Abstract
Hepatitis B virus (HBV) infection increases the risk of liver decompensation, cirrhosis and hepatocellular carcinoma (HCC). Apolipoprotein E (ApoE), one of the major cholesterol carriers, plays important role in the metabolism of lipoprotein and the regulation of immune response. The present study was aimed to explore whether the genetic variation in ApoE gene affected disease progression in HBV infected individuals. We collected sera samples from healthy volunteers (n=40), inactive HBV carriers (n=30), and patients with acute hepatitis (n=60), severe hepatitis (n=12), HBV-related liver cirrhosis (n=58) or primary HCC (n=39). We found that ApoE and interlukin-6 (IL-6) was progressively increased, while IL-2 was gradually decreased with the increasing grade of disease severity. Furthermore, high ApoE levels in HBV infected individuals were correlated with increased IL-6 and decreased IL-2 levels, indicating immune abnormalities in these patients. The frequency of E3/3 genotype was progressively increased from carriers group, hepatitis group to progressive group (cirrhosis and HCC). The serum levels of low-density lipoprotein cholesterol (LDL-C) differed among ApoE phenotypes, with E3/4, E4/4> E3/3>E2/3. Our study suggested that ApoE may have a role in the pathogenesis and progression of HBV-related liver disease and indicated the possible underlying mechanisms.
Keywords: Hepatitis B virus, apolipoprotein E, polymorphisms
Introduction
Hepatitis B, an infectious disease caused by hepatitis B virus (HBV), is widely prevalent in the world especially in developing country. HBV infection increases the risk of liver decompensation, cirrhosis and hepatocellular carcinoma (HCC). According to World Health Organization (WHO) statistics, more than 2 billion people have been infected with HBV and approximately 378 million are chronic carriers and at risk for HBV-related liver disease worldwide. HBV results in 500,000 to 700,000 deaths each year [1]. HBV infection has a wide clinical spectrum ranging from inactive HBV carrier state to active hepatitis, severe hepatitis, HBV-related liver cirrhosis and primary HCC associated with HBV infection [2,3]. A variety of risk factors, including older age, male gender, longer duration of HBV infection, high HBV-DNA levels and alcohol consumption, have been identified associated with the progressive course of this disease [4]. Furthermore, previous investigations suggest that host genetic polymorphism, which is responsible for differential immune response during HBV infection, may affect the individual susceptibility to infectious pathogens [5-7].
Apolipoprotein E (ApoE) is one of the major cholesterol carriers and synthesized principally in the liver [8]. ApoE plays important role in the metabolism of lipoprotein and the regulation of immune response [9]. The ApoE gene is mapped to chromosome 19 in band 19q13.2. The ApoE gene is polymorphic with three common codominant alleles (E2, E3 and E4), resulting in six different genotypes (E2/2, E2/3, E2/4, E3/3, E3/4 and E4/4) [10]. The three major isoforms, ApoE2 (cys112, cys158), ApoE3 (cys112, arg158) and ApoE4 (arg112, arg158), are encoded by E2, E3 and E4, respectively. Although these three major isoforms differ from one another by only one or two amino acids substitutions, these differences alter the structure and function of ApoE [11-13]. ApoE polymorphisms have been linked to the progression of cardiovascular disease [8] and Alzheimer’s disease [11,14]. Several studies demonstrated the association of ApoE genotypes with the outcomes of viral diseases, including herpex simplex virus (HSV) [15], hepatits C virus (HCV) [16-20] and HBV [7,21-23]. However, few studies have been performed on the correlation of ApoE genotypes with disease progression in people with HBV infection.
In the present study, we hypothesized that genetic variation in ApoE gene could affect disease progression in HBV infected individuals. We determined the ApoE genotype distribution in healthy volunteers, inactive HBV carriers, and patients with acute hepatitis, chronic hepatitis, HBV-related liver cirrhosis or primary HCC, and analyzed the association of serum levels of interleukin-2 (IL-2), IL-6, high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) with ApoE concentration or polymorphisms. Our study revealed the possible mechanisms of the involvement of ApoE in the pathogenesis and progression of HBV infected individuals.
