Abstract
Background: Congenital heart disease (CHD) is the most common birth abnormality, especially for sporadic CHD. However, the etiology of sporadic CHD is largely unknown. NKX2-5, the earliest sign of cardiac progenitor cell differentiation, plays a key role in cardiac morphogenesis, and the mutation of this gene can cause sporadic CHD. Purpose: To investigate the association of genetic variations of NKX2-5 with sporadic CHD in Chinese Bai people. Methods: The whole 2 coding exons and flanking intron sequences of NKX2-5 gene were screened using DNA sequencing in 70 Chinese Bai patients with sporadic CHD and 136 healthy controls. Results: A novel heterozygous DNA sequence variant (DSV), 1433A>G, was identified in one tetralogy of Fallot (TOF) patient and one persistent left superior vena cava (PLSVC) patient, but none in controls. The frequency of single nucleotide polymorphism (SNP) rs2277923 in CHD group was significantly higher than that in control group. The allele and genotype were associated with the occurrence of CHD. Conclusion: The novel DSV (1433A>G) may be relevant with TOF and PLSVC, and the SNP rs2277923 of NKX2-5 gene contributes to the risk of sporadic CHD in Chinese Bai people.
Keywords: Congenital heart disease, NKX2-5, single nucleotide polymorphism, Chinese Bai people
Introduction
Congenital heart disease (CHD) is the main cause of death in infants, which affects 0.4%-1% of live births worldwide [1]. CHD can be classified as syndromic CHD and non-syndromic CHD, based on whether the heart is associated with external cardiac abnormalities. In non-syndromic CHD, there is only a heart defect, without external cardiac abnormality. It can be divided into two subtypes, including familial CHD and sporadic CHD. The total number of patients with sporadic CHD approximately accounts for 80% of CHD patients [2-4].
CHD is considered to be caused by genetic and environmental factors. Recent reports [5,6] have stated that, an increasing number of genes, including cardiac transcription factor GATA4, NKX2-5 and TBX5, are associated with non-syndromic CHD. NKX2-5 is a homeobox transcription factor. It is the first known marker to be a genetic cause of CHD, and is initially validated in pedigrees with autosomal dominant inheritance of cardiac septal defects [7]. The dosage-sensitive interdependence between cardiac transcription factors and chromatin remodeling complex has been observed in the heart development [8]. NKX2-5 gene knockout at mid-embryonic stage can cause premature death and defective cardiac morphogenesis [9]. Approximately 38 missense and nonsense mutations in NKX2-5 gene have been found, and these mutations mainly exist in Caucasian population (only 4 mutations found in Chinese patients). A total of 7 mutations (rs17052019, rs2277923, rs3729938, rs3729753, rs3729754, rs703752 and rs11552707) have been highly proved to be involved in congenital heart malformation [10].
The epidemiological investigation shows that, the incidence of CHD has significant difference among different nationalities in Yunnan province of China. The incidence of CHD in Bai minority accounts for 8.12‰ of Yunnan people, with Dai of 5.39‰ and Han of 3.11‰ [11]. The Bai minority has a long history and culture in the Southwest border area of China, which mainly distributes in Dali Bai Minority Autonomous District, Yunnan, China. Historically, The Bai nationality is a mainstay group of the culture of Tibetan and Myanmar, which assimilate large numbers of people from different nationalities. Bai people descend from the ancient Baiman population within Kunming ethnic group (a branch of ancient Qiang nationality) living in Erhai lake area, and it also assimilates an array of Han people. Meanwhile, partial gene flow influence deriving from the central Asian region has existed in Bai people. This study investigated the distribution and frequency of above 7 mutations in NKX2-5 gene in Chinese Bai people with sporadic CHD. The objective is to clarify the correlation between the single nucleotide polymorphisms (SNPs) of NKX2-5 gene and sporadic CHD.
