Abstract
Background: The aim is to discuss the relationship of Line-1 methylation and the MDR1 expression in esophageal squamous cell carcinoma (ESCC). Methods: We analyzed the methylation level of Line-1 by quantitative real-time MSP, and the expression of MDR1 by real-time RT-PCR in 310 ESCC and corresponding non-tumor tissues. Results: We found that the methylation index (MI) of Line-1 decreased from 0.90 in non-tumor tissues toward 0.78 in ESCC. The cumulative survival was significantly shorter in ESCC patients with MI ≤ 0.78 (34 months) than that in patients with MI > 0.78 (43 months). There was a statistical difference between MI ≤ 0.78 and MI > 0.78 cases with these clinicopathologic parameters (age, AJCC stage, differentiation; P = 0.010, P < 0.0001, P = 0.015, respectively). These results implied that Line-1 hypomethylation could be more in ESCC patients with older, advanced tumor and poor differentiation group. Meanwhile, ESCC with demethylation of Line-1 were shown elevated MDR1 expression in tumor (Mean-∆∆Ct = 0.21), but ESCC with hypermethylation of Line-1 were considered to be decreased MDR1 expression in tumor (Mean-∆∆Ct = -0.86). Conclusions: Line-1 hypomethylation could be as a biomarker of poor prognosis in ESCC patients. MDR1 gene could be activated via epigenetic mechanisms with demethylation of Line-1 in ESCC, and enhance tumor progression.
Keywords: MDR1, Line-1, ESCC, hypomethylation
Introduction
Esophageal carcinoma is a serious malignancy with regards to mortality and prognosis, and the 5-year survival rate for all patients diagnosed with esophageal cancer ranges from 15% to 20% [1]. Esophageal squamous cell carcinoma (ESCC) is the fourth leading cause of cancer-related deaths, and the overall five-year survival rate is only 10% or so in China. ESCC incidence is 20.85/100 000 and the mortality is 16.24/100 000 [2]. ESCC evolves in all parts of the esophagus via a mere progression from normal squamous epithelial cells to dysplasia and malignant invasion, and multimodal therapies including surgery, chemotherapy, and chemoradiotherapy [3]. It should be clearly understand the pathogenesis for better clinical therapy. However, it remains unclear for molecular alterations in carcinogenesis of ESCC.
Global DNA hypomethylation and regional CpG hypermethylation in cancer tissue has been demonstrated and may be linked to carcinogenesis. DNA methylation has been widely characterized by analysis of specific CpG island promoter regions of tumor-related genes in patient’s specimens of carcinomas and/or associated precursor lesions [4,5]. Our previously studies suggested that ESCC show concurrent hypermethylation of a number of cell-cycle related genes, which has been defined, for CpG island methylator phenotype (CIMP). CIMP + may specifically define a subgroup of patients associated with unfavorable outcome in ESCC [6]. However, its presence and importance remain poorly understood in ESCC [7].
As Line-1 constitutes a substantial portion of the human genome, and the level of Line-1 methylation has been shown to be a surrogate marker of global DNA methylation level in various cancers, such as non-small cell lung cancer, colon cancer, breast cancer [8,9]. Line-1 hypomethylation have been also as a biomarker of poor prognosis in cancer patients [9]. A few studies reported Line-1 hypomethylation as a poor prognostic marker in ESCC, but results were still filled with controversy in detail. The chromosomal instability could be associated with Line-1 methylation levels in ESCC due to the percentage of copy number alterations in the whole genome [10]. Line-1 hypomethylation was significantly associated with a history of tobacco in noncancerous esophageal mucosae of ESCC patients [11]. Meanwhile, Line-1 methylation levels were significantly associated with CDK6 mRNA and CDK6 protein expression levels in ESCC specimens [12]. However, further studies are needed to validate the molecular basis for the association between Line-1 hypomethylation and patient prognosis in ESCC.