Materials and methods
Study patients
A total of 199 subjects, which included 30 HBV carriers, 60 patients with active hepatitis, 12 patients with severe hepatitis, 58 patients with HBV-related liver cirrhosis and 39 patients with HCC, admitted to Zhejiang Hospital (Hangzhou, China) from June 2012 to December 2013 were enrolled in this study. There were 107 male (32.14 ± 8.63 years, range 17-79 years) and 92 female (31.47 ± 7.16 years, range 20-73 years). Diagnosis was based on the criteria explained in viral hepatitis prevention and control program revised on the national infectious and parasitic Conference (Xi’an) in 2000. The studies excluded patients with other hepatitis virus infection, other malignant cancer, autoimmune liver disease, alcoholic liver disease, nonalcoholic fatty liver disease, hyperlipidemia, coronary heart disease or atherosclerosis. Sera samples were obtained from these patients before treatment. Samples from 40 healthy volunteer, without history of liver disease, were collected as a control. No significant differences were found in gender and age between the patients and normal control group. Ethical approval for the study was provided by the independent ethics committee, XX. Informed and written consent was obtained from each individual according to the ethics committee guidelines.
ApoE genotyping
To determine the ApoE genotype, we genotyped two single-nucleotide polymorphisms (SNPs; NCBI SNPs rs429358 and rs7412). Genomic DNA was extracted from whole blood samples using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s guidelines. ApoE genotyping was performed using the multiplexed ligation detection reaction technique (LDR) as described previously [24]. Firstly, PCR products containing the SNPs were generated using the primers as follows: rs429358, forward: 5’-GCCTACAAATCGGAACTGGA-3’ and reverse: 5’-CAGCTCCTCGGTGCTCTG-3’; rs7412, forward: 5’-TAAGCGGCTCCTCCGCGAT-3’ and reverse: 5’-GCCCCGGCCTGGTACACTG-3’. Secondly, PCR products were treated with proteinase K at 37°C for 15 min, 55°C for 10 min, and 90°C for 10 min, to destroy any remaining DNA polymerase activity before the LDR. LDRs were performed by using Taq DNA ligase (New England Biolabs, Beverly, MA, USA) with the following reaction conditions: 95°C for 2 minutes, followed by 40 cycles of ligation (94°C for 15s followed 50°C for 25S). The Probe sequences for LDR genotyping were listed in Table 1. Finally, LDR amplification product was run on an ABI prism 3730 sequencer (Applied Biosystems, Foster City, CA, USA) and genotypes analyzed using Genemapper software (Applied Biosystems).
Table 1.
The probes of ligase detection reactions
| Probe | Sequence | Size (bp) |
|---|---|---|
| rs429358_ modify | P-CACGTCCTCCATGTCCGCGCTTTTTTTTTTTTTTTTTTTTTTTTTTT-FAM | |
| rs429358_C | TTTTTTTTTTTTTTTTTTTTTTCGGTACTGCACCAGGCGGCCGCG | 92 |
| rs429358_T | TTTTTTTTTTTTTTTTTTTTTTTTCGGTACTGCACCAGGCGGCCGCA | 94 |
| rs7412_ modify | P-CTTCTGCAGGTCATCGGCATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-FAM | |
| rs7412_C | TTTTTTTTTTTTTTTTTTTTTTTTCCGGCCTGGTACACTGCCAGGCG | 97 |
| rs7412_T | TTTTTTTTTTTTTTTTTTTTTTTTTTCCGGCCTGGTACACTGCCAGGCA | 99 |
Measurement of serum indices
Serum ApoE levels were measured by a turbidimetric immunoassay with a commercial kit (Wako Pure Chemical Industries, Tokyo, Japan). Serum concentrations of HDL-C and LDL-C were determined by an enzymatic method according to the manufacturer’s instructions (Sekisui Medical, Tokyo, Japan). All were performed on an automatic analyzer (AU5400, Olympus, Japan).
