Abstract
Background: A new diagnostic and prognostic biomarker may be of value in cancer diseases. Our study aimed to evaluate the CDKN1A/p21 and TGFBR2 level measurable in a cohort of patients with breast cancer after mastectomy, and to confirm their suitability to serve as prognostic biomarkers of the cancer. Methods: The expression levels of CDKN1A/p21 and TGFBR2 were detected by reverse transcription-PCR (RT-PCR), western blot assay and immunohistochemical staining for 65 primary tumor samples and paired adjacent noncancerous breast tissues. Their relations to clinicopathologic parameters and to the prognosis of patients with breast cancer were analyzed. Results: We found the mRNA and protein expression levels of CDKN1A/p21 were significantly upregulated in breast cancer tissues compared with adjacent nontumorous breast tissues. Increased CDKN1A/p21 expression showed a significant correlation with larger tumor size (P=0.014), higher tumor dedifferentiation grade (P=0.021), lymph node metastasis (P=0.019) and a shorter disease-free survival (P=0.044). Contrarily, the expression levels of TGFBR2 mRNA and protein were significantly decreased in breast cancer tissues compared with adjacent nontumorous breast tissues. Underexpression of TGFBR2 in breast cancer was correlated with larger tumor size (P=0.034), lymph node metastasis (P=0.039) and a shorter disease-free survival (P=0.035). Statistical analysis suggested that there was no significant association between CDKN1A/p21 and TGFBR2 expression. Conclusions: in summary, our results suggested that high CDKN1A/p21 and low TGFBR2 expression was closely correlated with adverse pathological parameters and poor prognosis in breast cancer. Both CDKN1A/p21 and TGFBR2 are presented as possible candidates for breast cancer biomarkers.
Keywords: CDKN1A/p21, TGFBR2, breast cancer, biomarker
Introduction
It is well recognized that breast cancer is a heterogeneous disease. Although remarkable progress has been made in the early detection and treatment of breast cancer over the years, behavior is variable. Therefore, it is important to identify potential markers for the prognosis and also aid the selection of appropriate therapy, and it may be of value in the management of individual patients.
Cell cycle regulator p21, the protein product encoded by cyclin-dependent kinase inhibitor 1A (CDKN1A) gene, was first identified as acyclin-dependent kinase (Cdk) inhibitor with the ability to cause growth arrest through inhibition of Cdks, which are required for G1 to S transition [1]. In addition, by interaction with proliferating cell nuclear antigen (PCNA), CDKN1A/p21 was found to inhibit DNA replication [2]. p21 is widely expressed at low levels in most tissues under steady state, its expression is increased in response to DNA damage or other chemical or physical cellular stressors, plays a critical role in cell survive and genetic fidelity, by resulting in the activation of cell cycle checkpoints until repair has taken place. Because carcinogenesis closely related to cell cycle regulation, the roles of p21 in carcinoma progression have attracted great attention. Several studies have suggested CDKN1A/p21 promotes tumors, it may also mediate a drug-resistance phenotype [3,4], and clinical studies have indicated that high p21 expression was correlated with poor prognosis [5,6]. However, the functional role of CDKN1A/p21 in carcinogenesis remains controversial. Loss of expression or function of CDKN1A/p21 has been implicated in the genesis or progression of in a variety of carcinomas, including breast cancer [7,8], and it was correlated with poor prognosis clinically. These contrasting observations have undoubtedly increased the significance of p21 in the field of cancer biology. Moreover, to date, no consensus has been reached about the relationship between CDKN1A/p21 and clinicopathological parameters, and the characteristics of CDKN1A/p21 expression and its clinical/prognostic significance in human breast cancer remain unclear.
It is generally accepted that p21 expression is tightly controlled by the famous tumor suppressor p53, involved in mediate p53-dependent cell cycle arrest, DNA repair and apoptosis in response to various cellular stressors [9]. On the other hand, several studies have demonstrated that CDKN1A/p21 expression can be regulated by other p53-independent pathways, including transforming growth factor beta (TGF-β) signaling [7]. TGF-β belongs to TGF-β protein superfamily, plays an essential role in regulating cell growth, differentiation and apoptosis, and it is one of the important components of cellular microenvironment [10]. TGF-β plays an important role in the process of carcinogenesis, but its role remains complicated. At the early stage of tumorigenesis, TGF-β functions as a tumor suppressor through inhibiting the proliferation of tumor cells and promote apoptosis, whereas at advanced stages TGF-β involved in the process of cancer development, through promoting tumor cells invasion and metastasis, angiogenesis and immune escape [11]. Therefore, TGF-β pro-or anti-tumorigenesis depends on the cell and tissue contexts. The activation of TGF-β signal transduction begins with ligand binding to the TGF-β receptor type II (TGFBR2), the TGF-β receptor can regulate Smad or non-Smad signaling pathways, and then ultimately dictate TGF-β’s biological effects [12]. TGF-β ligands and their receptors are widely expressed in multiple human tissues; the regulatory role played by these growth factors is very importance in the occurrence and development of cancer. Previous researches found that the expression of TGFBR2 was often low in a wide variety of malignant tumor, including breast cancer. Moreover, it has been reported that p21 and TGF-β induced tumor cells apoptosis appear to depend on relatively high expression of TGFBR2, and then activate the MAPK-ERK and SMAD pathways [13]. Therefore, loss of TGF-β receptors expression could help tumor cells escape from TGF-β mediated inhibition may be a predictor of poor prognosis. However, there are few studies focused on the correction between TGFBR2 expression and prognosis in breast cancer, in addition, to our knowledge, no reports have investigated the correlation between CDKN1A/p21 and TGFBR2 in human breast cancer samples. Therefore, it is necessary to perform further investigation to understand the prognostic value of TGFBR2 in breast cancer.