Multidrugresistance (MDR) means the resistance of cancer cells to structurally and mechanistically unrelated classes of anticancer drugs, which leads to cancer relapse and death [13]. The overexpression of ATP-binding cassette (ABC) transporters has been as an important mechanisms underlying MDR. P-glycoprotein (ABCB1, MDR1) is the first ABC transporter, which is frequently overexpressed in cancer cells resulting in drugs being pumped out of the cells and reducing the effectiveness of the drugs in killing cancer cells [14,15]. Drug resistance has been a major obstacle that limits the effectiveness of chemotherapy in ESCC [16]. It should be noted that the histone deacetylase (HDAC) inhibitors might elicit the overexpression of ABC transporters. It should be resulted that epigenetic mechanisms, such as DNA methylation, could be involved in MDR1 expression, but the complex mechanisms of MDR in ESCC remain unclear [17,18]. Our studies confirmed that Line-1 hypomethylation could activate stem cell marker CD133 expression during carcinogenesis in HCC, which implied that global DNA hypomethylation could be as an important factor for tumor-related gene expression [19].
However, none of studies has examined the potential effect of MDR1 expression and Line-1 hypomethylation. We hypothesized that global DNA hypomethylation might be an important influential factor for MDR1 expression, leading to resistance of cancer cells. We therefore assessed the influence of Line-1 hypomethylation in relation to MDR1 expression alterations in 310 ESCC patients and matched non-tumor tissues.
Material and methods
Patients and tissue samples
Tumor samples from 310 cases of ESCC patients that underwent surgical resection between 2001 and 2014 were investigated, together with corresponding non-tumor tissue specimens. These specimens were collected from Changzhou Cancer Hospital and Nanyang Center Hospital in China. The samples were frozen in liquid nitrogen right after surgical resection and were kept in -196°C until processing for RNA/DNA extraction. The patients consisted of 180 men and 130 women, ranging in age from 36 to 82 years (mean ± SD, 52.47 ± 12.43 years). 88 cases were well differentiation, 142 cases were moderately differentiation, and 80 cases were poorly differentiation. The present study was in accordance with the ethics standards of the committee on Human Experimentation of the Soochow University. Written informed consent was obtained from each patient.
Quantitative methylation analysis of Line-1
We isolated specific tumor tissues and non-tumor tissues and removed tissues by surgical needles under direct microscopic visualization. After microdissection, the tissue samples were placed into Eppendorf tubes and were incubated with proteinase K at 37°C overnight. Then the tissue was extracted twice in phenol and twice in chloroform, followed by ethanol precipitation. Genomic DNA (3 μg) from tissues and cells was denatured with 0.3 M NaOH at 37°C for 10 min. Then freshly prepared (208 μl) 3 M sodium bisulphate (PH 5.0) and 12 μl fresh 100 mM hydroquinone solutions were added. Bisulfite treatment was carried out as described previously [19]. Line-1 methylation was measured using a methylation-specific real-time PCR assay as previously described [19]. The Line-1 primers used were: unmethylated forward primer, TGTGTGTGAGTTGAAGTAGGGT, reverse primer, ACCCAATTTTCCAAATACAACCATCA; methylated forward primer, CGCGAGTCGAAGTAGGGC, reverse primer, ACCCGATTTTCCAAATACGACCG. MSP were performed in a 25 μl reaction volume with bisulfite-modified DNA. Reactions step was followed by 40 cycles at 95°C 30 seconds, 55°C for 35 seconds, and 72°C for 40 seconds in the Mx3000P QPCR System (Stratagene, USA) with SYBR Premix Ex Taq (TaKaRa Bio, Dalian, China). The methylation index (MI) of Line-1 was calculated using the formula: 100 × methylated reaction/(unmethylated reaction + methylated reaction).
Quantitative real-time PCR analysis
RT-qPCR was performed using SYBR Green I chemistry (Invitrogen Life Technologies, Carlsbad, CA, USA). Total RNA was isolated from 310 ESCC, and adjacent normal tissues samples using TRIzol reagent, according to the protocol provided by the manufacturer. The quantity and quality of the RNA samples were measured carefully by spectrophotometer and electrophoresis. The first-strand cDNA was synthesized from a moloney murine leukemia virus-reverse transcriptase kit using 2 ug total RNA according to the manufacturer’s instructions (Takara Bio, Inc., Dalian, China). Primer sequences of MDR1 for reverse transcription-PCR (RT-PCR) reaction were forward (5’-CTGGTTTGATGTGCACGATGTTGG-3’) and reverse (5’-TGCCAAGACCTCTTCAGCTACTG-3’) [20]. Quantitative real-time PCR (qPCR) were carried out using the Mx3000P QPCR System (Stratagene, USA). The cDNA was then used for qPCR in a 20 μl SYBR Premix Ex Taq. qPCR for MDR1 mRNA expression was performed by 40 cycles of 30 seconds at 95°C, 40 seconds at 60°C, and 40 seconds at 72°C. As an internal control for qPCR, β-actin mRNA expression was amplified from the same cDNA samples. All results were normalized to β-actin amplification. CT values for triplicate reactions were averaged and relative MDR1 expression was determined with the comparative CT method, using average CT values for MDR1 and β-actin.