Subjects and methods
Subjects
A cohort of 70 CHD patients (29 male and 41 female; average age of 14.0±7.3 years), chosen based on ultrasound cardiogram imaging evidence of CHD, were recruited from Department of Cardiac Surgery, Yan’an Affiliated Hospital of Kunming Medical University (Kunming, China). The variety of CHD was listed in Table 1. CHD in all patients was confirmed by surgery. The 136 controls (57 male and 79 female; average age of 13.5±3.4 years) were selected from Department of Physical Examination, Yan’an Affiliated Hospital of Kunming Medical University (Kunming, China). Written informed consents were signed in all subjects. This study was approved by the Ethics Committee of Kunming Medical University. CHD patients and controls were all unrelated, and were the Chinese Bai people who settled in Dali Bai Minority Autonomous District in Yunnan, China. They had no migration history or history of marriage with other nationalities within three generations.
Table 1.
Distribution of congenital heart disease in patients
| Sort | Gender | Total (%) | |
|---|---|---|---|
|
| |||
| Male | Female | ||
| VSD | 5 | 8 | 13 (18.6) |
| ASD | 8 | 18 | 26 (37.1) |
| PFO | 3 | 3 | 6 (8.6) |
| TOF | 2 | 3 | 5 (7.1) |
| PDA | 0 | 4 | 4 (5.7) |
| VSD+ASD | 2 | 1 | 3 (4.3) |
| VSD+PFO | 1 | 1 | 2 (2.6) |
| ASD+PDA | 0 | 1 | 1 (1.4) |
| ASD+PFO | 1 | 0 | 1 (1.4) |
| RVOTS | 0 | 1 | 1 (1.4) |
| TECD | 1 | 0 | 1 (1.4) |
| TI | 1 | 0 | 1 (1.4) |
| BAV | 2 | 0 | 2 (2.6) |
| HCM | 1 | 0 | 1 (1.4) |
| PLSVC | 1 | 0 | 1 (1.4) |
| Else | 1 | 1 | 2 (2.6) |
| Total | 29 | 41 | 70 (100) |
PFO, patent foramen ovale; TOF, tetralogy of Fallot; PDA, patent ductus arteriosus; RVOTS, right ventricular outflow tract septum; TECD, complete type endocardial cushion defect; TI, tricuspid insufficiency; BAV, bicuspid aortic valve; HCM, hypertrophic cardiomyopathy; PLSVC, persistent left superior vena cava.
DNA extraction and genotyping
2mL of peripheral blood was collected from 70 patients and 136 healthy controls. The DNA was extracted using Axyprep Blood Genomic DNA Miniprep kit (Axygen Scientific Inc., CA, USA). 7 SNPs (rs17052019, rs2277923, rs3729938, rs3729753, rs3729754, rs703752 and rs11552707) were selected based on SNP database in NCBI and HapMap to examine the association between NKX2-5 gene and CHD. These SPNs were listed in Table 2. 3 primers (N1-N3) were designed by Prime 5.0 to amplify 7 SNPs in exon and 3’non-coding regions of NKX2-5 gene. N1 was used amplify rs17052019 and rs2277923. N2 was used amplify rs3729938, rs3729753 and rs3729754. N3 was used amplify rs703752 and rs115527072. Primers and their characteristics are listed in Table 3.
Table 2.
Information of 7 SNPs in NKX2-5
| Chromosome position | mRNA position | dbSNP rs# cluster ID | Function | dbSNP allele | Protein residue | Amino acid position | Region |
|---|---|---|---|---|---|---|---|
| 172662040 | 276 | rs17052019 | Missense/contig reference | C/A | Ala[A]/Asp[D] | 16 | Exon1 |
| 172662024 | 292 | rs2277923 | Synonymous/contig reference | G/A | Glu[E]/Glu[E] | 21 | Exon1 |
| 172660011 | 765 | rs3729938 | Missense/contig reference | G/C | Cys[C]/Ser[S] | 179 | Exon2 |
| 172659941 | 835 | rs3729753 | Synonymous/contig reference | C/G | Leu[L]/Leu[L] | 202 | Exon2 |
| 172659915 | 861 | rs3729754 | Missense/contig reference | T/C | Leu[L]/Pro[P] | 211 | Exon2 |
| 172659511 | 1265 | rs703752 | 3’UTR | A/G | 3’UTR | ||
| 172659237 | 1539 | rs11552707 | 3’UTR | G/T | 3’UTR |
Table 3.