In the current study, we sought to assess the expression levels of CDKN1A/p21 and TGFBR2 in a cohort of breast cancer patients from our institute, and evaluate their correlation with established pathological parameters and the prognosis of breast cancer patients.
Materials and methods
Patients and tissue samples
Breast tumor samples were obtained from 65 patients with histologically proven primary breast cancer who underwent either mastectomy or wide local excision with axillary surgery at The Affiliated Tumor Hospital of Guangxi University (Nanning City, China) between January 2011 and January 2012. Patient eligibility criteria were not having received preoperative chemotherapy or radiation therapy. All these patients were women, and their clinicopathologic data were retrieved from clinical records and pathological reports. Matched fresh specimens of breast cancer and adjacent noncancerous breast tissues were collected for reverse transcriptase-polymerase chain reaction (RT-PCR) and western blotting. This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University (Nanning City, China), and previous informed consent was obtained.
RNA extraction and RT-PCR analysis
Total RNA from the tissues were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RT-PCR was performed as described in previous studies. The CDKN1A primer sequences were 5’-GGAAGGGACACACAAGAAGAAG-3’ and 5’-AGCCTCTACTGCCACCATCTTA-3’, the TGFBR2 primer sequences were 5’-CCATTCTTCTCAAGTCCCAAAG-3’ and 5’-ATTTTTCTCCCACAAGGCAGTA-3. Primers for GAPDH were 5’-CAAGGTCATCCATGACAACTTT-3’ and 5’-GTCCACCACCCTGTTGCTGTAG-3’. All primers were obtained from Invitrogen. Primer Premier 5.0 software (Applied Biosystems) was used to design the primers. The intensity of CDKN1A, TGFBR2 or GAPDH bands was quantified by using the Molecular Imager VersaDoc MP 4000 (Bio-Rad, Hercules, CA), and analyzed with the Image J software (National Institutes of Health, USA). The intensity value for CDKN1A and TGFBR2 RNA was normalized to the in-lane value of GAPDH, and this normalized ratio from duplicate lanes was averaged.
Western blotting assay
Tissue Total protein was extracted tissue lysis buffer and resolved by SDS-PAGE. After blocking, blots were probed with the appropriate primary antibodies anti-p21 antibody (Cell Signaling Technology, Beverly, MA) or anti-TGFBR2 antibody (Cell Signaling Technology, Beverly, MA) overnight at 4°C, and then washed and incubated with horseradish peroxidase-conjugated secondary antibodies. β-actin protein levels (Santa Cruz, CA) were used as a loading control. Bands were detected and imaged using a LiCor Odyssey scanner. The near-infrared fluorescent values of bands were quantified by using Odyssey 3.0 analytical software (LiCor, Lincoln, NE). The near-infrared fluorescence value for p21 and TGFBR2 protein was normalized to the in-lane value of β-actin, and this normalized ratio from duplicate lanes was averaged.
Immunohistochemical analysis
The p21 and TGFBR2 expression were also detected by immunohistochemical analysis. Paraffin sections (4-μm thick) of tumor tissue were subjected to immunohistochemical staining using the standard streptavidin-perosidase (SP) methods. The tissue sections were stained with p21 antibody (1:50 dilution; Santa Cruz Biotechnology) and TGFBR2 polyclonal antibody (1:50 dilution; Sigma, St. Louis, Mo) respectively. The staining levels of p21 and TGFBR2 were assessed using a semi-quantitative staining index method. The percentage of positive cells was assessed quantitatively and scored as follows: 0, <5% of the total counted cells were stained; 1, 5% to 24% of the total counted cells were stained; 2, 25% to 50% of the total counted cells were stained; and 3, >50% of the total counted cells were stained. Those having positive staining in less than 3 score were regarded as “negative”, greater than 3 score as “positive”. All immunohistochemical analyses were carried out in a single reference laboratory and evaluated by light microscopy blindly and independently by two pathologists.
Follow-up and prognostic study
We obtained follow-up data by direct communication with all the included patients after surgery. All patients were followed-up until the date of death or when censored at the latest date (March 30th 2015). All patients were received standard treatment base on the postoperative pathologic types and stages, including chemotherapy, radiotherapy or endocrinotherapy. Disease relapse and metastasis were diagnosed by clinical examination, ultrasonography, computed tomography (CT) or Magnetic Resonance Imaging (MRI) scans. The primary endpoint was disease-free survival (DFS), defined as the time interval from breast cancer surgery to the first evidence of recurrence and metastasis.