Statistical analysis
All data were analyzed by SPSS 18.0 software (SPSS, Inc., Chicago, USA). Comparison was done with t test (unpaired or paired). All P values presented were two-sided, and a P value of less than 0.05 was considered statistically significant. Univariate analyses of the interaction between Line-1 methylation and clinical parameters were performed with Pearson’s Chi-square test or Fisher’s exact test. Survival curves were based on Kaplan-Meier estimates. Hazard ratios (HR) between two groups were calculated using Cox regression with the prognostic factors.
Results
Methylation index of Line-1 in ESCC and non-tumor tissues
Line-1 promoter methylation is prominent in the genome and is frequently used to be a marker of global methylation in several of cancers. The methylation status of the Line-1 promoter region was analyzed by a real-time methylation-specific polymerase chain reaction assay in 310 ESCC and their adjacent non-tumor tissues. The methylation index (MI) of Line-1 was calculated according to quantitative methylation data in ESCC and Non-tumor samples (Figure 1). The mean MI of Line-1 was 0.78 (95% CI, 0.77-0.79) in ESCC, and 0.91 (95% CI, 0.89-0.91) in Non-tumor samples. The MI level of Line-1 was significantly lower in ESCC samples compared with Non-tumor tissues (P < 0.0001). These results indicated a significant decrease in methylation levels of Line-1 promoter in ESCC compared with non-tumor samples.
Figure 1.
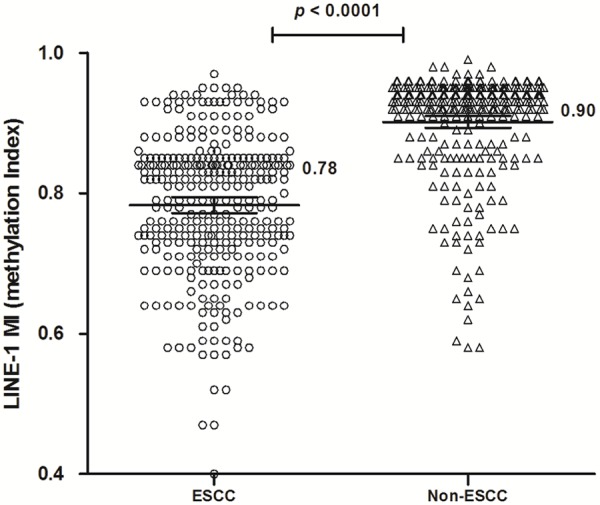
Line-1 methylation in ESCC and the matched non-tumor tissues. The methylation index (MI) of Line-1 was indicated by the mean and 95% CI in ESCC and Non-tumor tissues. The mean MI of Line-1 in ESCC (MI = 0.78) was lower than that in the matched non-tumor tissues (MI = 0.90, P < 0.0001). Statistical analyses were done using the paired t test.
Line-1 methylation levels and clinicopathologic features of ESCC
Demographic and clinical characteristics of the subjects of the present study are presented in Table 1. Using statistical analysis, we examined Line-1 methylation level with regard to ESCC patient clinicopathologic parameters of age, gender, tumor size, smoking history, drinking alcohol, AJCC stage, differentiation, and others (Table 1). The cutoff value 0.78 was set for MI and the patients were classified according to the mean MI of Line-1 in ESCC. There was a statistical difference between MI ≤ 0.78 and MI > 0.78 cases with these clinicopathologic parameters (age, AJCC stage, differentiation; P = 0.010, P < 0.0001, P = 0.015, respectively).
Table 1.