Sites of NKX2-5-specific primers
| Primer | SNPs | Forward primer (5’-3’) | Reverse primer (5’-3’) | Product (bp) | Annealing temperature (°C) |
|---|---|---|---|---|---|
| N1 | rs17052019 | TCTCCTGCCCCTTGTGCTCA | CGTAGGCCTCTGGCTTGAAGG | 332 | 61 |
| rs2277923 | |||||
| N2 | rs3729938 | TGGAGAAGACAGAGGCGGACAA | CGTAGGCGTTATAACCGTAGGGAT | 396 | 63 |
| rs3729753 | |||||
| rs3729754 | |||||
| N3 | rs703752 | ACAACAACTTCGTGAACTTCGG | ATCGTCATTTCTTACAGCAATAGGT | 542 | 58 |
| rs11552707 |
PCR was performed to amplify 7 SNPs using the following systems: 2 μL DNA sample, 0.15 μL Taq polymerase (5 U/μL), 1 μL forward and reverse primers (5 μM for each), 2 μL dNTP mixture, 2.5 μL 10 × PCR buffer (Mg2+ Plus), and 17.35 μL ddH2O to total volume of 25 μL. The 10 × PCR buffer (Mg2+ Plus) contained 100 mM Tris-HCL, 500 mM KCL and 15 mM MgCl2. The amplification conditions were as follows: 95°C for 5 min; 36 cycles at 95°C for 30 s, X°C for 30 s, 72°C for 40 s; final extension at 72°C for 5 min (X°C: N1, 61°C; N2, 63°C; N3, 58°C). PCR kits were purchased from TaKaRa Biotechnology (Dalian) Co., Ltd. (Dalian, China).
The genotypes of 7 SNPs of NKX2-5 gene were determined using DNA direct sequencing method (Beijing Liuhe Huada Gene Polytron Technologies Inc., Beijing, China). The DNAstar software (DNASTAR Corp., WI, USA) was used to compare DNA sequencing diagrams and find the related polymorphism sites.
Statistical analysis
All 7 SNPs were assessed for Hardy-Weinberg equilibrium (HWE) by χ2 statistics. The comparisons of genotype and allele frequencies were evaluated by χ2 test. The association of NKX2-5 gene polymorphisms with CHD risk was estimated by computing odds ratio (OR) and 95% confidence interval (CI) from the multivariate logistic regression analysis. All statistical analysis was performed by SPSS 17.0 software (SPSS Inc., Chicago, USA). In addition, haplotype constructions and Mendel linkage disequilibrium were analyzed using online computer platform SHEsis and Haploview software (http://analysis. bio-x.cn/myAnalysis.php) [12]. Haplotypes with frequencies > 3% in the combined cases and controls were examined. P < 0.05 was regarded as statistically significant.
Results
Sequencing results
In 7 SNPs in detection, rs2277923, rs3729753 and rs703752 existed polymorphism. A novel heterozygous DSV (1433A>G) in 3’UTR of NKX2-5 gene was identified in one tetralogy of Fallot (TOF) patient and one persistent left superior vena cava (PLSVC) patient. Its 1433th base A mutated into G. The sequencing results were shown in Figure 1. The rest rs17052019, rs3729938, rs3729754 and rs11552707 sites found no polymorphism, and the genotypes were AA, CC, CC and TT, respectively.
Figure 1.