Statistical analysis
All statistical analyses were carried out using SPSS version 16.0 for Windows (SPSS Inc., Chicago). The differences between the CDKN1A/p21 or TGFBR2 levels in breast cancer tissue and control tissue were evaluated using Chi-square or the Student’s t-test. The relationship between CDKN1A/p21 or TGFBR2 expression and clinicopathological parameters was assessed using the Spearman’s rank correlation test. Correlation between CDKN1A/p21 and TGFBR2 expression was calculated by Spearman’s rank correlation coefficients. Kaplan-Meier analysis was employed to evaluate the distribution of disease-free survival (DFS). Univariate and multivariate Cox regression analyses were performed to determine independent prognostic factors. Differences were considered significant when the associated P value was less than 0.05.
Results
Patient characteristics
Table 1 summarizes the characteristics of patients in this study. The median age of the included patients was 46 years (range 33-69 years), and 63.1% of these patients were premenopausal. The patients’ tumor stage range from stage II A to stage III B. Major pathological parameters were available, including tumor size, location, histological grade, lymph node status, and ER, PR, and HER2 status, as determined by conventional IHC.
Table 1.
Patient and baseline tumor characteristics
| Characteristic | Number (%) |
|---|---|
| Age (years) | |
| ≤50 | 44 (67.7) |
| >50 | 21 (32.3) |
| Menopausal status | |
| Premenopausal | 41 (63.1) |
| Postmenopausal | 24 (36.9) |
| Tumor size (cm) | |
| ≤5 cm | 53 (81.5) |
| >5 cm | 12 (18.5) |
| Clinical nodal status | |
| Negative | 34 (52.3) |
| Positive | 31 (47.7) |
| ER status | |
| Negative | 18 (27.7) |
| Positive | 47 (72.3) |
| PR status | |
| Negative | 21 (32.3) |
| Positive | 44 (67.7) |
| HER2 status | |
| Negative | 42 (64.6) |
| Positive | 23 (35.4) |
| Ki-67 index | |
| ≤14% | 13 (20) |
| >14% | 52 (80) |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal receptor 2.
The expression of CDKN1A/p21 and TGFBR2 in breast cancer and normal breast tissues
The intensity of CDKN1A/p21 and TGFBR2 mRNA expression were measured by RT-PCR in 65 breast tissues and adjacent noncancerous tissues. The average intensity value of CDKN1A mRNA was 0.81±0.08 in the breast cancer tissues and 0.13±0.04 in normal breast samples (Figure 1A and 1B), Suggesting that the transcript level of CDKN1A was upregulated in breast cancer. The difference was statistically significant (P<0.01). To determine whether CDKN1A upregulation was also apparent at the translational level, p21 protein expression was also analyzed in these tissues. Western blotting analysis showed that the p21 protein was highly expressed in breast cancer samples, compare with normal breast tissues (0.56±0.06 vs. 0.14±0.02), and the difference was statistically significant (P<0.01), and shown in Figure 1C and 1D. These results indicated that CDKN1A/p21 is upregulated in breast cancer tissues. Furthermore, TGFBR2 mRNA expression was also analyzed in these tissues. The results show that the average intensity value of TGFBR2 mRNA was 0.454±0.02 in the breast cancer tissues and 0.513±0.04 in normal breast samples, with a statistically significant difference (Figure 2A and 2B). For the protein level, TGFBR2 expression was significantly lower in breast cancer tissues compare with adjacent noncancerous tissues (0.315±0.04 vs. 0.457±0.07), as shown in Figure 2C and 2D, suggesting that TGFBR2 was downregulated from normal breast tissue to breast cancer.
Figure 1.

The differences in the CDKN1A/p21 levels between breast tumor tissue and normal breast tissue. Increased CDKN1A RNA expression was found in breast tumor tissue (A, B). Increased expression of p21 protein was seen in breast tumor tissue (C, D). Data are means ± SEM. T, tumor tissue; N, normal breast tissue. *P<0.05.
Figure 2.
