Abstract
Hepatic stellate cells (HSCs) play an important role in liver fibrosis. This study investigates the expression of hedgehog in HSC and the role of hedgehog signaling on activation and collagen secretion of HSC. Liver ex vivo perfusion with collagenase IV and density gradient centrifugation were used to isolate HSC. Expression of hedgehog signaling components Ihh, Smo, Ptc, Gli2 and Gli3 in HSC were detected by RT-PCR. Hedgehog siRNA vectors targeting Ihh, Smo and Gli2 were constructed and transfected into HSC respectively. Suppression of hedgehog signaling were detected by SYBR Green fluorescence quantitative RT-PCR. Effects of hedgehog signaling inhibition on HSC activation and collagen I secretion were analyzed. Hedgehog signaling components Ihh, Smo, Ptc, Gli2 and Gli3 were expressed in HSC. siRNA vectors targeting Ihh, Smo and Gli2 were successfully constructed and decreased target gene expression. Suppression of hedgehog signaling significantly decreased the expression of α-SMA in HSC (P<0.01). Collagen type I secretion of HSC were also significantly decreased (P<0.01). In summary, HSC activation and collagen secretion can be regulated by hedgehog signaling. Hedgehog may play a role in the pathogenesis of liver fibrosis.
Keywords: Hepatic stellate cell, hedgehog, liver fibrosis, activation, collagen secretion
Introduction
Hepatic fibrosis is a common result of chronic liver injury and may progress to liver cirrhosis and portal hypertension. It has been proved that hepatic stellate cells (HSCs) play a key role in hepatic fibrosis and portal hypertension [1-8]. HSCs reside in the space of Disse, in close contact with sinusoidal endothelial cells and hepatocytes. A characteristic feature of HSC is that they possess long, branching cytoplasmic processes. In normal liver, HSCs are responsible for the transport and storage of retinoids and for the production of basement membrane components. After liver injury, HSCs are activated. The maker of HSCs activated phenotype is the expression of the intracellular microfilament protein α-smooth muscle actin (α-SMA) [9-13]. Activated HSCs lose retinoids, produce increased level of extracellular matrix (ECM) e.g. collagen, vimentin, et al., and express cytokins and its receptors. Because HSC play a role in both hepatic fibrosis and portal hypertension, they are postulated therapeutic target in the prevention and treatment the complications of chronic liver disease.
Hedgehog signaling pathway is known to regulate critical cellular responses including proliferation, apoptosis, migration, and differentiation. It also modulates wound healing responses in a number of adult tissues and organs, including the liver during progression of chronic liver diseases [14]. Normal liver usually expresses quite low levels of hedgehog signaling pathway components. Moreover, hedgehog signaling has been reported to be progressively increased during the process of liver injury [15,16]. Hedgehog signaling may be contributes to the process of HSC activation and their ability to synthesize ECM components.
In this study, we examined the expression of hedgehog pathway components in HSC. And hedgehog signaling was suppressed in HSC, to observe the activation and collagen secretion of HSC.
Materials and methods
Isolation and culture of primary rat HSCs
Rat HSCs were isolated from normal Wistar rats by liver ex vivo perfusion of collagenase type IV and density gradient centrifugation methods [17-20]. Isolated HSCs were seeded on uncoated plastic tissue culture dishes and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, New York, USA) supplemented with 20% fetal bovine serum (FBS) (Hyclone, Logan, UT, USA). Cell viability was determined by Trypan blue exclusion staining. Cells were counted under fluorescence microscopy for auto-fluorescence at a wavelength of 328 nm. HSC purity was identified by immunocytochemistry detection of the expression of either desmin or glial fibrillary acidic proteins (GFAP). Details of the primary antibodies are shown in Table 1. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Peking University People’s Hospital (Permit Number: RDB2012-11). All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Table 1.