Enzyme-linked immunosorbent assay (ELISA) analysis
Serum concentrations of IL-2 and IL-6 were assessed by using ELISA assay (eBioscience, San Diego, CA, USA) following the instructions of the manufacturer. Plates were read at 450 nm using a microplate reader (Multiscan MS™, Labsystems, Helsinki, Finland).
Statistical analysis
All statistical analyses were performed with SPSS software version 16.0 (Chicago, IL, USA). Statistical comparisons between conditions were conducted using a chi-square or Fisher’s exact test, as appropriate. Data were reported as the means ± SD, and comparisons were conducted using ANOVA. The relationships between two factors were assessed by Pearson correlation analysis. Differences were considered statistically significant when P<0.05.
Results
Associations between ApoE polymorphisms and HBV infection or HBV disease progression
The frequency of ApoE allele and genotype in healthy volunteers, HBV carriers, and patients with active hepatitis, severe hepatitis, cirrhosis and HCC was shown in Tables 2 and 3. The ApoE E3 allele and E3/3 genotype were the most frequent in all groups.
Table 2.
Distributions of AopE alleles
| Group | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Normal | Carrier | Active hepatitis | Severe hepatitis | Cirrhosis | HCC | All | |
| E2 | 3 (3.8%) | 4 (5.7%) | 6 (5.0%) | 0 (0) | 1 (0.9%) | 3 (3.9%) | 17 |
| E3 | 70 (87.5%) | 56 (80.0%) | 105 (87.5%) | 20 (83.3%) | 111 (97.4%) | 67 (88.2%) | 429 |
| E4 | 7 (8.8%) | 10 (14.3%) | 9 (7.5%) | 4 (16.7%) | 2 (1.8%) | 6 (7.9%) | 38 |
| All | 80 | 70 | 120 | 24 | 114 | 76 | 484 |
Table 3.
Distributions of ApoE genotypes
| Group | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Normal (n=40) | Carrier (n=30) | Active hepatitis (n=60) | Severe hepatitis (n=12) | Cirrhosis (n=58) | HCC (n=39) | All (n=239) | |
| E2/3 | 3 (7.5%) | 4 (13.3%) | 6 (10.0%) | 0 (0) | 1 (1.8%) | 2 (5.3%) | 16 |
| E2/4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.6%) | 1 |
| E3/3 | 30 (75.0%) | 19 (63.3%) | 45 (75.0%) | 8 (66.7%) | 54 (94.7%) | 31 (81.6%) | 187 |
| E3/4 | 7 (17.5%) | 4 (13.3%) | 9 (17.5%) | 4 (33.3%) | 2 (3.5%) | 3 (7.9%) | 29 |
| E4/4 | 0 (0) | 3 (10.0%) | 0 (0) | 0 (0) | 0 (0) | 1 (2.6%) | 4 |
| NA | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.7%) | 1 (2.6%) | 2 |
We then evaluated whether ApoE polymorphism was related to HBV infection or disease progression in HBV infection. We could not find any association between normal control and HBV-infected group (Table 4), indicating that ApoE genotypes did not affect the susceptibility of human hepatitis B virus. Hepatitis group and progressive group (cirrhosis and HCC) were found significantly different from carriers group in the distribution of ApoE polymorphism. A significant difference was also found between hepatitis group and progressive group (Table 5).
Table 4.
Association between ApoE polymorphism and HBV infection
| Normal (n=30) | HBV persistence (n=199) | P | |
|---|---|---|---|
| Genotype | |||
| E2/3 | 3 (7.5%) | 13 (6.5%) | 0.7525 |
| E2/4 | 0 (0) | 1 (0.5%) | |
| E3/3 | 30 (75.0%) | 157 (78.9%) | |
| E3/4 | 7 (17.5%) | 22 (11.1%) | |
| E4/4 | 0 (0) | 4 (2.0%) | |
| NA | 0 (0) | 2 (1.0%) |
Note: Statistical analysis refers to a chi-square or Fisher’s exact test, as appropriate.
Table 5.