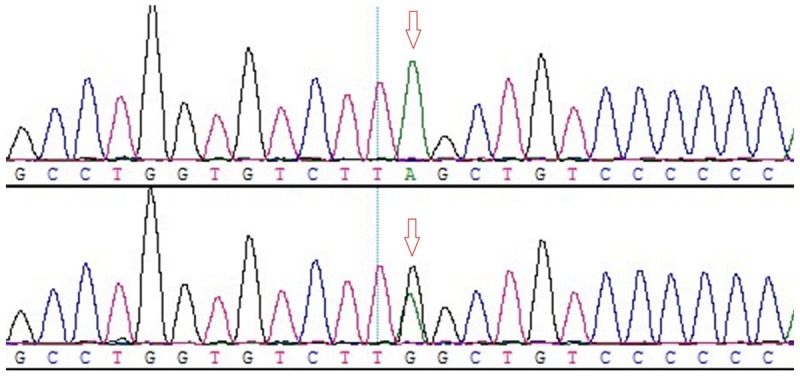
Sequence diagram of novel heterozygous DSV (1433A>G) site. Arrows indicated the mutations. Genotype: Above, AA; below, AG. Green, black, red and blue represented A allele, G allele, T allele and C allele, respectively.
Results of statistical analysis
rs2277923, rs3729753, rs703752 and novel DSV (1433A>G) SNPs of CHD patients and controls were in line with the genetic balance Hardy-Weinberg test. GG, GA and AA genotypes were detected in rs2277923 site, and the frequency distribution of 3 genotypes had significant difference between CHD and control groups (P = 0.032). Logistic regression analysis revealed that, the GG genotype was negatively correlated with CHD (P = 0.018; OR = 0.432; 95% CI: 1.152-4.655). The frequency distribution of two alleles had significant difference between two groups (P = 0.009). Logistic regression analysis revealed that, A allele was positively correlated with CHD (P = 0.014; OR = 1.719; 95% CI: 1.140-2.594). AA and AG genotypes were detected in novel DSV (1433A>G) site. The frequency distribution of two genotypes had significant difference between two groups (P = 0.048), but the relative risk analysis showed no statistical significance (P = 0.999). The differences of rs3729753 and rs703752 polymorphism distribution had no statistical significance between groups (P > 0.05) (Table 4).
Table 4.
The genotype and allele frequency distribution of all SNPs sites between CHD and control groups
| SNPs | Genotype/allele | Frequency | P | Logistic regression analysis | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| CHD N = 70 | Con N = 136 | OR, 95% CI | P | |||
| rs2277923 | GG | 13 (0.186) | 47 (0.346) | 0.032 | 0.432, [1.152-4.655] | 0.018 |
| GA | 37 (0.529) | 65 (0.478) | 0.817, [0.458-1.455] | 0.491 | ||
| AA | 20 (0.286) | 24 (0.176) | 0.536, [0.271-1.058] | 0.072 | ||
| G | 63 (0.450) | 159 (0.585) | 0.009 | 1.719, [1.140-2.594] | 0.014 | |
| A | 77 (0.550) | 113 (0.415) | ||||
| rs3729753 | GG | 68 (0.971) | 131 (0.963) | 0.759 | 0.771, [0.202-2.933] | 0.702 |
| GC | 2 (0.029) | 5 (0.037) | 1.298, [0.245-6.865] | 0.759 | ||
| G | 138 (0.986) | 267 (0.982) | 0.760 | 0.773, [0.148-4.041] | 0.763 | |
| C | 2 (0.014) | 5 (0.018) | ||||
| rs703752 | GG | 52 (0.743) | 103 (0.757) | 0.879 | 1.080, [0.556-2.099] | 0.819 |
| GT | 17 (0.243) | 30 (0.221) | 0.882, [0.447-1.742] | 0.718 | ||
| TT | 1 (0.014) | 3 (0.022) | 1.556, [0.159-15.243] | 0.704 | ||
| G | 121 (0.864) | 236 (0.868) | 0.924 | 0.971, [0.534-1.765] | 0.812 | |
| T | 19 (0.136) | 36 (0.132) | ||||
| 1433A>G | AA | 68 (0.971) | 136 (1.00) | 0.048 | 1.616E9, [0.000-] | 0.999 |
| AG | 2 (0.029) | 0 (0.000) | 0.000, [0.000-] | 0.999 | ||
| A | 138 (0.986) | 272 (1.000) | 0.115 | 0.000, [0.000-] | 0.999 | |
| G | 2 (0.014) | 0 (0.000) | ||||
Haplotype and linkage disequilibrium analysis
The linkage disequilibrium figure was constructed according to the distribution of rs2277923, rs3729753, rs703752 and novel DSV (1433A>G) SNPs sites (Figure 2). Only 2 SNPs (rs2277923 and rs703752) of this section were linked (D’ = 1.0; r2 = 0.18). There were 7 haplotypes according to haplotype analysis of 4 SNPs. One of the most common haplotypes, GGGA, showed obvious protective effects in CHD patients (P = 0.015; OR = 0.596; 95% CI: 0.393-0.904). AGGA haplotype showed obvious risky effects in CHD occurrence (P = 0.005; OR = 1.835; 95% CI: 1.193-2.824) (Table 5).