Antibodies used for immunocytochemistry and Western blot
| Primary antibody | Source | Company, Country |
|---|---|---|
| desmin | Mouse, monoclonal | DAKO, Denmark |
| gliad fibrillary acidic proteins (GFAP) | Mouse, monoclonal | DAKO, Denmark |
| α-smooth muscle actin (α-SMA) | Mouse, monoclonal | Santa Cruz, CA, USA |
| collagen type I | Rabbit, polyclonal | Santa Cruz, CA, USA |
| α-tubulin | Goat, polyclonal | Santa Cruz, CA, USA |
Expression of hedgehog signaling pathway in HSC
Total RNA of HSC was isolated from subconfluent cells using Trizol Reagent (Gibco BRL). Hedgehog signaling pathway components Ihh, Smo, Ptc, Gli2 and Gli3 expression were detected by RT-PCR. Primer sequences and product lengths are shown in Table 2. β-actin was used as a control. All PCR reaction was performed in a total volume of 25 μl. PCR mixture contained 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1.5 mM MgCl, 0.01% gelatin, 200 pmol each primer, 2 U Taq DNA polymerase, and 0.5 μl cDNA. Amplification was performed by denaturation at 94°C for 2 min, followed by 28 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s with a final extension at 72°C for 5 min. PCR products were electrophoresed on 2% agarose gel.
Table 2.
RT-PCR primers and products lengths
| Primers | Sequences | Length of the Products |
|---|---|---|
| Ihh | 5-AAACTCGTGCCTCTTGCCTA-3 | 229 bp |
| 5-TGACAGAGATGGCCAGTGAG-3 | ||
| Smo | 5-AATTGGCCTGGTGCTTATTG-3 | 183 bp |
| 5-CTGAAGGTGATGAGCACGAA-3 | ||
| Ptc | 5-ATTTCTTGCCCTTGGTGTTG-3 | 166 bp |
| 5-GAAGGCAGTGACATTGCTGA-3 | ||
| Gli2 | 5-ACGCTAAGTGGCAGTCCTGT-3 | 246 bp |
| 5-TGGGGCAGCGAGACTAAATA-3 | ||
| Gli3 | 5-GGGGACAAAGATGAAAGCAA-3 | 194 bp |
| 5-GCTTTGAACGGTTTCTGCTC-3 | ||
| β-actin | 5-TGGGACGATATGGAGAAGAT-3 | 523 bp |
| 5-ATTGCCGATAGTGATGACCT-3 |
Construction of hedgehog signaling siRNA vector
According to the RT-PCR results of hedgehog signaling pathway components expression, Ihh, Smo and Gli2 were selected for RNA interference. Using Qiagen software to design targeting sequences. Each interference gene designs 3 to 5 interference targets. Targeting sequences were synthesized and then connected into the linear eukaryotic expression vector. The connected products were transfected into the prepared JM109 bacterial competent cells, to prepare recombinant plasmid pEGFP-N1 (purchased from Shanghai GeneChem Co., Ltd.) containing hedgehog signaling siRNA sequence. The clonal was confirmed by sequencing.
Suppression of hedgehog signaling in HSC
Recombinant hedgehog siRNA vectors were transfected into HSCs. Transfection steps were in accordance with the Lipofectamine 2000 manufacturer’s instructions. Suppression of hedgehog signaling were detected by SYBR Green fluorescence quantitative RT-PCR. Total cellular RNA was extracted using RNeasy Mini Kits (Qiagen, Hilden, GA) and reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed via ABI PRISM 7500 Real-Time PCR Systerm (Applied Biosystems) with 1× SYBR Green Universal PCR Mastermix (Takara). Quantitative PCR was performed by pre-denaturation at 95°C for 1 min, followed by 40 cycles at denaturation 95°C for 15 s, annealing at 60°C for 15 s, and extending at 72°C for 20 s. Transcript levels were calculated according to the ΔΔCt method. According to quantitative PCR results, each interference gene selects one interference target (Table 3).
Table 3.