Association between ApoE polymorphism and disease progression in the HBV infection group
| Carrier (n=30) | Hepatitis (n=72) | P | Progressive (CH+HCC) (n=97) | P | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| VS carrier | VS carrier | VS hepatitis | ||||
| Genotype | ||||||
| E2/3 | 4 (13.3%) | 6 (8.3%) | 0.0395* | 3 (3.2%) | 0.0106* | 0.028* |
| E2/4 | 0 (0) | 0 (0) | 1 (1.1%) | |||
| E3/3 | 19 (63.3%) | 53 (73.6%) | 85 (89.5%) | |||
| E3/4 | 4 (13.3%) | 13 (18.1%) | 5 (5.3%) | |||
| E4/4 | 3 (10.0%) | 0 (0) | 1 (1.1%) | |||
| NA | 0 (0) | 0 (0) | 2 (2.1%) | |||
Note: Statistical analysis refers to a chi-square or Fisher’s exact test, as appropriate.
P<0.05.
ApoE serum concentration in study subjects
In order to investigate the functional significance of ApoE at protein level, we measured ApoE serum concentrations of the 239 subjects by a turbidimetric immunoassay. There was no significant difference in ApoE serum levels between healthy volunteers and HBV carriers. Compared with healthy volunteers, the ApoE serum levels of patients with active hepatitis, severe hepatitis, cirrhosis and HCC increased to 2.72, 3.82, 4.21 and 4.58 fold, respectively (Figure 1A). We then analyzed whether ApoE serum levels correlated with ApoE genotypes (Figure 1B). No significant differences were detected in ApoE serum levels among the four ApoE genotypes, E2/E3, E3/E3, E3/E4 and E4/E4.
Figure 1.
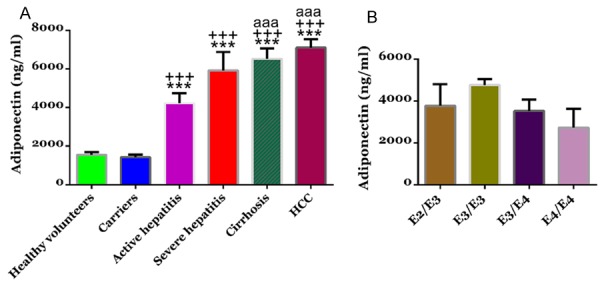
Apolipoprotein E (APOE) serum levels in study subjects. A. Serum was collected from healthy volunteers (n=40), HBV carriers (n=30), patients with active hepatitis (n=60), patients with severe hepatitis (n=12), patients with HBV-related liver cirrhosis (n=58), and patients with HCC (n=39) and APOE concentrations were determined by using a turbidimetric immunoassay. *P<0.05, ***P<0.001 VS healthy volunteers; ++P<0.01, +++P<0.001 VS HBV carriers; aaaP<0.001 VS patients with active hepatitis; sssP<0.001 VS patients with severe hepatitis; cccP<0.001 VS patients with HBV-related liver cirrhosis. B. Serum APOE levels in subjects with different APOE genotypes.
Serum IL-6 and IL-2 levels
To explore the changes of IL-6 and IL-2 with the progression of HBV-related liver disease, we detected their levels in healthy volunteers and patients with HBV-related liver disease (Figure 2). There were no significant difference in serum levels of IL-6 and IL-2 between healthy volunteers and HBV carriers. Comparing with healthy volunteers, the IL-6 serum levels of patients with active hepatitis, severe hepatitis, cirrhosis and HCC increased to 1.54, 2.56, 3.36 and 4.72 fold, respectively (Figure 2A), while the IL-2 serum levels decreased to 76%, 80%, 64% and 56%, respectively (Figure 2D).
Figure 2.
