Abstract
We conducted a study to investigate the association between the clinical outcome and GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms in advanced NSCLC patients with cisplatin-based chemotherapy. Between January 2010 and December 2012, a total of 206 patients with advanced NSCLC were histopathologically confirmed were included into analysis. By logistic regression analysis, individuals carrying the AG and GG genotypes of GSTP1 Ile105Val were associated with better response to chemotherapy when compared with the AA genotype, and the adjusted Ors (95% CI) were 2.06 (1.10-3.86) and 4.89 (1.52-18.33), respectively. The TT genotype of XRCC1 Arg194Trp was correlated with better response to chemotherapy compared to the CC genotype, and the adjusted OR (95% CI) was 3.23 (1.20-9.30). By Cox Hazard Proportional Model, the GG genotype of GSTP1 Ile105Val and the TT genotype of XRCC1 Arg194Trp were found to be associated with lower risk of death from all causes when compared with the wide-type genotype, and the adjusted HRs (95% CI) were 0.05 (0.01-0.18) and 0.20 (0.07-0.62), respectively. Moreover, individuals carrying both the G/A+G/G genotype of GSTP1 Ile105Val and the G/A+A/A of XRCC1 Arg194Trp were associated with heavy greater CR+PR response to chemotherapy (OR=2.98, 95% CI=1.39-6.42), and also correlated with longer overall survival of advanced NSCLC (HR=0.19, 95% CI=0.05-0.61). In conclusion, we found that the GSTP1 Ile105Val and XRCC1 Arg194Trp were associated with better response to chemotherapy and longer survival of advanced NSCLC, compared to the wide-type genotype.
Keywords: GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His, Arg399Gln, NSCLC, clinical outcome
Introduction
Lung cancer is one of the most common malignant tumors worldwide, and lung cancer is the leading cause of cancer-related deaths [1]. There are two types of lung cancer, including small cell lung cancer and non-small cell lung cancer (NSCLC). It is estimated that NSCLC accounts for nearly 80% of all lung cancers, and NSCLC mainly includes squamous cell carcinoma and adenocarcinoma lung cancer [2]. NSCLC is both frequently diagnosed in men and women, and it is reported that about 1.8 million people are diagnosed with NSCLC annually worldwide [1]. The NSCLC are always diagnosis at their advanced stage [3]. Although combination chemotherapy, chemoradiation and radiation therapy are common used for advanced NSCLC, the five-year survival of NSCLC is very low with a 5-year overall survival only around 15% for all stages [3]. TNM stage is a crucial factor for the prognosis of NSCLC, but not all patients with the same TNM stage present similar treatment outcome of NSCLC. Therefore, genetic factors may play an important role in the development of NSCLC.
Glutathione S-transferase P1 (GSTP1) is a glutathione S-transferase class member, and it is widely expressed in different human tissues. GSTP1 plays a role in the phase II metabolism of xenobiotics, and contributes to the process of cisplatin biothransformation [4]. X-ray repair cross-complementing group 1 (XRCC1) is an important member of base excision repair pathway, and it is responsible for the single-strand break repair and base excision repair [5]. It is reported that XRCC1 cooperates ligase and poly (ADP-ribose) polymerase, and thus efficiently repair DNA damage including cisplatin-induced damage [6]. In our study, we conducted a study to investigate the association between the clinical outcome and GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms in advanced NSCLC patients with cisplatin-based chemotherapy.
Patients and methods
Patients
A total of 206 patients with advanced NSCLC were histopathologically confirmed and recruited between March 2010 and March 2013. All patients with advanced NSCLC did not receive any anticancer therapies before surgery and had adequate hematology, renal and liver function with the Eastern Cooperative Oncology Group (ECOG) performance status 0 or 1. During and after adjuvant chemotherapy.