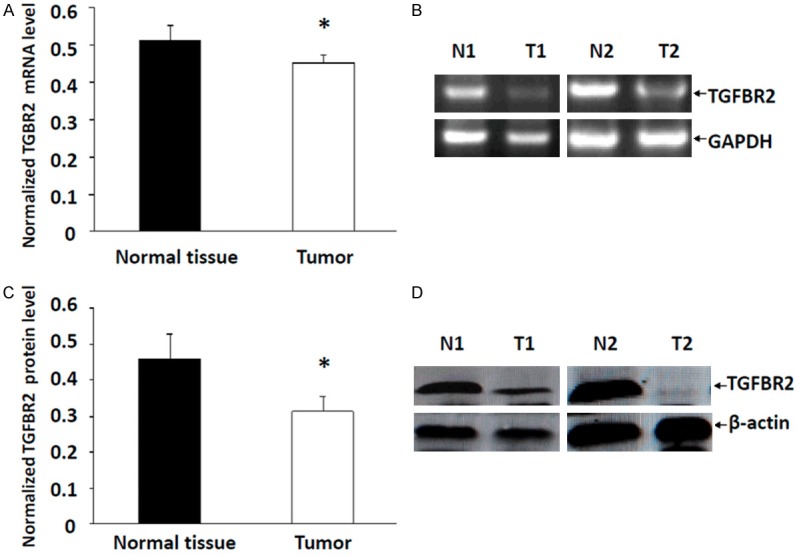
The differences of TGFBR2 levels between breast tumor tissue and normal breast tissue. Decreased TGFBR2 RNA expression was found in breast tumor tissue (A, B). Decreased expression of TGFBR2 protein was seen in breast tumor tissue (C, D). Data are means ± SEM. T, tumor tissue; N, normal breast tissue. *P<0.05.
Immunohistochemical staining for p21 and TGFBR2
In 65 breast cancer samples, 66.2% (43/65) of samples were positive for p21 expression, while 44.6% (29/65) were positive in the adjacent noncancerous samples, which was significantly different (P=0.021). As for TGFBR2 expression, the positive rate were 38.5% (25/65) in breast cancer samples and 72.3% (47/65) in adjacent noncancerous samples respectively, and the differences were statistical significance (P=0.000) (Table 2).
Table 2.
p21 and TGFBR2 protein expression in breast cancer and adjacent noncancerous tissue detected by immunohistochemical analysis
| Status | Breast cancer tissue (n) | Adjacent noncancerous tissue (n) | Χ2 | P | |
|---|---|---|---|---|---|
| p21 | + | 43 | 29 | 6.01 | 0.021 |
| - | 22 | 36 | |||
| TGFBR2 | + | 25 | 47 | 15.07 | 0.000 |
| - | 40 | 18 |
+, positive; -, negative.
Association of p21 and TGFBR2 protein expression with clinicopathological parameters of breast cancer patients
To investigate the role of CDKN1A/p21 and TGFBR2 in the clinical progression of breast cancer, the expression levels of the proteins were analyzed against the clinicopathological variables of the breast cancer patients. The results indicated that p21 protein expression was significantly associated with larger tumor size (P=0.014), higher tumor dedifferentiation grade (P=0.021) and lymph node metastasis (P=0.019). However, no association with age (P=0.142), menstrual status (P=0.082), ER status (P=0.122), or HER2 status (P=0.059) was identified. By contrast, a loss of TGFBR2 protein expression was closely associated with larger tumor size (P=0.034) and lymph node metastasis (P=0.039). There was no association with age (P=0.142), tumor dedifferentiation grade (P=0.055), menstrual status (P=0.082), ER status (P=0.154) or HER2 status (P=0.063) (Table 3).
Table 3.
Spearman analysis of correlation between p21, TGFBR2 protein expression and clinicopathological characteristics
| Variables | p21 expression level | TGFBR2 expression level | ||
|---|---|---|---|---|
|
|
||||
| Spearman correlation | P value | Spearman correlation | P value | |
| Age | 0.002 | 0.142 | 0.002 | 0.142 |
| Menstrual status | 0.014 | 0.082 | 0.014 | 0.082 |
| Tumor size | 0.216 | 0.014 | 0.186 | 0.034 |
| Tumor dedifferentiation grade | 0.193 | 0.021 | 0.028 | 0.055 |
| Lymph node status | 0.203 | 0.019 | 0.178 | 0.039 |
| ER status | 0.005 | 0.122 | 0.011 | 0.154 |
| HER2 status | 0.033 | 0.059 | 0.017 | 0.063 |
| TGFBR2 expression level | -0.041 | 0.067 | ||
ER, estrogen receptor; HER-2, human epidermal receptor.
Association between p21 and TGFBR2 expression in breast cancer
The expression of p21 was compared with TGFBR2 expression in breast cancer, and the results were showed in Table 3. Spearman’s rank correlation test showed that the expression of p21 protein was not significantly associated with TGFBR2 expression (P=0.067).
Prognostic implications of p21 and TGFBR2 expression
To detect the relation between p21 or TGFBR2 levels and breast cancer prognosis, 65 patients were divided into different groups according to p21 or TGFBR2 protein expression. A Kaplan-Meier analysis showed that p21-positive expression was strongly associated with decreased DFS (Figure 3A, P=0.044), and patients with TGFBR2-negative expression had shorter DFS than patients with positive TGFBR2 expression (Figure 3B, P=0.035). To test whether p21 or TGFBR2 expression is the independent factors predicting prognosis, univariate and multivariate Cox regression analyses were performed to identify independent prognostic factors. The multivariate analysis showed that p21, TGFBR2, lymph node metastasis and tumor dedifferentiation grade were identified as independent predictive factors for DFS (Table 4).
Figure 3.
