamada T. MicroRNA expression and functional profiles of osteosarcoma. Oncology. 2014;86:94–103. doi: 10.1159/000357408. [DOI] [PubMed] [Google Scholar]
- 10.Shimbo K, Miyaki S, Ishitobi H, Kato Y, Kubo T, Shimose S, Ochi M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem Biophys Res Commun. 2014;445:381–7. doi: 10.1016/j.bbrc.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Gougelet A, Pissaloux D, Besse A, Perez J, Duc A, Dutour A, Blay JY, Alberti L. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int J Cancer. 2011;129:680–90. doi: 10.1002/ijc.25715. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Pang Y, Moses HL. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–7. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Upregulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–9. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 14.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–9. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 16.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. MiR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T, Ito H, Oshimura M, Ochiya T. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19:1123–30. doi: 10.1038/mt.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q, Dan ZL, Tian DA, Zhang P. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol. 2013;19:7758–65. doi: 10.3748/wjg.v19.i43.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, Avancès C, Villalba M, Culine S, Fajas L. MiR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anninga JK, Gelderblom H, Fiocco M, Kroep JR, Taminiau AH, Hogendoorn PC, Egeler RM. Chemotherapeutic adjuvant treatment for osteosarcoma: where do we stand? Eur J Cancer. 2011;47:2431–45. doi: 10.1016/j.ejca.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Hu G, Zhou J. Repression of versican expression by microRNA-143. J Biol Chem. 2010;285:23241–50. doi: 10.1074/jbc.M109.084673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schedlich LJ, Yenson VM, Baxter RC. TGF-beta-induced expression of IGFBP-3 regulates IGF1R signaling in human osteosarcoma cells. Mol Cell Endocrinol. 2013;377:56–64. doi: 10.1016/j.mce.2013.06.033. [DOI] [PubMed] [Google Scholar]