Figure 2.
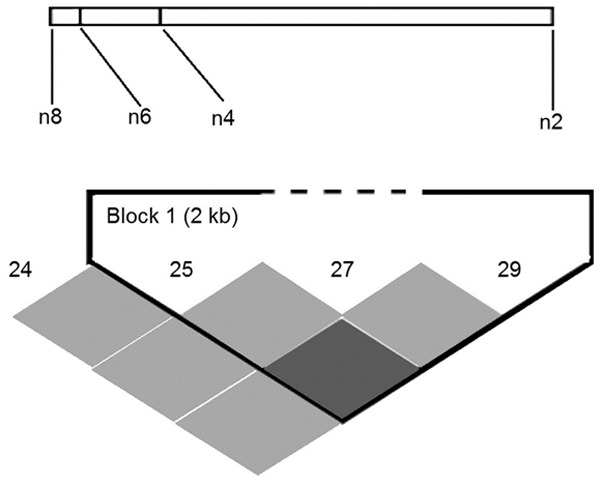
Linkage disequilibrium map (n2: rs2277923; n4: rs3729753; n6: rs703752; n8: 1433A>G).
Table 5.
Haplotype analysis results of rs2277923, rs3729753, rs703752 and 1433A>G SNPs site
| Haplotype | CHD (frequency) | Con (frequency) | P | OR, 95% CI |
|---|---|---|---|---|
| GCGA | 2.00 (0.014) | 4.95 (0.018) | / | / |
| AGGA | 58.01 (0.414) | 77.00 (0.283) | 0.005 | 1.835, [1.193-2.824] |
| GGGA | 60.99 (0.436) | 154.05 (0.566) | 0.015 | 0.596, [0.393-0.904] |
| ACTA | 0.00 (0.000) | 0.05 (0.000) | / | / |
| AGTA | 16.99 (0.121) | 35.95 (0.132) | 0.785 | 0.918, [0.495-1.703] |
| GGTA | 0.01 (0.000) | 0.00 (0.000) | / | / |
| AGTG | 2.00 (0.014) | 0.00 (0.000) | / | / |
Discussion
CHD is seriously harmful to the health of infants and young children. It arises from abnormal heart development during embryogenesis [13,14]. The regulation morphogenesis of CHD has been extensively studied, which confirms that some mutated genes such as GATA4 and NKX2-5 can cause CHD [15].
NKX2-5 protein is the important transcription factor of the heart. The human NKX2-5 gene locates at chromosome 5q35, and contains two exons encoding protein of 324 amino acids and 5’UTR as well as 3’UTR regions [7]. It is the fifth found NK-2 family gene, so named NKX2-5. NKX2-5 gene is a highly conserved homeobox protein gene and expressed in the developing heart, as well as in adult heart. In the embryonic heart, NKX2-5 plays essential roles in cardiac progenitor determination, cardiomyocyte differentiation, cardiac morphogenesis and conduction system development [16-20]. In adult heart, NKX2-5 is required for cardiomyocyte homeostasis and postnatal formation of ventricular conduction system [21,22]. Approximately 38 mutations in NKX2-5 gene have been identified in diverse types of CHD, including atrial septal defect, ventricular septal defect and TOF. In comparison with other population, these mutations have not been found often in mongoloid CHD patients. Analysis of NKX2-5 gene in subjects from southern China with sporadic CHD shows that, there is no any nonsynonymous mutation in 224 CHD patients, excepting 3 reported SNPs in 128 patients. It is suggested that the variant of the NKX2-5 gene may differ between races and regions [23,24].