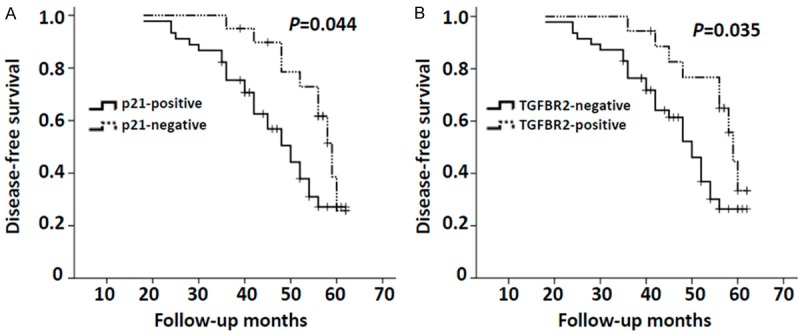
A. Kaplan-Meier curves of disease-free survival according to the p21 expression status (p21-positive and p21-negative). Log-rank tests indicated that there were significant differences in the disease-free survival of two subgroups (P=0.044). B. Kaplan-Meier curves of disease-free survival according to the TGFBR2 expression status (TGFBR2-negative and TGFBR2-positive). Log-rank tests indicated that there were significant differences in the disease-free survival of two subgroups (P=0.035).
Table 4.
Cox regression analyses of p21, TGFBR2, clinical variables and DFS
| Characteristic | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
||||
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| Age | 1.41 (0.65-2.48) | 0.287 | ||
| Menstrual status | 1.21 (0.63-2.59) | 0.412 | ||
| Tumor size | 1.58 (0.73-3.24) | 0.059 | ||
| Tumor dedifferentiation grade | 2.37 (0.92-5.74) | 0.022 | 1.85 (0.77-6.39) | 0.036 |
| Lymph node status | 3.28 (1.26-5.41) | 0.000 | 2.73 (1.18-5.76) | 0.001 |
| HER2 status | 1.62 (0.46-3.52) | 0.043 | 1.42 (0.65-4.23) | 0.071 |
| p21 | 2.64 (1.26-5.87) | 0.008 | 2.52 (1.14-5.34) | 0.015 |
| TGFBR2 | 1.89 (0.75-3.17) | 0.025 | 1.54 (0.71-3.66) | 0.048 |
DFS, disease-free survival; HER-2, human epidermal receptor.
Discussion
Although the significance of CDKN1A/p21 has been investigated in several different cancers, including lung [14], gastric [5], oesophageal [15], gallbladder [16], pancreatic [17], ovarian [18], and colorectal [19] carcinomas, the association between CDKN1A/p21 and cancer clinically has not been documented. When it comes to breast cancer, multiple studies have yielded conflicting results. Our study provided the evidence that CDKN1A/p21 was highly elevated in breast cancer tissues at both the mRNA and protein levels, compared with adjacent noncancerous tissues. High expression of p21 protein is significantly correlated with larger tumor size, poorly differentiated grade and lymph node metastasis. Adverse pathological parameters are associated with poor prognosis; therefore, the value of p21 in predicting breast carcinoma prognosis was evaluated. Our data revealed that p21-positive expression was strongly associated with shorter disease-free survival, and multivariate Cox regression analysis showed that p21 was an independent prognostic factor for disease-free survival. These data suggest that p21 is likely to be of particular utility as a prognostic marker for breast cancer patients. These ex vivo data are consistent with the results of previous studies. In previous reports, investigators showed that p21 overexpression was associated with positive nodal status, larger tumor size and a worse prognosis in breast cancer patients [20,21]. In addition, CDKN1A/p21 has been identified involvement in drug resistance, including chemotherapy drugs, tamoxifen and trastuzumab resistance [3], thus affects the prognosis of patients. On the contrary, others have shown that patients with p21-/p53+ IHC-labeled tumors had a worse 5-year survival rate than p21+/p53+-tumor-bearing patients [22], suggesting that loss of p21 correlates with poor patient prognosis, likely due to the loss of its growth-inhibitory functions and ability to suppress oncogenes. There is growing evidence that the function of p21 is related to its localization in cells. When localized to the cytoplasm, p21 functions as an oncogene, therefore promoting cell proliferation and progression through the cell cycle, whereas nuclear p21 was reported be involved in the pro-differentiating and senescence-promoting effects [4,23]. The molecular regulation and cellular function of p21 are being researched at present. Reports have shown that in breast cancer cells, HER2 may contribute to relocalization of p21 from the nucleus to the cytoplasm, resulting in a loss of p21 tumor suppressor functions [23,24]. Others found that HER2 overexpression was positively correlated with p21 in breast tumors and there was significant correlation of p21 positivity with worse disease free survival [25]. However, in our study, p21 expression was not correlated with HER2 significantly. This result could be interpreted that HER2 only affect the localization of p21, without affecting its expression.
The exact mechanisms of p21’s effect on oncogenesis and development were still unclear. Studies have reported that p21 is required for TGF-β-mediated cell migration and invasion, and TGF-β’s biological responses is dependent on the expression of TGFBR2 directly [26]. However, no studies have analyzed the correlation between CDKN1A/p21 and TGFBR2 expression in breast cancer. Therefore, in the current study, we sought to investigate expression of the TGFBR2 in breast cancer specimens and define the prognostic significance, and analyze the correlation between TGFBR2 and CDKN1A/p21 in breast cancer.