Correlation of clinicophthologic variables with Line-1 hypomethylation in ESCC
| Variable | No. | MI ≤ 0.78 (N = 143) | MI > 0.78 (N = 167) | Odds ratio (95% CI) | P value* | |
|---|---|---|---|---|---|---|
| Gender | Male | 180 | 89 | 91 | 1.38 (0.87-2.17) | 0.168 |
| Female | 130 | 54 | 76 | |||
| Age (y) | ≥ 50 | 262 | 129 | 133 | 2.36 (1.21-4.59) | 0.010 |
| < 50 | 48 | 14 | 34 | |||
| Size | < 3 cm | 172 | 86 | 86 | 1.42 (0.90-2.23) | 0.127 |
| ≥ 3 cm | 138 | 57 | 81 | |||
| Tobacco | Yes | 168 | 83 | 85 | 1.34 (0.85-2.09) | 0.208 |
| No | 142 | 60 | 82 | |||
| Alcohol | Yes | 147 | 74 | 73 | 1.38 (0.88-2.16) | 0.158 |
| No | 163 | 69 | 94 | |||
| Depth of invasion | T1-2 | 224 | 106 | 118 | 1.19 (0.72-1.96) | 0.497 |
| T3-4 | 86 | 37 | 49 | |||
| AJCC Stage | I-II | 125 | 39 | 86 | 0.35 (0.22-0.57) | < 0.0001 |
| III-IV | 185 | 104 | 81 | |||
| Lymph node metastasis | N0-1 | 148 | 60 | 88 | 1.47 (0.97-2.24) | 0.071 |
| N2-3 | 262 | 83 | 179 | |||
| Distant metastasis | M0 | 251 | 123 | 128 | 1.40 (0.75-2.59) | 0.294 |
| M1 | 59 | 20 | 29 | |||
| Differentiation | G1 | 88 | 36 | 52 | 0.015 | |
| G2 | 142 | 59 | 83 | |||
| G3 | 80 | 48 | 32 |
Comparison was done with Pearson’s Chi-square test or Fisher’s exact test.
The next, we analyzed Line-1 methylation level to age, AJCC stage, and differentiation in ESCC patients (Figure 2). The results found that Line-1 MI were 0.84 (95% CI, 0.81-0.86), 0.81 (95% CI, 0.79-0.83), 0.77 (95% CI, 0.76-0.79), 0.74 (95% CI, 0.71-0.76) in ESCC patients with < 50 years, 50~59 years, 60~69 years, and ≥ 70 years groups, respectively. And, Line-1 MI were 0.85 (95% CI, 0.83-0.87), 0.82 (95% CI, 0.79-0.84), 0.77 (95% CI, 0.75-0.78), 0.72 (95% CI, 0.68-0.75) in ESCC patients with AJCC stage I, II, III, IV groups, respectively. Line-1 MI were 0.78 (95% CI, 0.76-0.81), 0.80 (95% CI, 0.78-0.81), 0.75 (95% CI, 0.74-0.78) in ESCC patients with G1, G2, G3 groups, respectively. These results implied that Line-1 hypomethylation could be more in ESCC patients with older, advanced tumor and poor differentiation group.
Figure 2.
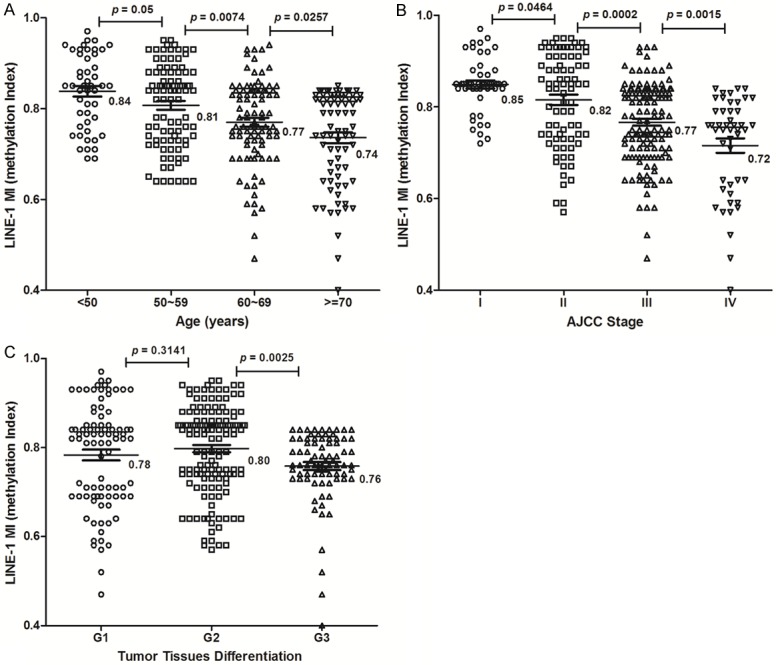
The level of Line-1 methylation associated with age, AJCC stage, and differentiation in ESCC. The methylation index (MI) of Line-1 was indicated by the mean and 95% CI in ESCC tissues. A. The mean MI of Line-1 in different age groups. B. The mean MI of Line-1 in different AJCC stage groups. C. The mean MI of Line-1 in different differentiation groups. Statistical analyses were done using the unpaired t test.