In this study, 4 SNPs of the NKX2-5 gene in 70 Chinese Bai patients with CHD are screened for genetic mutations. One SNP (rs2277923) is found to be significantly associated with the risk of CHD, and A allele of rs2277923 is likely a risk factor for CHD. When further exploring the genotype, it is found that people with GG genotype of CHD are at lower risk than people with AA or AG genotype, but AA genotype is not associated with the occurrence of CHD independently after the logistic regression analysis, It is believed that the allele frequency and genotype frequency are not consistently attributed to the existence of heterozygous. In addition, a novel mutation (1433A>G) in 3’UTR region of NKX2-5 gene may be associated with the incidence of TOF and PLSVC in Chinese Bai people. Analysis of the linkage disequilibrium shows that, there is no high-degree linkage at this region, and only 2 SNPs (rs2277923 and rs703752) have a linkage (D’ = 1.0; r2 = 0.18). Moreover, analysis of haplotype has provided evidence that the GGGA haplotype of NKX2-5 gene has a protective effect in the incidence of CHD, and the AGGA has a risky effect.
The rs2277923 SNPs of NKX2-5 gene is the 63th base G mutates into A in the coding chain, causing 21st codon GAG converting into GAA. As the third codon is degenerate codon, it still codes Glu after base substitution. So it is synonymous mutation (Glu21Glu). Results of this study suggest that, A allele of rs2277923 in NKX2-5 gene is likely a risk factor for CHD, which is similar to previous study [25] which investigates the genetic variations of NKX2-5 associated with sporadic CHD in 125 cases and 105 controls of Chinese Han people. In that study, the A allele frequency distribution of rs2277923 has significant difference between CHD and control group (P = 0.04; OR = 1.48; 95% CI: 1.01-2.14), which indicates that A allele of rs2277923 in NKX2-5 gene contributes to the risk of CHD. That study also finds that, the transcription activity of rs2277923 SNPs encoding protein is lower than the wild type by 20%, which may account for the contributor to CHD. Besides, a meta-analysis regarding the associations between two genetic variants in NKX2-5 and risk of CHD reveals that, the rs2277923 variant in NKX2-5 gene may contribute to CHD risk for Chinese [26]. However, there are some differences from our study. Above study [26] does not provide evidence that the rs2277923 SNPs in NKX2-5 gene is associated with sporadic CHD in Chinese Han people. The different research conclusions may be related to the racial and geographic differences. The objects of this study are the Bai minority people. They have no migration history or history of marriage with other nationalities within three generations, and settle in Dali Bai minority autonomous district. Additionally, the sample size of this study is small, and we have not respectively divided subtypes of CHD for analysis. It is necessary to enlarge sample for further research as to the association of NKX2-5 gene mutations and sporadic CHD in Chinese Bai people.