Our result showed that TGFBR2 expression was decreased in breast cancer. In addition, the low expression of TGFBR2 protein was closely associated with larger tumor size and lymph node metastasis, but no association with other clinicopathological features. It is noteworthy that survival analysis (Kaplan-Meier) showed TGFBR2-negative expression had a shorter disease-free survival, and Cox regression analysis indicated that TGFBR2 was an independent prognostic factor. These data suggested that TGFBR2 plays as a tumor suppressor in our single center study group, which may offer a potential tumor biomarker. Our results were consistent with previously published studies [27,28]. However, it is normally accepted that TGF-β signaling plays a different role in the development of tumor depend on the different stages of tumors, and rely on the cell type and context of the cells [29]. As the key element in the TGF-β signaling pathways, TGFBR2 may also play different biological effects according to the different status of the tumor. Gobbi et al. reported that in human breast neoplasms, down-regulation of TGFBR2 is correlated with progression, invasion and metastasis of both in situ and invasive breast carcinomas [30]. Paiva et al. reported that absence of TGFBR2 expression predicts bone and lung metastasis in breast carcinomas, and is associated with poor prognosis [31]. These data demonstrate the suppressive role of TGFBR2 in breast tumorigenesis and tumor progression, and may serve as a prognostic marker. In contrast, there are several reports showed that TGFBR2 act as a tumor promoter in late stages of breast cancer progression. Figueroa et al. reported that TGFBR2 expression is related with earlier age at onset and adverse tumor characteristics in invasive breast cancers [32]. It is speculated that the decreased expression of TGFBR2 in breast cancer in our study may be due to the included patients were most in relatively early stage, therefore show a tumor-suppressor trend. What is particularly intriguing is studies have reported that TGFBR2 expression in breast cancer was inversely correlated with overall survival, but only in patients with ER-negative tumors [33]. Loss of TGFBR2 expression among ER-negative tumors was associated with better overall survival. However, in our study, TGFBR2 expression was not correlated with ER status. Besides, TGFBR2 was not correlated with HER2 expression in our study, while in vitro studies have shown that high expression of HER2 could convert TGF-β from a neutral or even anti-migratory factor to a strongly pro-migratory and -invasive factor in untransformed MCF-10A human mammary epithelial cells [34,35], and others demonstrated that human HER2+ breast cancer associated with decreased TGF-β signaling [36]. The differences could be accounted for heterogeneity of tumor, and more studies are needed to confirm the correlation between TGFBR2 expression and other pathological features.
The action of TGF-β as a tumor suppressor is shown by functional inactivation of its receptors and Smads, and elevated expression of TGF-β signaling in human carcinoma [13]. TGF-β controls cell proliferation mainly by inducing or activating cdk inhibitors such as, p16, p15, p21 and/or p27, and inhibiting cell cycle progression through G1-arrest [10,37,38]. Andres Rojas et al. reported that the regulation of TGFBR2 expression levels affects the TGF-β mediated expression of CDKN1A/p21 [13]. In the present study, we have analyzed the correlation between TGFBR2 and p21 expression in breast cancer. However, the result showed that the expression of TGFBR2 protein was not significantly associated with p21 protein expression, suggesting that p21 was not activated by TGF-β signaling constantly. It was possible that the difference could be attributed to the different biological behavior of different individuals, at the same time suggest that the network of protein activation were broad and complex in tumor cells.
In summary, the expression of CDKN1A/p21 was significantly upregulated, TGFBR2 were significantly decreased in breast cancer tissues, compared with adjacent nontumorous breast tissues. The expression of both CDKN1A/p21 and TGFBR2 were found to be prognostic factors for patients with breast cancer. Therefore, they may serve as novel prognostic markers in addition to conventional factors. However, due to the limited quantity of patients in our study, a larger case population is needed to further verify our results. In addition, further investigation of the cell biology of CDKN1A/p21 and TGFBR2, and their relationship is necessary.
Acknowledgements
This project is supported by National Natural Science Foundation of China (No. 81360396) and the Key Projects of Health Department in Guangxi Zhuang Autonomous Region (No. 2010079).
Disclosure of conflict of interest
None.