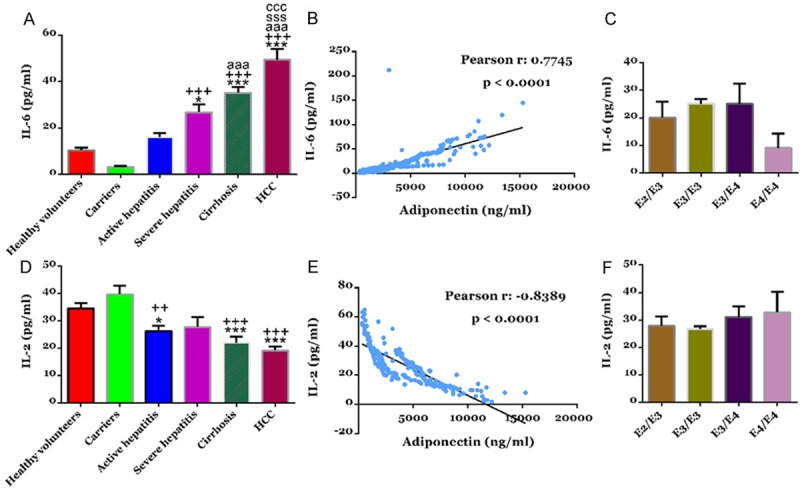
Serum levels of cytokines, IL-6 and IL-2 in study subjects. Serum IL-6 (A) and IL-2 (D) concentration were determined by using ELISA. ***P<0.001 VS healthy volunteers; +++P<0.001 VS HBV carriers; aaaP<0.001 VS patients with active hepatitis. The serum levels of APOE showed a significant positive correlation with IL-6 (B) and a significant negative correlation with IL-2 (E). Serum IL-6 (C) or IL-2 (F) levels in subjects with different APOE genotypes.
ApoE is known to be involved in the regulation of immune response. We then analysis the correlation of ApoE with both cytokines by Pearson’s analysis. ApoE serum levels were positively correlated with serum levels of IL-6 (r=0.7745, P<0.0001, Figure 2B), while negatively correlated with serum levels of IL-2 (r=-0.8389, P<0.0001, Figure 2E).
Further, we analyzed whether ApoE genotypes correlated with serum levels of IL-6 (Figure 2C) and IL-2 (Figure 2F). No significant differences were detected in the serum levels of both cytokines among the four ApoE genotypes, E2/E3, E3/E3, E3/E4 and E4/E4.
Serum HDL-C and LDL-C concentrations
ApoE is well known cholesterol transporter and plays an important role in lipid metabolism. We then detected serum concentrations of HDL-C and LDL-C in study subjects. As shown in Figure 3A, there were no significant difference in serum concentrations of HDL-C between healthy volunteers and HBV carriers. The HDL-C concentrations of patients with active hepatitis, severe hepatitis, cirrhosis and HCC were significantly lower than that of healthy volunteers. LDL-C concentrations showed no difference among healthy volunteers, HBV carriers, patients with active hepatitis and patients with severe hepatitis, while notable decrease of LDL-C concentrations was observed in the serum of patients with cirrhosis and HCC (Figure 3D). Pearson analysis revealed no correlation between the serum levels of ApoE and HDL-C (Figure 3B) or LDL-C (Figure 3E).
Figure 3.
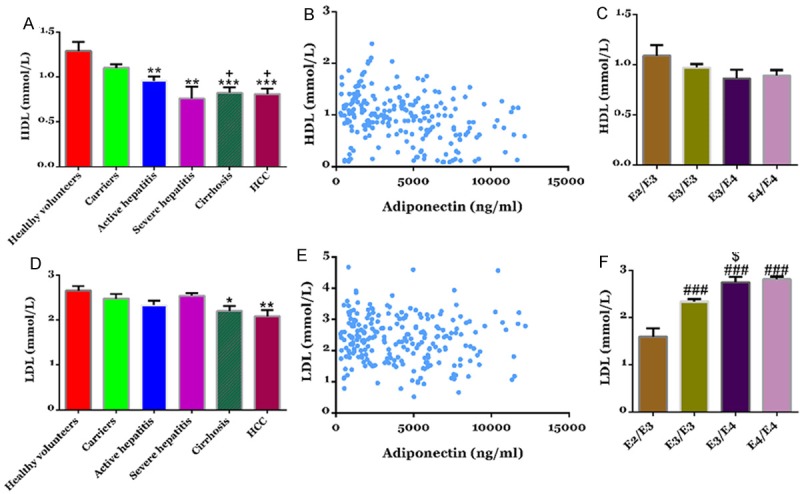
Serum levels of HDL-C and LDL-C in study subjects. Serum IL-6 (A) and IL-2 (D) concentration were determined by using ELISA. **P<0.01, ***P<0.001 VS healthy volunteers; +P<0.05 VS HBV carriers. No correlation was observed between the serum levels of APOE and HDL-C (B) or LDL-C (E). Effects of APOE Phenotype on HDL-C (C) or LDL-C (F).