The patients underwent a physical examination, computed tomography, and magnetic resonance imaging before adjuvant chemotherapy, and the patients were treated for one adjuvant chemotherapy regimen for at least four cycles or until having a progressive disease. Chemotherapy response was evaluated based on RECIST criteria [7]. Patients who showed complete response (CR) and partial response (PR) to chemotherapy were considered to be good response to chemotherapy, and patients who showed stable disease (SD) and progressive disease (PD) were considered poor response to chemotherapy. The survival time of NSCLC was evaluated by overall survival, and the overall survival time was calculated by the time from the date of chemotherapy and the data of death from any cause or the end of follow-up. Routine follow-ups of all patients were ended up to March 2015, and patients were followed up every four weeks through telephone or attending the clinics.
DNA extraction and genotyping
2 ml peripheral blood was collected from each patient with NSCLC and control subject, and the collected blood was stored in refrigerator at -70°C until use. Genomic DNA was isolated from peripheral blood lymphocytes using Qiagen blood mini kit (Qiagen, Germany) according to the manufacturer’s instruction. The GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln genotypes were analyzed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) based on the manufacturer’s instruction. The primers for GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms were designed using the Sequenom Assay Design 3.1 software. The PCR was set as follows: started at 95°C for 5 min for the initial denaturation, following 30 cycles of denaturation at 95°C for 30 s, annealing at 62°C for 45 s, extension at 72°C for 30 s and final extension at 72°C for 5 mins. The PCR products were digested by the BsmA1, PvuII, RsaI and NciI for GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln, respectively. These digested products were stained with ethidium bromide and separated on 3.5% agarose gel, and visualized under UA light after ethidium staining.
Statistical analysis
The demographic and clinical characteristics of patients with advanced NSCLC were expressed by frequencies and percentage. The genotype distributions of GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His and Arg399Gln were tested for deviation from Hardy-Weinberg equilibrium. The association between response to chemotherapy and polymorphisms in GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His and Arg399Gln was estimated odds ratio (OR) and 95% confidence interval (95% CI) in logistic regression analysis with adjustments of potential confounding factors. Kaplan-Meier method was used to calculate overall survival of advanced NSCLC. Log-rank test was used to compare the differences of overall survival time among different genotypes. Hazard ratio (HR) and 95% CI of significant genotypes were estimated Cox Hazard Proportional Model with adjustments of potential confounding factors. All P values were based on two-tails test and P<0.05 was set as significant level. All analyses were performed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
The demographical and clinical characteristics of selected patients with advanced NSCLC were summarized in Table 1. The mean age of patients with advanced NSCLC was 56.07±8.85 years. There were 82 (39.81%) females and 124 (60.19%) males in the present study. Of 206 patients with advanced NSCLC, 135 (65.53%) were smokers, 77 (37.38%) were drinkers, 74 (35.92%) were at III TNM stage, 132 (64.08%) were at IV TNM stage, 94 (45.63%) were adenocarcinoma type, 112 (54.37%) were squamous carcinoma type, 106 (51.46%) showed CR+PR to chemotherapy and 100 (48.54%) showed SD+PD to chemotherapy. By Log-rank test, we found that the overall survival time showed significantly difference between the response to chemotherapy (P<0.001).
Table 1.