References
- 1.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 2.Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol. 2001;21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawthorne VS, Huang WC, Neal CL, Tseng LM, Hung MC, Yu D. ErbB2-mediated Src and signal transducer and activator of transcription 3 activation leads to transcriptional up-regulation of p21Cip1 and chemoresistance in breast cancer cells. Mol Cancer Res. 2009;7:592–600. doi: 10.1158/1541-7786.MCR-08-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng X, Xia W, Yang JY, Hsu JL, Chou CK, Sun HL, Wyszomierski SL, Mills GB, Muller WJ, Yu D, Hung MC. Activation of p21(CIP1/WAF1) in mammary epithelium accelerates mammary tumorigenesis and promotes lung metastasis. Biochem Biophys Res Commun. 2010;403:103–107. doi: 10.1016/j.bbrc.2010.10.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Yu H, Cai H, Wang Y. Expression of CD24, p21, p53, and c-myc in alpha-fetoprotein-producing gastric cancer: Correlation with clinicopathologic characteristics and survival. J Surg Oncol. 2014;109:859–864. doi: 10.1002/jso.23599. [DOI] [PubMed] [Google Scholar]
- 6.Taghavi N, Biramijamal F, Sotoudeh M, Moaven O, Khademi H, Abbaszadegan MR, Malekzadeh R. Association of p53/p21 expression with cigarette smoking and prognosis in esophageal squamous cell carcinoma patients. World J Gastroenterol. 2010;16:4958–4967. doi: 10.3748/wjg.v16.i39.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Hu Y, Hu W, Xie X, Ela Bella A, Fu J, Rao D. Expression and prognostic relevance of p21WAF1 in stage III esophageal squamous cell carcinoma. Dis Esophagus. 2012;25:67–71. doi: 10.1111/j.1442-2050.2011.01217.x. [DOI] [PubMed] [Google Scholar]
- 9.Cmielova J, Rezacova M. p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected] . J Cell Biochem. 2011;112:3502–3506. doi: 10.1002/jcb.23296. [DOI] [PubMed] [Google Scholar]
- 10.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 12.Joshi A, Cao D. TGF-beta signaling, tumor microenvironment and tumor progression: the butterfly effect. Front Biosci (Landmark Ed) 2010;15:180–194. doi: 10.2741/3614. [DOI] [PubMed] [Google Scholar]
- 13.Rojas A, Padidam M, Cress D, Grady WM. TGF-beta receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-beta. Biochim Biophys Acta. 2009;1793:1165–1173. doi: 10.1016/j.bbamcr.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie D, Lan L, Huang K, Chen L, Xu C, Wang R, Shi Y, Wu X, Wang L, Liu Y, Lu B. Association of p53/p21 expression and cigarette smoking with tumor progression and poor prognosis in non-small cell lung cancer patients. Oncol Rep. 2014;32:2517–2526. doi: 10.3892/or.2014.3538. [DOI] [PubMed] [Google Scholar]
- 15.Shiozaki A, Nakashima S, Ichikawa D, Fujiwara H, Konishi H, Komatsu S, Kubota T, Okamoto K, Iitaka D, Shimizu H, Nako Y, Takemoto K, Kishimoto M, Otsuji E. Prognostic significance of p21 expression in patients with esophageal squamous cell carcinoma. Anticancer Res. 2013;33:4329–4335. [PubMed] [Google Scholar]
- 16.Puhalla H, Wrba F, Kandioler D, Lehnert M, Huynh A, Gruenberger T, Tamandl D, Filipits M. Expression of p21(Wafl/Cip1), p57(Kip2) and HER2/neu in patients with gallbladder cancer. Anticancer Res. 2007;27:1679–1684. [PubMed] [Google Scholar]
- 17.Han J, Wang F, Yuan SQ, Guo Y, Zeng ZL, Li LR, Yang J, Wang DS, Liu MY, Zhao H, Liu KY, Liao JW, Zou QF, Xu RH. Reduced expression of p21-activated protein kinase 1 correlates with poor histological differentiation in pancreatic cancer. BMC cancer. 2014;14:650. doi: 10.1186/1471-2407-14-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skirnisdottir I, Seidal T. Association of p21, p21 p27 and p21 p53 status to histological subtypes and prognosis in low-stage epithelial ovarian cancer. Cancer Genomics Proteomics. 2013;10:27–34. [PubMed] [Google Scholar]
- 19.Liu KH, Huynh N, Patel O, Shulkes A, Baldwin G, He H. P21-activated kinase 1 promotes colorectal cancer survival by up-regulation of hypoxia-inducible factor-1alpha. Cancer Lett. 2013;340:22–29. doi: 10.1016/j.canlet.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 20.Caffo O, Doglioni C, Veronese S, Bonzanini M, Marchetti A, Buttitta F, Fina P, Leek R, Morelli L, Palma PD, Harris AL, Barbareschi M. Prognostic value of p21(WAF1) and p53 expression in breast carcinoma: an immunohistochemical study in 261 patients with long-term follow-up. Clin Cancer Res. 1996;2:1591–1599. [PubMed] [Google Scholar]