To investigate the association between the level of Line-1 promoter methylation status and outcomes after post-resection of ESCC, the survival of these patient groups was compared using the Kaplan-Meier method and the log-rank test (Figure 3). Results showed a significantly longer median cumulative survival (43 months) was seen in ESCC with MI > 0.78 group compared with 34 months in the ESCC with MI ≤ 0.78 group (log-rank P < 0.0001). These results suggested that Line-1 MI level could be an independent predictor for prognostic factor in ESCC.
Figure 3.
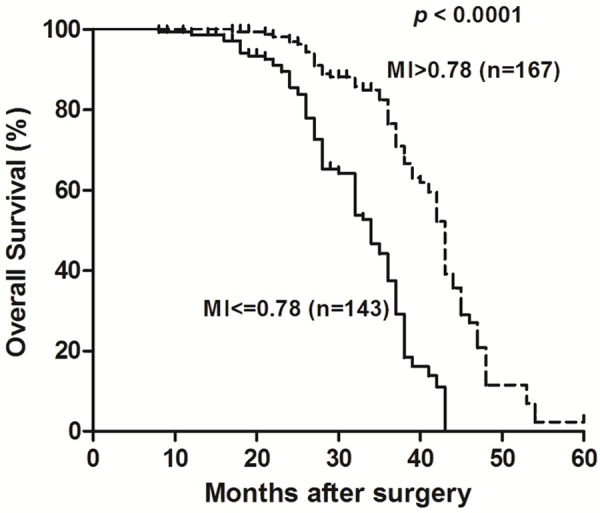
Line-1 hypomethylation confers poor prognosis in ESCC. The overall survival of these patients was compared using the Kaplan-Meier method and the log-rank test. Results showed a significantly longer median cumulative survival (43 months) was seen in ESCC with MI > 0.78 group compared with 34 months in the ESCC with MI ≤ 0.78 group (log-rank P < 0.0001).
Elevated MDR1 expression and Line-1 hypomethylation in ESCC
MDR1 has been a major difficulty for the failure of chemotherapy in ESCC. To investigate the association between Line-1 methylation and multidrug resistance, MDR1 expression was analyzed by real-time RT-PCR in 310 ESCC samples and the matched non-tumor tissues (Figure 4A). We found that MDR1 mRNA expression were higher in ESCC tissues with MI ≤ 0.78 (Mean-∆∆Ct = 0.21; 95% CI, -0.58-0.477) than that in ESCC tissues with MI > 0.78 (Mean-∆∆Ct = -0.86; 95% CI, -1.20--0.517); P < 0.0001). There was an elevated tendency for MDR1 expression from Line-1 hypermethylation to hypomethylation ESCC tissues, and more ESCC samples with Line-1 hypomethylation showed higher MDR1 expression. The elevated MDR1 expression was significantly associated with the demethylation status of Line-1 in ESCC patients (R2 = 0.059, P < 0.0001; Figure 4B).
Figure 4.