Demographical and clinical characteristics of included patients and control subjects
| Variables | Patients N=206 | % | Survival time | P for Log-rank test |
|---|---|---|---|---|
| Age, years | 56.07±8.85 | |||
| Gender | ||||
| Female | 82 | 39.81 | 21.65±1.40 | |
| Male | 124 | 60.19 | 22.15±1.32 | 0.06 |
| Smoking status | ||||
| Never | 71 | 34.47 | 21.84±1.41 | |
| Ever | 135 | 65.53 | 22.32±1.29 | 0.42 |
| Drinking status | ||||
| Never | 129 | 62.62 | 21.16±1.20 | |
| Ever | 77 | 37.38 | 22.48±1.63 | 0.45 |
| TNM stage | ||||
| III | 74 | 35.92 | 21.52±1.53 | |
| IV | 132 | 64.08 | 22.51±1.25 | 0.50 |
| Histology | ||||
| Adenocarcinoma | 94 | 45.63 | 20.60±1.57 | |
| Squamous carcinoma | 112 | 54.37 | 22.27±2.64 | 0.59 |
| Response to chemotherapy | ||||
| CR+PR | 106 | 51.46 | 26.75±1.36 | |
| SD+PD | 100 | 48.54 | 17.29±1.21 | <0.001 |
We found that the genotype distributions of GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His and Arg399Gln were in line with from Hardy-Weinberg equilibrium, and the P values were 0.80, 0.19, 0.18 and 0.08, respectively. By logistic regression analysis, we found that individuals carrying the AG and GG genotypes of GSTP1 Ile105Val were associated with better response to chemotherapy when compared with the AA genotype, and the adjusted Ors (95% CI) were 2.06 (1.10-3.86) and 4.89 (1.52-18.33), respectively (Table 2). Moreover, we observed that the TT genotype of XRCC1 Arg194Trp was correlated with better response to chemotherapy compared to the CC genotype, and the adjusted OR (95% CI) was 3.23 (1.20-9.30). However, no significant correlations between XRCC1 Arg280His and XRCC1 Arg399Gln polymorphisms and response to chemotherapy were found in the present study.
Table 2.
Association between GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His and Arg399Gln and response to chemotherapy in advanced NSCLC
| Genes | N | % | HWE | Good response | % | Poor response | % | OR (95% CI)1 | P value |
|---|---|---|---|---|---|---|---|---|---|
| GSTP1 Ile105Val | |||||||||
| AA | 91 | 44.17 | 36 | 33.96 | 55 | 55.00 | 1.0 (Ref.) | - | |
| AG | 94 | 45.63 | 54 | 50.94 | 40 | 40.00 | 2.06 (1.10-3.86) | 0.02 | |
| GG | 21 | 10.19 | 0.80 | 16 | 15.09 | 5 | 5.00 | 4.89 (1.52-18.33) | 0.002 |
| XRCC1 Arg194Trp | |||||||||
| CC | 94 | 45.63 | 41 | 38.68 | 53 | 53.00 | 1.0 (Ref.) | - | |
| CT | 84 | 40.78 | 45 | 42.45 | 39 | 39.00 | 1.49 (0.79-2.81) | 0.18 | |
| TT | 28 | 13.59 | 0.19 | 20 | 18.87 | 8 | 8.00 | 3.23 (1.20-9.30) | 0.01 |
| XRCC1 Arg280His | |||||||||
| AA | 104 | 50.49 | 51 | 48.11 | 53 | 53.00 | 1.0 (Ref.) | - | |
| AG | 79 | 38.35 | 40 | 37.74 | 39 | 39.00 | 1.07 (0.57-2.00) | 0.83 | |
| GG | 23 | 11.17 | 0.18 | 15 | 14.15 | 8 | 8.00 | 1.95 (0.70-5.76) | 0.16 |
| XRCC1 Arg399Gln | |||||||||
| GG | 91 | 44.17 | 44 | 41.51 | 47 | 47.00 | 1.0 (Ref.) | - | |
| GA | 83 | 40.29 | 41 | 38.68 | 42 | 42.00 | 1.04 (0.55-1.97) | 0.89 | |
| AA | 32 | 15.53 | 0.08 | 21 | 19.81 | 11 | 11.00 | 2.04 (0.82-5.23) | 0.09 |
Ajusted for age, gender, smoking status, drinking status, TNM stage and histology.