Sequence of siRNA
| Target | Loop | ||
|---|---|---|---|
| Ihh | 5’-ACCTTCAGCGATGTGCTCAT | TTCAAGAGA | ATGAGCACATCGCTGAAGGT-3’ |
| Smo | 5’-TACCAAGAAGCCCATTCCTGA | TTCAAGAGA | TCAGGAATGGGCTTCTTGGTA-3’ |
| Gli2 | 5’-AGCAGCAACTGTATAAGTGA | TTCAAGAGA | TCACTTATACAGTTGCTGCT-3’ |
Effect of hedgehog signaling inhibition on expression of α-SMA in HSC
HSC cells were allocated into five groups, group 1 as control, group 2: the blank plasmid (Non) group, group 3: siRNA-Ihh group, group 4: siRNA-Smo group, and group 5: siRNA-Gli2 group. Group 2 was transfected with blank plasmid not containing targeting sequence. Group 3 to 5 were transfected with siRNA vectors containing Ihh, Smo and Gli2 targeting sequences respectively. After 72 h transfection, cells were collected and cellular total proteins were extracted, expression of α-SMA was detected by Western blot. The gray scale ratio of α-SMA/α-tubulin represents the relative expression amount.
Effect of hedgehog signaling inhibition on secretion of collagen type I in HSC
Grouping method as mentioned, effect of Ihh, Smo and Gli2 suppression on secretion of type I collagen in HSCs were measured. Cell culture supernatant were collected after 72 h transfection, ELSIA method was used to determination of the content of collagen type I, differences were compared between groups.
Statistical analysis
The final value of data was expressed as mean ± standard deviation (x̅ ± s) and statistical analyses included the univariate variance analysis and t-test. P<0.05 was considered statistically significant.
Results
Primary culture of rat HSC
The yield rate of HSC was 2.82×107 with the cell viability exceeding 98%. The purity of HSC identified by double staining with antibodies against desmin and GFAP was higher than 97%. (Figure 1).
Figure 1.
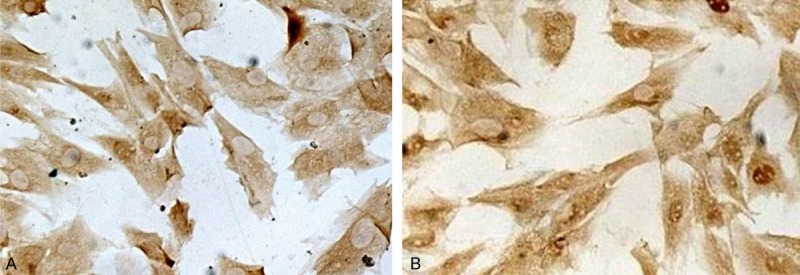
Primary HSC identified by immunocytochemistry staining of desmin and GFAP. A: HSC were positive of desmin (original magnification, ×200); B: HSC were positive of GFAP (original magnification, ×200).
Hedgehog signaling pathway components expressed in HSC
Hedgehog signaling pathway components, Ihh, Ptc, Smo, Gli2 and Gli3 gene were expressed in HSC, as detected by RT-PCR and shown in Figure 2A.
Figure 2.