Further, there was no significant difference in the serum levels of HDL-C among the four ApoE genotypes, E2/E3, E3/E3, E3/E4 and E4/E4 (Figure 3C). LDL-C was progressively increased from E2/3 to E3/3 to E3/4 (Figure 3F).
Discussion
In a previous study, serum levels of ApoE were found significantly higher in patients with HBV-related liver cirrhosis and HCC compared with that of the healthy controls [22]. Here, we collected serum samples from healthy volunteers, HBV carriers, and patients with active hepatitis, severe hepatitis, cirrhosis and HCC to further explored the changes of ApoE serum concentration with the progression of HBV-relative liver diseases. Our data demonstrated that ApoE was gradually increased with the increasing grade of disease severity (Figure 1A). E3 and E3/3 are the most common allele and genetic phenotype of APOE, respectively [22,23]. Ahn et al. revealed that significantly more patients carrying the E3/3 genotype had developed liver cirrhosis [22]. Here, the frequency of E3/3 genotype was progressively increased from carriers group, hepatitis group to progressive group (cirrhosis and HCC) (Table 5). Our data further suggested that the serum levels and genotypes of ApoE were related with the progression of HBV-related liver diseases.
Cytokines are low-molecular-weight proteins regulate various inflammatory responses. It has been demonstrated that cytokine networks perform multiple functions in physiology and pathology [25]. Increased IL-6 [26] and decreased IL-2 [27,28] were observed in chronic liver disease. In the present study, a progressive increase of IL-6 and a gradual down-regulation of IL-2 was observed with the increasing grade of disease severity (Figure 2). Furthermore, ApoE levels were positively correlated with IL-6 levels, but negatively related with IL-2 levels. IL-6 and IL-2 levels were not significantly different among ApoE phenotypes. These data suggested that high serum levels of ApoE, correlating with high IL-6 and low IL-2 could represent one of the factors leading to immune abnormalities in HBV-related diseases.
The liver plays a critical role in the synthesis and degradation of lipid and lipoprotein [29]. In various liver diseases, liver functions are impaired, leading to the broken of lipid and lipoprotein homeostasis. Here, we find a notable reduction of HDL-C in patients with hepatitis, cirrhosis and HCC (Figure 3A), and a decrease of LDL-C in patients with cirrhosis and HCC (Figure 3D). These results may be ascribed to hepatocellular dysfunction and has been confirmed earlier in other studies [30,31]. Some reports have indicated that the E4 allele is associated with higher LDL-C and the E2 with lower LDL-C [32,33]. In the present study, although the serum levels of ApoE was not correlated with the levels of HDL-C or LDL-C (P<0.05), LDL-C differed among ApoE phenotypes, with E3/4, E4/4> E3/3>E2/3 (Figure 3F), while no difference was observed in HDL-C among ApoE phenotypes (Figure 3C). Our data demonstrated the close relation of LDL-C levels with ApoE phenotypes.
Taken together, we demonstrated the association of serum concentration and polymorphisms of ApoE with the progression of HBV-related liver disease. High serum levels of ApoE in patients with HBV-related liver disease may be related with immune abnormalities manifested by increased IL-6 and decreased IL-2. Moreover, ApoE genotypes influenced LDL-C concentration, which indicated the association of ApoE genotypes and lipid metabolism in these patients.