The mean overall survival time was 22.16±0.97 months, and a total of 166 patients were died from all causes during the follow-up period. The GG genotypes of GSTP1 Ile105Val was correlated with a longer overall survival time when compared with the AA genotype (GG vs. AA: 42.76±4.28 months vs. 19.43±1.62 months, P value for Log-rank test <0.001; Figure 1). The TT genotype of XRCC1 Arg194Trp was associated with a longer overall survival time compared to the CC genotype (TT vs. CC: 30.73±3.02 months vs. 22.39±1.63 months, P value for Log-rank test =0.01; Figure 2). After adjusting for potential confounding factors, the GG genotype of GSTP1 Ile105Val and the TT genotype of XRCC1 Arg194Trp were found to be associated with lower risk of death from all causes when compared with the wide-type genotype, and the adjusted HRs (95% CI) were 0.05 (0.01-0.18) and 0.20 (0.07-0.62), respectively (Table 3). We did not observe a significant association between XRCC1 Arg280His and Arg399Gln and overall survival of patients with advanced NSCLC.
Figure 1.
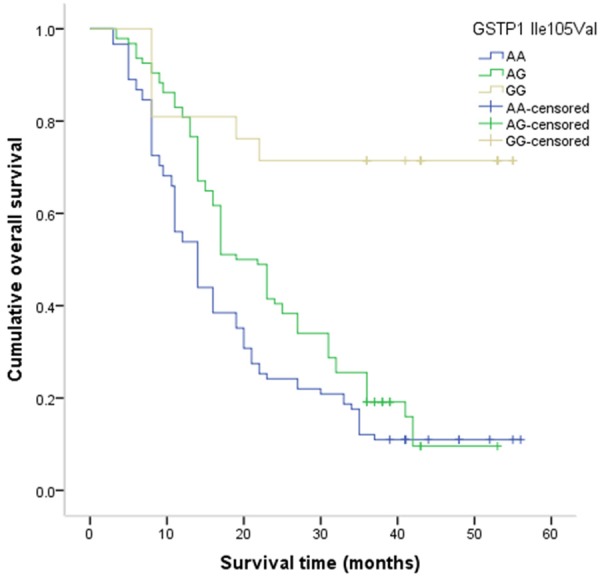
Kaplan-Meier method for the overall survival of advanced NSCLC by GSTP1 Ile105Val.
Figure 2.
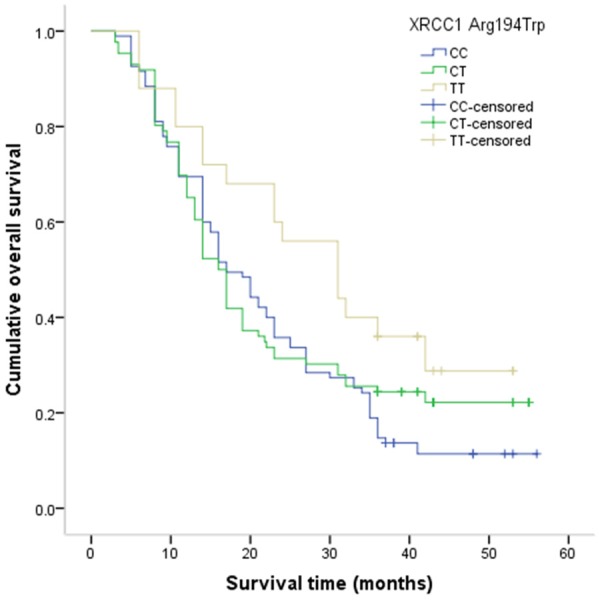
Kaplan-Meier method for the overall survival of advanced NSCLC by XRCC1 Arg194Trp.
Table 3.