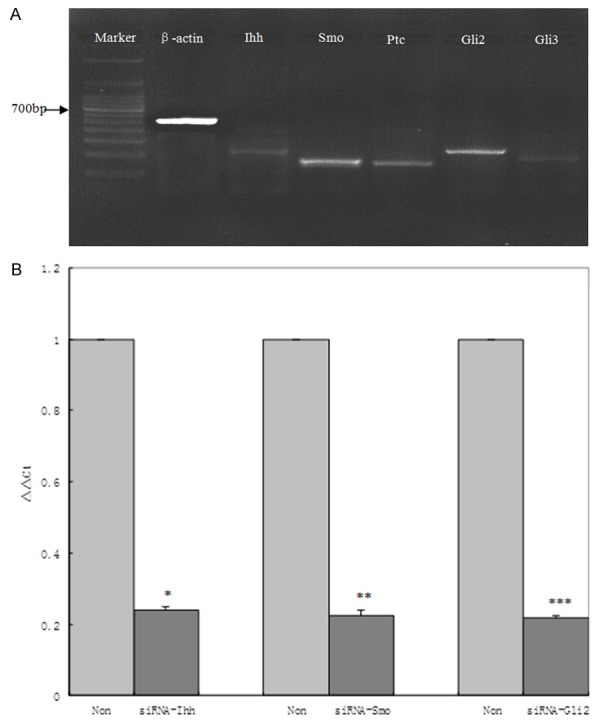
A: Hedgehog signaling components, Ihh, Ptc, Smo, Gli2 and Gli3 gene were expressed in HSC, marker: 100bp DNA ladder. B: After RNA interference, the expression of Ihh, Smo and Gli2 decreased. Compared with non-interference group, *,**,***P<0.01.
Down-regulation of hedgehog signaling inhibit HSC activation and collagen secretion
After RNA interference, the hedgehog signaling expression of Ihh, Smo and Gli2 decreased. Fluorescence quantitative RT-PCR results showed that, when the expression of non-interference group was set to 1, the expression of Ihh, Smo and Gli2 gene were 0.261 ± 0.103, 0.247 ± 0.115 and 0.228 ± 0.098 respectively in siRNA-Ihh, siRNA-Smo and siRNA-Gli2 groups (Figure 2B). Differences between non-interference and interference groups were significant (P<0.01).
When hedgehog signaling was suppressed in HSC, expression of α-SMA was inhibited (Figure 3A). The scale ratio of α-SMA/α-tubulin in siRNA-Ihh, siRNA-Smo and siRNA-Gli2 group were 0.203 ± 0.009, 0.298 ± 0.017 and 0.096 ± 0.013 respectively, significant lower than that of the control and blank group (Figure 3B, 1.069 ± 0.018 and 1.352 ± 0.009 respectively, P<0.01).
Figure 3.
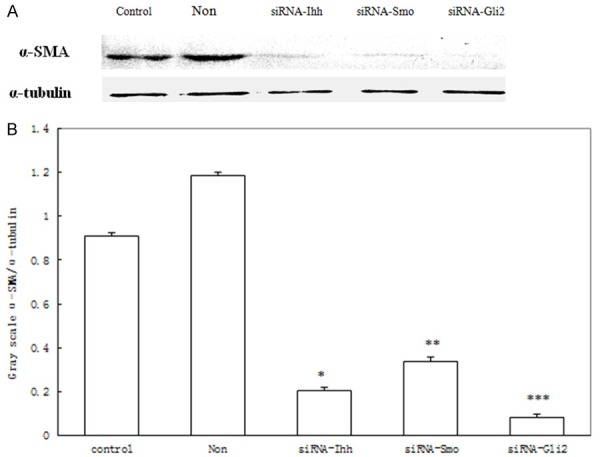
A: When hedgehog was suppressed in HSC, expression of α-SMA was inhibited. B: The scale ratio of α-SMA/α-tubulin in siRNA-Ihh, siRNA-Smo and siRNA-Gli2 group were significant lower than that of the control and blank group, *,**,***P<0.01.
Collagen type I secretion of HSC was also inhibited by hedgehog signaling pathway suppression. As shown in Figure 4, collagen type I content of cell culture supernatant in siRNA-Ihh, siRNA-Smo and siRNA-Gli2 group were 20.88 ± 1.5 ng/mL, 17.93 ± 0.9 ng/mL and 18.69 ± 1.2 ng/mL respectively, significant lower than that of the control and blank group (57.36 ± 3.1 ng/mL and 51.75 ± 2.5 ng/mL respectively, P<0.01).
Figure 4.