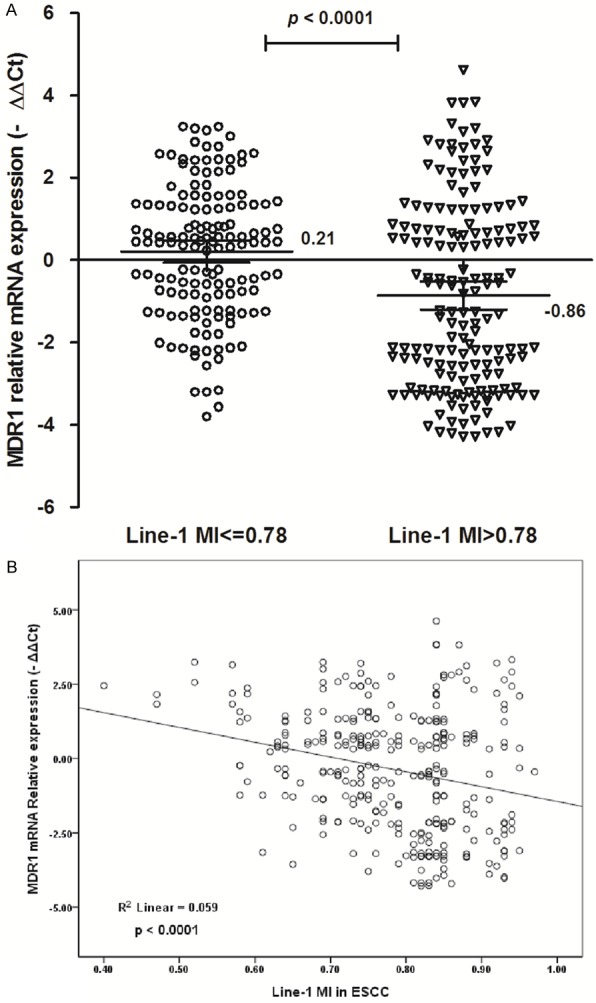
Elevated MDR1 expression associated with Line-1 demethylation in ESCC patients. A. The MDR1 expressions were increased in ESCC with Line-1 MI ≤ 0.78 than that in ESCC with Line-1 MI > 0.78 (P < 0.0001). -∆∆Ct, -(∆CtESCC-∆CtNon-tumor). Statistical analyses were done using the unpaired t test, while error bars represent 95% CI. B. The scatter plots summarized relative expression levels of MDR1 (-∆∆CT) associated with Line-1 methylation index in ESCC patients (P < 0.0001).
Furthermore, multivariate analysis using the Cox proportional hazards model revealed that the status of age, Line-1 MI, and -∆∆CtMDR1 mRNA were significant factors affecting cumulative survival after surgery in ESCC (Table 2). These results implied that Line-1 demethylation could play an important role in elevated MDR1 expression during tumorigenesis of ESCC.
Table 2.
Results of Cox regression analysis summarizing significant independent prognostic factors
| Factor | β | SE | Hazard ratio (95% CI) | P |
|---|---|---|---|---|
| Survival after surgery | ||||
| Age | 0.043 | 0.026 | 1.232 (1.024-1.082) | 0.034 |
| MI | 1.172 | 0.482 | 2.843 (1.408-7.023) | 0.026 |
| -∆∆CtMDR1 mRNA > 0 | 1.326 | 0.563 | 2.536 (1.327-5.534) | 0.014 |
CI, confidence interval; MI, methylation index.
Discussion
Esophageal cancer is the eighth most common cancer worldwide, and is the sixth most common cause of death from cancer. The predominant histological types of esophageal cancer are ESCC, which tends to localize in the East, such as Japan and China. Aberrant genome-wide hypomethylation and hypermethylation in cancer has been thought to be related to tumorigenesis [21]. DNA hypomethylation occurs at many genomic sequences, including repetitive elements and retrotransposons, thus resulting in genomic instability [22]. But the mechanism of global DNA hypomethylation during cancer development is still not fully understood [23].
As a good indicator of global DNA methylation level, we also found that Line-1 hypomethylation is an important feature in ESCC. Firstly, the MI of Line-1 decreased from 0.90 in non-tumor samples toward 0.78 in ESCC. There was a significant demethylation trend of Line-1 in ESCC, which implied that Line-1 hypomethylation is one of the most common molecular abnormalities during the carcinogenic process. Secondly, Line-1 hypomethylation were significantly associated with clinicopathologic features, such as age, AJCC stage, and differentiation. And our results implied that the Line-1 hypomethylation have been shown to be significantly associated with poor survival in ESCC. It could be some important factors resulting in Line-1 hypomethylation including age, AJCC stage, and differentiation. S. Iwagami et al. demonstrated that 30 ESCC tissues show far lower Line-1 methylation levels than matched normal mucosa by bisulfite pyrosequencing assay [24]. Line-1 hypomethylation was significantly associated with disease-free survival in their large samples study using 217 curatively resected ESCC specimens [25]. Our results indicated that Line-1 hypomethylation in ESCC with MI ≤ 0.78 is associated with a shorter survival (34 months) than those with MI > 0.78 (43 months), thus suggesting that it has potential for use as a prognostic biomarker.