Association between GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His and Arg399Gln and overall survival of patients with advanced NSCLC
| Genes | N | % | Death N=166 | % | Alive | % | Mean survival time | P value for Log-rank test | Adjusted HR (95% CI)1 | P value |
|---|---|---|---|---|---|---|---|---|---|---|
| GSTP1 Ile105Val | ||||||||||
| AA | 91 | 44.17 | 81 | 48.80 | 10 | 25.00 | 19.43±1.62 | 1.0 (Ref.) | - | |
| AG | 94 | 45.63 | 79 | 47.59 | 15 | 37.50 | 21.27±1.49 | 0.65 (0.25-1.66) | 0.32 | |
| GG | 21 | 10.19 | 6 | 3.61 | 15 | 37.50 | 42.76±4.28 | <0.001 | 0.05 (0.01-0.18) | <0.001 |
| XRCC1 Arg194Trp | ||||||||||
| CC | 94 | 45.63 | 83 | 50.00 | 11 | 27.50 | 22.39±1.63 | 1.0 (Ref.) | - | |
| CT | 84 | 40.78 | 66 | 39.76 | 18 | 45.00 | 24.07±2.02 | 0.49 (0.19-1.18) | 0.08 | |
| TT | 28 | 13.59 | 17 | 10.24 | 11 | 27.50 | 30.73±3.02 | 0.01 | 0.20 (0.07-0.62) | <0.001 |
| XRCC1 Arg280His | ||||||||||
| AA | 104 | 50.49 | 84 | 50.60 | 20 | 50.00 | 23.63±1.65 | 1.0 (Ref.) | - | |
| AG | 79 | 38.35 | 66 | 39.76 | 13 | 32.50 | 21.20±1.77 | 1.21 (0.53-2.85) | 0.63 | |
| GG | 23 | 11.17 | 16 | 9.64 | 7 | 17.50 | 25.12±3.17 | 0.25 | 0.54 (0.18-1.79) | 0.23 |
| XRCC1 Arg399Gln | ||||||||||
| GG | 91 | 44.17 | 73 | 43.98 | 18 | 45.00 | 22.48±1.87 | 1.0 (Ref.) | - | |
| GA | 83 | 40.29 | 71 | 42.77 | 12 | 30.00 | 24.55±1.74 | 1.45 (0.61-3.57) | 0.35 | |
| AA | 32 | 15.53 | 22 | 13.25 | 10 | 25.00 | 26.24±1.95 | 0.18 | 0.54 (0.20-1.52) | 0.18 |
Ajusted for age, gender, smoking status, drinking status, TNM stage and histology.
We performed gene-gene interaction between GSTP1 Ile105Val and XRCC1 Arg194Trp in the response to chemotherapy of advanced NSCLC (Table 4), and we found that individuals carrying both the GA+GG genotype of GSTP1 Ile105Val and the CT+TT of XRCC1 Arg194Trp were associated with better response to chemotherapy in advanced NSCLC when compared to those carrying with wide-type genotypes (OR=2.98, 95% CI=1.39-6.42). Moreover, individuals carrying both the GA+GG genotype of GSTP1 Ile105Val and the CT+TT of XRCC1 Arg194Trp were correlated with long overall survival of advanced NSCLC, and the adjusted OR (95% CI) was 0.19 (0.05-0.61) (Table 5).
Table 4.
Interaction between GSTP1 Ile105Val and XRCC1 Arg194Trp in the response to chemotherapy
| Gene | N | % | Good response | % | Poor response | % | OR (95% CI)1 | P value | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| GSTP1 Ile105Val | XRCC1 Arg194Trp | ||||||||
| AA | CC | 58 | 28.16 | 21 | 19.81 | 37 | 37.00 | 1.0 (Reference) | - |
| GA+GG | CC | 37 | 17.96 | 21 | 19.81 | 16 | 16.00 | 2.31 (0.92-5.86) | 0.05 |
| GA+GG | CT+TT | 78 | 37.86 | 49 | 46.23 | 29 | 29.00 | 2.98 (1.39-6.42) | 0.002 |
| AA | CT+TT | 33 | 16.02 | 15 | 14.15 | 18 | 18.00 | 1.47 (0.56-3.82) | 0.39 |
Ajusted for age, gender, smoking status, drinking status, TNM stage and histology.
Table 5.