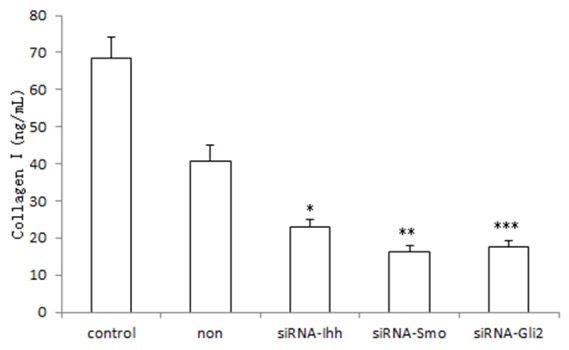
Collagen type I secretion of siRNA-Ihh, siRNA-Smo and siRNA-Gli2 group were significant lower than that of the control and blank group, *,**,***P<0.01.
Discussion
Research on HSCs has made great advances since Knook [17] successfully isolated and cultured HSC in 1982. Subsequently, numerous studies have shown that HSC plays a fundamental role in liver fibrogenesis [1-8]. HSC has become a focus in understanding the mechanisms involved in hepatic fibrogenesis.
There is emerging evidence that hedgehog, a master developmental regulator [21,22], becomes reactivated during adult wound healing [23]. Hedgehog signaling is initiated by the interaction of a family of ligands (Sonic hedgehog-Shh, Indian hedgehog-Ihh, and Desert hedgehog-Dhh) which interact with the Patched (Ptc) specific cell surface receptor that is expressed on the plasma membrane of responsive cell. Ligand-receptor interaction de-represses activity of another molecule, Smoothened (Smo), responsible for the propagation of the signal that leads to nuclear translocation of members of Glioblastoma (Gli1, Gli2, Gli3) family transcription factors, that regulate the expression of a number of critical Gli-target genes. In the absence of hedgehog ligands, Ptc represses Smo and leads to Gli ubiquitination and subsequent proteasomal degradation. Although the exact role of hedgehog in adult tissue repair remains somewhat obscure, the pathway seems to control critical cell fate decisions that are required for reconstruction of healthy tissue because fibrosis typically result when hedgehog signaling becomes deregulated [21]. Like several of the key cell types involved in liver repair, HSCs are hedgehog responsive [22,23]. The current study focused on HSC activation and collagen secretion. We found that hedgehog signaling pathway components were expressed in HSC. When hedgehog signaling was suppressed in HSC, expression of α-SMA and Collagen type I secretion of HSC were inhibited. Hedgehog signaling directly and significantly contributes to the process of HSC activation; drive quiescent HSC towards the activated phenotype. Our results suggested that the activation of HSC can be regulated by hedgehog signaling. The activation of hedgehog signaling in HSCs maybe contributes and sustains liver fibrogenesis. Hedgehog signaling promotes the ability of HSC synthesis and secretion of ECM [21].
In summary, our results suggested that HSC express hedgehog signaling pathway components. HSC activation and collagen secretion can be regulated by hedgehog signaling.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (No. 30872498) and Peking University People’s Hospital Research and Development Funds (No. RDB2014-08). This study was approved by the Ethics Committee of the Peking University People’s Hospital (No. RDB-2014-08).
Disclosure of conflict of interest
None.