As the mutations in the coding region can affect the protein function or make the protein cannot express, most of relative researches are focused on this genetic regions. In recent years, it is found that the 3’UTR, 5’UTR, promoter region and enhancer region also play an important role in the occurrence of CHD. Mutations in these regions can lower the protein physiological levels of gene encoding by affecting the efficiency of protein transcription, changing the methylation status near the site, affecting RNA folding, or interfering the combination of transcription factors and cofactors [27-31]. Our study finds a novel DSV in the 3’UTR of NKX2-5 gene in one TOF patient and one PLSVC, but none in controls. TOF is considered to be a neural crest cell and/or second heart field related conotruncal heart defects that occur during embryonic development [32]. In the embryonic period, the right and left anterior cardinal veins constitute the main venous drainage of the cephalic portion. As a result of the development of left innominate vein that bridges the anterior cardinal veins at 8 weeks gestation, the proximal part of the left cardinal vein regresses, and only a small portion of it remains as the left superior intercostal vein (LSVC). A defect in the regression of this vein is thought to be the cause of persistence of LSVC [33]. TOF and PLSVC have the similar formation time in the embryonic period. Dinesh et al. [34] find that, SNP 1212G>T in 3’UTR of NKX2-5 is observed in 40% of CHD cases in Mysore, South India. Similarly, a research on the role of related gene variants in the development of hypoplastic right heart syndrome (HRHS) shows that, 4 HRHS cases present a low frequency variation in the 3’UTR of NKX2-5 gene [35]. Notably, recent research demonstrates that, the influence of low frequency variation is significantly greater than the common variants in CHD [36]. Therefore, the noncoding regions need to be further studied in the future.
In conclusion, the Chinese Bai people with CHD have NKX2-5 gene mutations. This study may provide a foundation for a more integrated understanding of the molecular basis of human CHD. Further studies are required to confirm the risk factors of SNP rs2277923 in NKX2-5 gene in relation to CHD occurrence, and the function of novel DSV in 3’UTR should be verified.
Acknowledgements
This work was supported by National Natural Science Foundation of China (31160230 and 81560060), Special and Joint Program of Yunnan Province (2011FB166) and Doctorial Innovation Fund of Kunming Medical University.
Disclosure of conflict of interest
None.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics-2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Goede J, Geleijnse JM, Boer JM, Kromhout D, Verschuren WM. Linoleic acid intake, plasma cholesterol and 10-year incidence of CHD in 20,000 middle-aged men and women in the Netherlands. Br J Nutr. 2012;107:1070–1076. doi: 10.1017/S0007114511003837. [DOI] [PubMed] [Google Scholar]
- 3.Winkleby M, Sundquist K, Cubbin C. Inequities in CHD incidence and case fatality by neighborhood deprivation. Am J Prev Med. 2007;32:97–106. doi: 10.1016/j.amepre.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oude Griep LM, Verschuren WM, Kromhout D, Ocké MC, Geleijnse JM. Variety in fruit and vegetable consumption and 10-year incidence of CHD and stroke. Public Health Nutr. 2012;15:2280–2286. doi: 10.1017/S1368980012000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97–121. doi: 10.1146/annurev.physiol.68.040104.113828. [DOI] [PubMed] [Google Scholar]
- 6.Nermer M. Genetic insights into normal and abnormal heart development. Cardiovasc Pathol. 2008;17:48–54. doi: 10.1016/j.carpath.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital Heart Disease Caused by Mutations in the Transcription Factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguín P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, Zhou YQ, Yeh RF, Henkelman RM, Harvey RP, Metzger D, Chambon P, Stainier DY, Pollard KS, Scott IC, Bruneau BG. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terada R, Warren S, Lu JT, Chien KR, Wessels A, Kasahara H. Ablation of Nkx2-5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc Res. 2011;91:289–299. doi: 10.1093/cvr/cvr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng T, Wang L, Zhou SF, Li X. Mutations of the GATA4 and NKX2.5 genes in Chinese pediatric patients with non-familial congenital heart disease. Genetica. 2010;138:1231–1240. doi: 10.1007/s10709-010-9522-4. [DOI] [PubMed] [Google Scholar]