Our current study revealed a relationship between MDR1 expression and Line-1 hypomethylation, suggesting that aberrant expression of MDR1 might be epigenetically regulated in that aggressive type of esophageal cancer. The multidrug resistance protein 1 encoded by ABCC1 was originally discovered as a cause of multidrug resistance in tumor cells. However, MDR1 serves a broader role than simply mediating the ATP-dependent efflux of drugs from cells, which likely influences the etiology and progression of tumorigenesis and clinical chemotherapy [26].
Increased MDR1 expression is an important factor in patients with ESCC. Molecular correlates with MDR1 activation may be important for better understanding of Line-1 hypomethylation. There was a significant correlation between Line-1 hypomethylation and chromosomal aberrations in gastrointestinal stromal tumors. Generation of genomic instability by hypomethylation seems to depend on the propensity of hypomethylated DNA [27,28]. Our results indicated that elevated expression of MDR1 is significantly associated with Line-1 demethylation in ESCC. The MDR1 expression was higher in ESCC with Line-1 demethylation than that with Line-1 hypermethylation. Results of Cox regression analysis indicated that Line-1 demethylation and elevated MDR1 expression were significantly associated with patient survival in ESCC. These results implied that epigenetic changes in Line-1 are accompanied by the alter expression of MDR1, and demethylation of Line-1 was associated with elevated expression of MDR1 mRNA in ESCC.
Conclusions
Our paper demonstrates that Line-1 hypomethylation was associated with advance cancer-specific and overall mortality in ESCC. Meanwhile, Line-1 demethylation could play an important role in elevated MRD1 expression during carcinogenesis of ESCC. Associations have been drawn between MDR1 expression, Line-1 hypomethylation and prognosis in esophageal cancer; thus, our findings may provide new insights into the biological mechanisms of ESCC tumorigenesis and progression.
Acknowledgements
This study was supported by Jiangsu Provincial Special Program of Medical Science (Grant No: BL2013012); Changzhou Sci&Tech Program, China (Grant No: CE20155043, CJ20159023); the Advanced Medical Innovation Talents Project of Changzhou.
Disclosure of conflict of interest
None.
References
- 1.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 2.Shang L, Wang M. Molecular alterations and clinical relevance in esophageal squamous cell carcinoma. Front Med. 2013;7:401–410. doi: 10.1007/s11684-013-0286-y. [DOI] [PubMed] [Google Scholar]
- 3.Napier KJ, Scheerer M, Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112–120. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahrens TD, Werner M, Lassmann S. Epigenetics in esophageal cancers. Cell Tissue Res. 2014;356:643–655. doi: 10.1007/s00441-014-1876-y. [DOI] [PubMed] [Google Scholar]
- 6.Ling Y, Huang G, Fan L, Wei L, Zhu J, Liu Y, Zhu C, Zhang C. CpG island methylator phenotype of cell-cycle regulators associated with TNM stage and poor prognosis in patients with oesophageal squamous cell carcinoma. J Clin Pathol. 2011;64:246–251. doi: 10.1136/jcp.2010.082875. [DOI] [PubMed] [Google Scholar]
- 7.Toh Y, Egashira A, Yamamoto M. Epigenetic alterations and their clinical implications in esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg. 2013;61:262–269. doi: 10.1007/s11748-013-0235-3. [DOI] [PubMed] [Google Scholar]
- 8.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M, Agodi A. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PLoS One. 2014;9:e109478. doi: 10.1371/journal.pone.0109478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano H, Saeki H, Kitao H, Tsuda Y, Otsu H, Ando K, Ito S, Egashira A, Oki E, Morita M, Oda Y, Maehara Y. Chromosomal instability associated with global DNA hypomethylation is associated with the initiation and progression of esophageal squamous cell carcinoma. Ann Surg Oncol. 2014;21(Suppl 4):S696–702. doi: 10.1245/s10434-014-3818-z. [DOI] [PubMed] [Google Scholar]