Interaction between GSTP1 Ile105Val and XRCC1 Arg194Trp in the overall survival of advanced NSCLC
| Genes | N | % | Death | % | Alive | % | Mean survival time | Adjusted HR (95% CI)1 | P value | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| GSTP1 Ile105Val | XRCC1 Arg194Trp | |||||||||
| AA | CC | 58 | 28.16 | 54 | 32.53 | 4 | 10.00 | 20.27±1.16 | 1.0 (Reference) | - |
| GA+GG | CC | 37 | 17.96 | 29 | 17.47 | 8 | 20.00 | 22.92±1.07 | 0.27 (0.06-1.12) | 0.06 |
| GA+GG | CT+TT | 78 | 37.86 | 56 | 33.73 | 22 | 55.00 | 23.57±1.14 | 0.19 (0.05-0.61) | 0.002 |
| AA | CT+TT | 33 | 16.02 | 27 | 16.27 | 6 | 15.00 | 21.56±1.11 | 0.33 (0.06-1.56) | 0.10 |
Ajusted for age, gender, smoking status, drinking status, TNM stage and histology.
Discussion
Increasing evidences have shown that the genetic polymorphisms of drug metabolizing enzymes, drug transporters and drug targets play an important role in the inter-individual differences in the efficacy of chemotherapy treatment. In this study, we evaluated the association between the GSTP1 Ile105Val, XRCC1 Arg194Trp, Arg280His and Arg399Gln polymorphisms and the response to chemotherapy and overall survival of patients with NSCLC in a Chinese population. We observed that the GG genotype of GSTP1 IIe105Val and the TT genotype of XRCC1 Arg194Trp were associated with better response to chemotherapy and more overall survival time of patients with advanced NSCLC, and the two genes have synergistic effect in the response to chemotherapy.
It is reported that the GST enzymes has a crucial role in metabolizing many xenobiotics and cytotoxic cancer chemotherapeutic agents [8]. GSTP1 is the most isoenzyme of GSTs and overexpression in cancer or precancerous tissues [9]. Polymorphisms in the gene GSTP1 Ile105Val cause the amino acid substitution changes and hydroliphobicity of amino acid, and consequently influence the enzymatic stability and catalytic capability. The wild-type homogeneous AA genotype of GSTP1 Ile105Val shows the highest enzymatic activities [10]. Previous studies have reported the association between GSTP1 Ile105Val polymorphism and efficacy of chemotherapy on various cancers, but these studies have shown inconsistent results [11-16].
Recent studies have investigated the association of genetic polymorphism in the GSTP1 IIe105Val region with prognosis of advanced NSCLC. Individuals with GSTP1 gene polymorphism display a reduced ability to detoxify drug metabolites, and thus promote the overall survival of NSCLC [14,15,17-19]. Deng et al. also conducted a study in a Chinese population and found that the AA genotype of GSTP1 IIe105Val had better overall survival of advanced NSCLC [14]. Han et al., conducted a study in a Chinese population, and they found that the AA and GA genotypes were associated with the response to cisplatin-based chemotherapy and the treatment outcome of advanced NSCLC [15]. Another three studies have reported that the GSTP1 IIe105Val polymorphism could modify the treatment efficacy of platinum-based chemotherapy in advanced NSCLC [17-19]. However, some studies reported inconsistent results [20-21]. Kalikaki et al. conducted a study with 119 NSCLC patients, and they reported that GSTP1 IIe105Val polymorphism was not associated with the response to treatment and clinical outcome of advanced NSCLC [20]. Joerger et al. performed a study with 37 patients with advanced NSCLC, and they reported that GSTP1 IIe105Val polymorphism did not contribute to the clinical outcome of advanced NSCLC [21]. The discrepancies of the above results may be attributed to the differences in ethnicities and source of patients, as well as the disease stages and sample size.
XRCC1 is considered as a scaffold factor in base excision repair, and the XRCC1 Arg194Trp, Arg280His and Arg399Gln polymorphisms are associated with the change of XRCC1 protein activity to impact DNA repair capacity [22]. The severity of DNA damage could be influenced by cisplatin, which are associated with efficiency and toxicity of cisplatin-based chemotherapy. Many previous studies have reported that XRCC1 gene polymorphisms were associated with the prognosis of cancers treated with cisplatin-based chemotherapy, such as hepatocellular carcinoma, gastric cancer, colorectal cancer and non-small cell lung cancer [23-25]. Our study also found that XRCC1 Arg194Trp polymorphism contributed to the response to chemotherapy. However, previous studies reported inconsistent results regarding the association between XRCC1 Arg194Trp polymorphism and response to chemotherapy in advanced NSCLC [14,15,23,24,26,27]. Therefore, further studies with large sample size study are greatly needed to confirm our findings.