References
- 1.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockey DC. The cell and molecular biology of hepatic fibrogenesis. Clinical and therapeutic implications. Clin Liver Dis. 2000;4:319–355. doi: 10.1016/s1089-3261(05)70113-6. [DOI] [PubMed] [Google Scholar]
- 3.Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 4.Marra F, Pinzani M. Role of hepatic stellate cells in the pathogenesis of portal hypertension. Nefrologia. 2002;22(Suppl 5):34–40. [PubMed] [Google Scholar]
- 5.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novo E, Cannito S, Paternostro C, Bocca C, Miglietta A, Parola M. Cellular and molecular mechanisms in liver fibrogenesis. Arch Biochem Biophys. 2014;548:20–37. doi: 10.1016/j.abb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Povero D, Busletta C, Novo E, di Bonzo LV, Cannito S, Paternostro C, Parola M. Liver fibrosis: a dynamic and potentially reversible process. Histol Histopathol. 2010;25:1075–1091. doi: 10.14670/HH-25.1075. [DOI] [PubMed] [Google Scholar]
- 8.Friedman SL. Liver fibrosis: from mechanisms to treatment. Gastroenterol Clin Biol. 2007;31:812–814. doi: 10.1016/s0399-8320(07)73970-2. [DOI] [PubMed] [Google Scholar]
- 9.Rockey DC, Boyles JK, Gabbiani G, Friedman SL. Rat hepatic lipocytes express smooth muscle actin upon activation in vivo and in culture. J Submicrosc Cytol Pathol. 1992;24:193–203. [PubMed] [Google Scholar]
- 10.Reynaert H, Thompson MG, Thomas T, Geerts A. Hepatic stellate cells: role in microcirculation and pathophysiology of portal hypertension. Gut. 2002;50:571–581. doi: 10.1136/gut.50.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rockey DC. Hepatic fibrosis, stellate cells, and portal hypertension. Clin Liver Dis. 2006;10:459–479. vii–viii. doi: 10.1016/j.cld.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Rockey DC. Hepatic blood flow regulation by stellate cells in normal and injured liver. Semin Liver Dis. 2001;21:337–349. doi: 10.1055/s-2001-17551. [DOI] [PubMed] [Google Scholar]
- 13.Friedman SL, Rockey DC, McGuire RF, Maher JJ, Boyles JK, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology. 1992;15:234–243. doi: 10.1002/hep.1840150211. [DOI] [PubMed] [Google Scholar]
- 14.Omenetti A, Choi S, Michelotti G, Diehl AM. Hedgehog signaling in the liver. J Hepatol. 2011;54:366–373. doi: 10.1016/j.jhep.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Choi SS, Michelotti GA, Chan IS, Swiderska-Syn M, Karaca GF, Xie G, Moylan CA, Garibaldi F, Premont R, Suliman HB, Piantadosi CA, Diehl AM. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology. 2012;143:1319–1329. e1–11. doi: 10.1053/j.gastro.2012.07.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sicklick JK, Li YX, Melhem A, Schmelzer E, Zdanowicz M, Huang J, Caballero M, Fair JH, Ludlow JW, McClelland RE, Reid LM, Diehl AM. Hedgehog signaling maintains resident hepatic progenitors throughout life. Am J Physiol Gastrointest Liver Physiol. 2006;290:G859–870. doi: 10.1152/ajpgi.00456.2005. [DOI] [PubMed] [Google Scholar]
- 17.Knook DL, Seffelaar AM, de Leeuw AM. Fatstoring cells of the rat liver. Their isolation and purification. Exp Cell Res. 1982;139:468–471. doi: 10.1016/0014-4827(82)90283-x. [DOI] [PubMed] [Google Scholar]
- 18.Friedman SL, Roll FJ. Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal Biochem. 1987;161:207–218. doi: 10.1016/0003-2697(87)90673-7. [DOI] [PubMed] [Google Scholar]
- 19.Alpini G, Phillips JO, Vroman B, LaRusso NF. Recent advances in the isolation of liver cells. Hepatology. 1994;20:494–514. [PubMed] [Google Scholar]
- 20.Friedman SL, Rockey DC, McGuire RF, Maher JJ, Boyles JK, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology. 1992;15:234–243. doi: 10.1002/hep.1840150211. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins D. Hedgehog signaling: emerging evidence for non-canonical pathways. Cell Signal. 2009;21:1023–1034. doi: 10.1016/j.cellsig.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Omenetti A, Syn WK, Jung Y, Francis H, Porrello A, Witek RP, Choi SS, Yang L, Mayo MJ, Gershwin ME, Alpini G, Diehl AM. Repair-related activation of hedgehog signaling promotes cholangiocyte chemokine production. Hepatology. 2009;50:518–527. doi: 10.1002/hep.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]