- 11.Shigaki H, Baba Y, Watanabe M, Iwagami S, Miyake K, Ishimoto T, Iwatsuki M, Baba H. LINE-1 hypomethylation in noncancerous esophageal mucosae is associated with smoking history. Ann Surg Oncol. 2012;19:4238–4243. doi: 10.1245/s10434-012-2488-y. [DOI] [PubMed] [Google Scholar]
- 12.Baba Y, Watanabe M, Murata A, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Iwagami S, Yoshida N, Oki E, Sakamaki K, Nakao M, Baba H. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clin Cancer Res. 2014;20:1114–1124. doi: 10.1158/1078-0432.CCR-13-1645. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 14.Chang XB. Molecular mechanism of ATPdependent solute transport by multidrug resistance-associated protein 1. Methods Mol Biol. 2010;596:223–249. doi: 10.1007/978-1-60761-416-6_11. [DOI] [PubMed] [Google Scholar]
- 15.Cascorbi I, Haenisch S. Pharmacogenetics of ATP-binding cassette transporters and clinical implications. Methods Mol Biol. 2010;596:95–121. doi: 10.1007/978-1-60761-416-6_6. [DOI] [PubMed] [Google Scholar]
- 16.Gillet JP, Gottesman MM. Mechanisms of multidrug resistance in cancer. Methods Mol Biol. 2010;596:47–76. doi: 10.1007/978-1-60761-416-6_4. [DOI] [PubMed] [Google Scholar]
- 17.Tabe Y, Konopleva M, Contractor R, Munsell M, Schober WD, Jin L, Tsutsumi-Ishii Y, Nagaoka I, Igari J, Andreeff M. Up-regulation of MDR1 and induction of doxorubicin resistance by histone deacetylase inhibitor depsipeptide (FK228) and ATRA in acute promyelocytic leukemia cells. Blood. 2006;107:1546–1554. doi: 10.1182/blood-2004-10-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kathawala RJ, Gupta P, Ashby CR Jr, Chen ZS. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist Updat. 2015;18:1–17. doi: 10.1016/j.drup.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Xu Y, Zhao J, Fan L, Jiang G, Li R, Ling Y, Wu M, Wei L. Elevated expression of the stem cell marker CD133 associated with Line-1 demethylation in hepatocellular carcinoma. Ann Surg Oncol. 2011;18:2373–2380. doi: 10.1245/s10434-011-1599-1. [DOI] [PubMed] [Google Scholar]
- 20.Fan T, Zhang C, Zong M, Zhao Q, Yang X, Hao C, Zhang H, Yu S, Guo J, Gong R, Fan S, Wei L, Fan L. Peptidylarginine deiminase IV promotes the development of chemoresistance through inducing autophagy in hepatocellular carcinoma. Cell Biosci. 2014;4:49. doi: 10.1186/2045-3701-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sincic N, Herceg Z. DNA methylation and cancer: ghosts and angels above the genes. Curr Opin Oncol. 2011;23:69–76. doi: 10.1097/CCO.0b013e3283412eb4. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba Y, Watanabe M, Baba H. Review of the alterations in DNA methylation in esophageal squamous cell carcinoma. Surg Today. 2013;43:1355–1364. doi: 10.1007/s00595-012-0451-y. [DOI] [PubMed] [Google Scholar]
- 24.Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ida S, Nagai Y, Ishimoto T, Iwatsuki M, Sakamoto Y, Miyamoto Y, Baba H. Pyrosequencing assay to measure LINE-1 methylation level in esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19:2726–2732. doi: 10.1245/s10434-011-2176-3. [DOI] [PubMed] [Google Scholar]
- 25.Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Sakamaki K, Ohashi Y, Baba H. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg. 2013;257:449–455. doi: 10.1097/SLA.0b013e31826d8602. [DOI] [PubMed] [Google Scholar]
- 26.Cole SP. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J Biol Chem. 2014;289:30880–30888. doi: 10.1074/jbc.R114.609248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi S, Suzuki H, Niinuma T, Shimizu H, Nojima M, Iwaki H, Nobuoka T, Nishida T, Miyazaki Y, Takamaru H, Yamamoto E, Yamamoto H, Tokino T, Hasegawa T, Hirata K, Imai K, Toyota M, Shinomura Y. A novel correlation between LINE-1 hypomethylation and the malignancy of gastrointestinal stromal tumors. Clin Cancer Res. 2010;16:5114–5123. doi: 10.1158/1078-0432.CCR-10-0581. [DOI] [PubMed] [Google Scholar]
- 28.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]