There were two limitations to our study. First, patients with advanced NSCLC were selected from one hospital only, which may induce selection bias. Second, the sample size for analyzing the association between gene polymorphism and treatment outcome of advanced NSCLC in the present study was relatively small; therefore, some of the findings may be undervalued due to the same sample size. Therefore, studies with larger sample sizes must be performed to confirm our findings.
In conclusion, we found that the GSTP1 Ile105Val and XRCC1 Arg194Trp could influence the response to chemotherapy and overall survival of patients with advanced NSCLC. Further prospective studies with larger sample sizes are required to validate this association.
Disclosure of conflict of interest
None.
References
- 1.International Agency for Research on Cancer. Lung Cancer. Estimated Incidence, Mortality and Prevalence Worldwide in 2012. 2012. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. Access in 2015-08-01.
- 2.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henley SJ, Richards TB, Underwood JM, Eheman CR, Plescia M, McAfee TA Centers for Disease Control and Prevention (CDC) Lung cancer incidence trends among men and women--united states, 2005-2009. MMWR Morb Mortal Wkly Rep. 2014;63:1–5. [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer TD. The glutathione S-transferases: an update. Hepatology. 1989;9:486–96. doi: 10.1002/hep.1840090324. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 6.Zhu G, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry. 2009;48:4916–25. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffaud F, Therasse P. New guidelines to evaluate the response to treatment in solid tumors. Bull Cancer. 2000;87:881–886. [PubMed] [Google Scholar]
- 8.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–20. [PubMed] [Google Scholar]
- 9.Tiseo M, Bordi P, Bortesi B, Boni L, Boni C, Baldini E, Grossi F, Recchia F, Zanelli F, Fontanini G, Naldi N, Campanini N, Azzoni C, Bordi C, Ardizzoni A Bio-FAST trial group. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108:1695–703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booton R, Ward T, Heighway J, Ashcroft L, Morris J, Thatcher N. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. 2006;1:679–83. [PubMed] [Google Scholar]
- 11.Djukic TI, Savic-Radojevic AR, Pekmezovic TD, Matic MG, Pljesa-Ercegovac MS, Coric VM, Radic TM, Suvakov SR, Krivic BN, Dragicevic DP, Simic TP. Glutathione S-transferase T1, O1 and O2 polymorphisms are associated with survival in muscle invasive bladder cancer patients. PLoS One. 2013;8:e74724. doi: 10.1371/journal.pone.0074724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kap EJ, Richter S, Rudolph A, Jansen L, Ulrich A, Hoffmeister M, Ulrich CM, Brenner H, Chang-Claude J. Genetic variants in the glutathione S-transferase genes and survival in colorectal cancer patients after chemotherapy and differences according to treatment with oxaliplatin. Pharmacogenet Genomics. 2014;24:340–7. doi: 10.1097/FPC.0000000000000059. [DOI] [PubMed] [Google Scholar]
- 13.Goričar K, Kovač V, Jazbec J, Zakotnik B, Lamovec J, Dolžan V. Genetic variability of DNA repair mechanisms and glutathione-S-transferase genes influences treatment outcome in osteosarcoma. Cancer Epidemiol. 2015;39:182–8. doi: 10.1016/j.canep.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Deng JH, Deng J, Shi DH, Ouyang XN, Niu PG. Clinical outcome of cisplatin-based chemotherapy is associated with the polymorphisms of GSTP1 and XRCC1 in advanced non-small cell lung cancer patients. Clin Transl Oncol. 2015;17:720–6. doi: 10.1007/s12094-015-1299-6. [DOI] [PubMed] [Google Scholar]