- 21.Hinnis AR, Luckett JC, Walker RA. Survivin is an independent predictor of short-term survival in poor prognostic breast cancer patients. Br J Cancer. 2007;96:639–645. doi: 10.1038/sj.bjc.6603616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domagala W, Welcker M, Chosia M, Karbowniczek M, Harezga B, Bartkova J, Bartek J, Osborn M. p21/WAF1/Cip1 expression in invasive ductal breast carcinoma: relationship to p53, proliferation rate, and survival at 5 years. Virchows Arch. 2001;439:132–140. doi: 10.1007/s004280100410. [DOI] [PubMed] [Google Scholar]
- 23.Diaz Flaque MC, Vicario R, Proietti CJ, Izzo F, Schillaci R, Elizalde PV. Progestin drives breast cancer growth by inducing p21(CIP1) expression through the assembly of a transcriptional complex among Stat3, progesterone receptor and ErbB-2. Steroids. 2013;78:559–567. doi: 10.1016/j.steroids.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Winters ZE, Leek RD, Bradburn MJ, Norbury CJ, Harris AL. Cytoplasmic p21WAF1/CIP1 expression is correlated with HER-2/ neu in breast cancer and is an independent predictor of prognosis. Breast Cancer Res. 2003;5:R242–249. doi: 10.1186/bcr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y, Zhou BP, Hung MC. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res. 2004;10:3815–3824. doi: 10.1158/1078-0432.CCR-03-0527. [DOI] [PubMed] [Google Scholar]
- 26.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 27.Busch S, Acar A, Magnusson Y, Gregersson P, Ryden L, Landberg G. TGF-beta receptor type-2 expression in cancer-associated fibroblasts regulates breast cancer cell growth and survival and is a prognostic marker in pre-menopausal breast cancer. Oncogene. 2015;34:27–38. doi: 10.1038/onc.2013.527. [DOI] [PubMed] [Google Scholar]
- 28.Paiva CE, Drigo SA, Rosa FE, Moraes Neto FA, Caldeira JR, Soares FA, Domingues MA, Rogatto SR. Absence of transforming growth factor-beta type II receptor is associated with poorer prognosis in HER2-negative breast tumours. Ann Oncol. 2010;21:734–740. doi: 10.1093/annonc/mdp518. [DOI] [PubMed] [Google Scholar]
- 29.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 30.Gobbi H, Arteaga CL, Jensen RA, Simpson JF, Dupont WD, Olson SJ, Schuyler PA, Plummer WD Jr, Page DL. Loss of expression of transforming growth factor beta type II receptor correlates with high tumour grade in human breast in-situ and invasive carcinomas. Histopathology. 2000;36:168–177. doi: 10.1046/j.1365-2559.2000.00841.x. [DOI] [PubMed] [Google Scholar]
- 31.Paiva CE, Serrano SV, Paiva BS, Scapulatempo-Neto C, Soares FA, Rogatto SR, Marques ME. Absence of TGF-betaRII predicts bone and lung metastasis and is associated with poor prognosis in stage III breast tumors. Cancer Biomark. 2012;11:209–217. doi: 10.3233/CBM-2012-00281. [DOI] [PubMed] [Google Scholar]
- 32.Figueroa JD, Flanders KC, Garcia-Closas M, Anderson WF, Yang XR, Matsuno RK, Duggan MA, Pfeiffer RM, Ooshima A, Cornelison R, Gierach GL, Brinton LA, Lissowska J, Peplonska B, Wakefield LM, Sherman ME. Expression of TGF-beta signaling factors in invasive breast cancers: relationships with age at diagnosis and tumor characteristics. Breast Cancer Res Treat. 2010;121:727–735. doi: 10.1007/s10549-009-0590-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buck MB, Fritz P, Dippon J, Zugmaier G, Knabbe C. Prognostic significance of transforming growth factor beta receptor II in estrogen receptor-negative breast cancer patients. Clin Cancer Res. 2004;10:491–498. doi: 10.1158/1078-0432.ccr-0320-03. [DOI] [PubMed] [Google Scholar]
- 34.Seton-Rogers SE, Brugge JS. ErbB2 and TGF-beta: a cooperative role in mammary tumor progression? Cell Cycle. 2004;3:597–600. [PubMed] [Google Scholar]
- 35.Ueda Y, Wang S, Dumont N, Yi JY, Koh Y, Arteaga CL. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J Biol Chem. 2004;279:24505–24513. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
- 36.Novitskiy SV, Forrester E, Pickup MW, Gorska AE, Chytil A, Aakre M, Polosukhina D, Owens P, Yusupova DR, Zhao Z, Ye F, Shyr Y, Moses HL. Attenuated transforming growth factor beta signaling promotes metastasis in a model of HER2 mammary carcinogenesis. Breast Cancer Res. 2014;16:425. doi: 10.1186/s13058-014-0425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai M, Al-Odaini AA, Arakelian A, Rabbani SA, Ali S, Lebrun JJ. A novel function for p21Cip1 and acetyltransferase p/CAF as critical transcriptional regulators of TGFbeta-mediated breast cancer cell migration and invasion. Breast Cancer Res. 2012;14:R127. doi: 10.1186/bcr3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HP, Kim TY, Lee MS, Jong HS, Lee JW, Bang YJ. TGF-beta1-mediated activations of c-Src and Rac1 modulate levels of cyclins and p27(Kip1) CDK inhibitor in hepatoma cells replated on fibronectin. Biochim Biophys Acta. 2005;1743:151–161. doi: 10.1016/j.bbamcr.2004.09.014. [DOI] [PubMed] [Google Scholar]