Acknowledgements
This study was supported by Zhejiang medical and health science and technology program (Grant No. 2013KYA005).
Disclosure of conflict of interest
None.
References
- 1.Franco E, Bagnato B, Marino MG, Meleleo C, Serino L, Zaratti L. Hepatitis B. Epidemiology and prevention in developing countries. World J Hepatol. 2012;4:74–80. doi: 10.4254/wjh.v4.i3.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iino S. Natural history of hepatitis B and C virus infections. Oncology. 2002;62(Suppl 1):18–23. doi: 10.1159/000048271. [DOI] [PubMed] [Google Scholar]
- 3.European Association For The Study of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–85. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507–39. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 5.Kwiatkowski D. Genetic dissection of the molecular pathogenesis of severe infection. Intensive Care Med. 2000;26:S089–S97. doi: 10.1007/s001340051124. [DOI] [PubMed] [Google Scholar]
- 6.Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, Kaslow RA. HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology. 2004;39:978–88. doi: 10.1002/hep.20142. [DOI] [PubMed] [Google Scholar]
- 7.Wang FS. Current status and prospects of studies on human genetic alleles associated with hepatitis B virus infection. World J Gastroenterol. 2003;9:641–4. doi: 10.3748/wjg.v9.i4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minihane AM, Jofre-Monseny L, Olano-Martin E, Rimbach G. ApoE genotype, cardiovascular risk and responsiveness to dietary fat manipulation. Proc Nutr Soc. 2007;66:183–97. doi: 10.1017/S0029665107005435. [DOI] [PubMed] [Google Scholar]
- 9.Mahley RW, Rall SC Jr. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–37. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg DT, Kuzawa CW, Hayes MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. 2010;143:100–11. doi: 10.1002/ajpa.21298. [DOI] [PubMed] [Google Scholar]
- 11.Zuo L, Van Dyck CH, Luo X, Kranzler HR, Yang BZ, Gelernter J. Variation at APOE and STH loci and Alzheimer’s disease. Behav Brain Funct. 2006;2:13. doi: 10.1186/1744-9081-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghebranious N, Ivacic L, Mallum J, Dokken C. Detection of ApoE E2, E3 and E4 alleles using MALDI-TOF mass spectrometry and the homogeneous mass-extend technology. Nucleic Acids Res. 2005;33:e149. doi: 10.1093/nar/gni155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenberg DT, Kuzawa CW, Hayes MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. 2010;143:100–11. doi: 10.1002/ajpa.21298. [DOI] [PubMed] [Google Scholar]
- 14.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 15.Lin WR, Wozniak MA, Esiri MM, Klenerman P, Itzhaki RF. Herpes simplex encephalitis: involvement of apolipoprotein E genotype. J Neurol Neurosurg Psychiatry. 2001;70:117–9. doi: 10.1136/jnnp.70.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wozniak MA, Itzhaki RF, Faragher EB, James MW, Ryder SD, Irving WL. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology. 2002;36:456–63. doi: 10.1053/jhep.2002.34745. [DOI] [PubMed] [Google Scholar]
- 17.Price DA, Bassendine MF, Norris S, Golding C, Toms GL, Schmid ML, Morris CM, Burt AD, Donaldson PT. Apolipoprotein E3 allele is associated with persistent hepatitis C virus infection. Gut. 2006;55:715–8. doi: 10.1136/gut.2005.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller T, Gessner R, Sarrazin C, Graf C, Halangk J, Witt H, Köttgen E, Wiedenmann B, Berg T. Apolipoprotein E4 allele is associated with poor treatment response in hepatitis C virus (HCV) genotype 1. Hepatology. 2003;38:1592. doi: 10.1016/j.hep.2003.09.042. author reply 1592-3. [DOI] [PubMed] [Google Scholar]