- 15.Han B, Guo Z, Ma Y, Kang S, Wang Y, Wei Q, Wu X. Association of GSTP1 and XRCC1 gene polymorphisms with clinical outcome of advanced non-small cell lung cancer patients with cisplatin-based chemotherapy. Int J Clin Exp Pathol. 2015;8:4113–9. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Huang A, Zhang D, Yao J, Zhang Y, Li X. Genetic variability of glutathione S-transferases influences treatment outcome of breast cancer. Tumour Biol. 2015;36:5925–9. doi: 10.1007/s13277-015-3266-9. [DOI] [PubMed] [Google Scholar]
- 17.Lv H, Han T, Shi X, Yao Y, Yao Y, Qiu W, Yue L, Liang J. Genetic polymorphism of GSTP1 and ERCC1 correlated with response to platinumbased chemotherapy in non-small cell lung cancer. Med Oncol. 2014;31:86. doi: 10.1007/s12032-014-0086-5. [DOI] [PubMed] [Google Scholar]
- 18.Ke HG, Li J, Shen Y, You QS, Yan Y, Dong HX, Liu JH, Shen ZY. Prognostic significance of GSTP1, XRCC1 and XRCC3 polymorphisms in nonsmall cell lung cancer patients. Asian Pac J Cancer Prev. 2012;13:4413–6. doi: 10.7314/apjcp.2012.13.9.4413. [DOI] [PubMed] [Google Scholar]
- 19.Lu C, Spitz MR, Zhao H, Dong Q, Truong M, Chang JY, Blumenschein GR Jr, Hong WK, Wu X. Association between glutathione Stransferase pi polymorphisms and survival in patients with advanced nonsmall cell lung carcinoma. Cancer. 2006;106:441–7. doi: 10.1002/cncr.21619. [DOI] [PubMed] [Google Scholar]
- 20.Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, Mavroudis D. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:118–23. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 21.Joerger M, Burgers JA, Baas P, Doodeman VD, Smits PH, Jansen RS, Vainchtein LD, Rosing H, Huitema AD, Beijnen JH, Schellens JH. Gene polymorphisms, pharmacokinetics, and hematological toxicity in advanced non-small-cell lung cancer patients receiving cisplatin/gemcitabine. Cancer Chemother Pharmacol. 2012;69:25–33. doi: 10.1007/s00280-011-1670-4. [DOI] [PubMed] [Google Scholar]
- 22.Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ Health Perspect. 2003;111:1843–50. doi: 10.1289/ehp.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Zhao J. Effect of APE1 and XRCC1 gene polymorphism on susceptibility to hepatocellular carcinoma and sensitivity to cisplatin. Int J Clin Exp Med. 2015;8:9931–6. [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan I, Salazar J, Majem M, Pallarés C, Del Río E, Páez D, Baiget M, Barnadas A. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Lett. 2014;353:160–6. doi: 10.1016/j.canlet.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Wu H, Xu C, Chen G, Wang J. X-ray repair crosscomplementing 1 polymorphism and prognosis of platinum-based chemotherapy in gastric and colorectal cancer: a meta-analysis. J Gastroenterol Hepatol. 2014;29:926–33. doi: 10.1111/jgh.12444. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Huang XE, Jin GF, Shen HB, Xu L. Lack of any relationship between chemotherapy toxicity in non-small cell lung cancer cases and polymorphisms in XRCC1 codon 399 or XPD codon 751. Asian Pac J Cancer Prev. 2011;12:739–42. [PubMed] [Google Scholar]
- 27.Lee SY, Kang HG, Yoo SS, Kang YR, Choi YY, Lee WK, Choi JE, Jeon HS, Shin KM, Oh IJ, Kim KS, Lee J, Cha SI, Kim CH, Kim YC, Park JY. Polymorphisms in DNA repair and apoptosisrelated genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer. 2013;82:330–9. doi: 10.1016/j.lungcan.2013.07.024. [DOI] [PubMed] [Google Scholar]