- 19.Owen DM, Huang H, Ye J, Gale M. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheridan DA, Bridge SH, Felmlee DJ, Crossey MM, Thomas HC, Taylor-Robinson SD, Toms GL, Neely RD, Bassendine MF. Apolipoprotein-E and hepatitis C lipoviral particles in genotype 1 infection: evidence for an association with interferon sensitivity. J Hepatol. 2012;57:32–8. doi: 10.1016/j.jhep.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z, Xiong C, Wang Y, Zhou X, Yan SK. Investigation of the relationship between apolipoprotein E gene polymorphisms and hepatitis B virus infection in northern China. Clin Chem Lab Med. 2010;48:1803–7. doi: 10.1515/CCLM.2010.354. [DOI] [PubMed] [Google Scholar]
- 22.Ahn SJ, Kim DK, Kim SS, Bae CB, Cho HJ, Kim HG, Kim YJ, Lee JH, Lee HJ, Lee MY, Kim KB, Cho JH, Cho SW, Cheong JY. Association between apolipoprotein E genotype, chronic liver disease, and hepatitis B virus. Clin Mol Hepatol. 2012;18:295–301. doi: 10.3350/cmh.2012.18.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toniutto P, Fattovich G, Fabris C, Minisini R, Burlone M, Pravadelli C, Peraro L, Falleti E, Caldera F, Bitetto D, Pirisi M. Genetic polymorphism at the apolipoprotein E locus affects the outcome of chronic hepatitis B. J Med Virol. 2010;82:224–331. doi: 10.1002/jmv.21642. [DOI] [PubMed] [Google Scholar]
- 24.Hennig BJ, Hellier S, Frodsham AJ, Zhang L, Klenerman P, Knapp S, Wright M, Thomas HC, Thursz M, Hill AV. Association of low-density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun. 2002;3:359–67. doi: 10.1038/sj.gene.6363883. [DOI] [PubMed] [Google Scholar]
- 25.Ramadori G, Armbrust T. Cytokines in the liver. Eur J Gastroenterol Hepatol. 2001;13:777–84. doi: 10.1097/00042737-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Tangkijvanich P, Vimolket T, Theamboonlers A, Kullavanijaya P, Suwangool P, Poovorawan Y. Serum interleukin-6 and interferon-gamma levels in patients with hepatitis B-associated chronic liver disease. Asian Pac J Allergy Immunol. 2000;18:109–14. [PubMed] [Google Scholar]
- 27.Williams R. Interleukin-1 and interleukin-2 activity in chronic hepatitis B virus infection. Gastroenterology. 1988;94:999–1005. doi: 10.1016/0016-5085(88)90559-8. [DOI] [PubMed] [Google Scholar]
- 28.Saxena S, Nouri-Aria KT, Anderson M, Eddleston A, Williams R. Interleukin 2 activity in chronic liver disease and the effect of in vitro alpha-interferon. Clin Exp Immunol. 1986;63:541. [PMC free article] [PubMed] [Google Scholar]
- 29.Tietge UJ, Boker KH, Bahr MJ, Weinberg S, Pichlmayr R, Schmidt HH, Manns MP. Lipid parameters predicting liver function in patients with cirrhosis and after liver transplantation. Hepatogastroenterology. 1998;45:2255–60. [PubMed] [Google Scholar]
- 30.Booth S, Clifton PM, Nestel PJ. Lack of effect of acute alcohol ingestion on plasma lipids. Clin Chem. 1991;37:1649. [PubMed] [Google Scholar]
- 31.Ghadir MR, Riahin AA, Havaspour A, Nooranipour M, Habibinejad AA. The relationship between lipid profile and severity of liver damage in cirrhotic patients. Hepat Mon. 2010;10:285–8. [PMC free article] [PubMed] [Google Scholar]
- 32.Eto M, Watanabe K, Ishii K. Reciprocal effects of apolipoprotein E alleles (epsilon 2 and epsilon 4) on plasma lipid levels in normolipidemic subjects. Clin Genet. 1986;29:477–84. doi: 10.1111/j.1399-0004.1986.tb00547.x. [DOI] [PubMed] [Google Scholar]
- 33.Sing CF, Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet. 1985;37:268–85. [PMC free article] [PubMed] [Google Scholar]