- 11.Jiang LH, Duan CQ, Ma ZQ, Zhu LJ, Yin WJ, Zou HL, Li P, Wu J, Wei J, Na ZH, Chen WM. The prevalence survey of congenital heart disease for 3-18 years old in parts of Yunnan province. Chin J Epidemiol. 2005;26:182–186. (In Chinese) [PubMed] [Google Scholar]
- 12.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 13.Misra C, Sachan N, McNally CR, Koenig SN, Nichols HA, Guggilam A, Lucchesi PA, Pu WT, Srivastava D, Garg V. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet. 2012;8:e1002690. doi: 10.1371/journal.pgen.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Zhou S, Chen Q, Xie X, Huang G, Wang J, Zhoua S, Ma X. Hairy-related transcription factor 2 is not potentially related to congenital heart disease in Chinese patients. Int J Cardiol. 2011;146:415–416. doi: 10.1016/j.ijcard.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Ng KM, Chan YC, Lee YK, Lai WH, Au KW, Fung ML, Siu CW, Li RA, Tse HF. Cobalt chloride pretreatment promotes cardiac differentiation of human embryonic stem cells under atmospheric oxygen level. Cell Reprogram. 2011;13:527–537. doi: 10.1089/cell.2011.0038. [DOI] [PubMed] [Google Scholar]
- 16.Habets PE, Moorman AF, Clout DE, Roon MA, Lingbeek M, Lohuizen MV, Campione M, Christoffels V. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O’Brien TX, Gourdie RG, Berul CI, Izumo S. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman C. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 20.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meysen S, Marger L, Hewett KW, Jarry-Guichard T, Agarkova I, Chauvin JP, Perriard JC, Izumo S, Gourdie RG, Mangoni ME, Nargeo Jt, Gros D. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev Biol. 2007;303:740–753. doi: 10.1016/j.ydbio.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 22.Toko H, Zhu W, Takimoto E, Shiojima I, Hiroi Y, Zou Y, Oka T, Akazawa H, Mizukami M, Sakamoto M, Terasaki F, Kitaura Y, Takano H, Nagai T, Nagai R, Komuro I. Csx/Nkx2-5 is required for homeostasis and survival of cardiac myocytes in the adult heart. J Biol Chem. 2002;277:24735–24743. doi: 10.1074/jbc.M107669200. [DOI] [PubMed] [Google Scholar]
- 23.Xiong F, Li Q, Zhang C, Chen Y, Li P, Wei XF, Li Q, Zhou WJ, Li L, Shang X, Xu XM. Analyses of GATA4, NKX2-5, AND TFAP2B genes in subjects from southern China with sporadic congenital heart disease. Cardiovasc Pathol. 2013;22:141–145. doi: 10.1016/j.carpath.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Sun M, Qiu GR, Sun GF, Yuan YH, Sun KL. Mutation and expression of NKX2.5 gene in human simple congenital heart disease. J China Medical University. 2001;30:321–324. [Google Scholar]
- 25.Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. Screening NKX2.5 mutation in a Sample of 230 Han Chinese children with congenital heart diseases. Genet Test Mol Biomarkers. 2009;13:159–162. doi: 10.1089/gtmb.2008.0044. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Qiao Y, Meng H, Pang S, Huang W, Zhang H, Yan B. Genetic analysis of the TBX3 gene promoter in ventricular septal defects. Gene. 2013;512:185–188. doi: 10.1016/j.gene.2012.10.066. [DOI] [PubMed] [Google Scholar]
- 27.Limsuwan A, Choubtum L, Wattanasirichaigoon D. 5’UTR Repeat polymorphisms of the BMPR2 gene in children with pulmonary hypertension associated with congenital heart disease. Heart Lung Circ. 2013;22:204–210. doi: 10.1016/j.hlc.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Reamon-Buettner SM, Cho SH, Borlak J. Mutations in the 3’-untranslated region of GATA as molecular hotspots for congenital heart disease (CHD) BMC Med Genet. 2007;8:38. doi: 10.1186/1471-2350-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012;21:3255–3263. doi: 10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao JY, Yang XY, Gong XH, Gu ZY, Duan WY, Wang J, Ye ZZ, Shen HB, Shi KH, Hou J, Huang GY, Jin L, Qiao B, Wang HY. Functional variant in methionine synthase reductase intron-1 significantly increases the risk of congenital heart disease in the Han Chinese population. Circulation. 2012;125:482–490. doi: 10.1161/CIRCULATIONAHA.111.050245. [DOI] [PubMed] [Google Scholar]
- 31.Zhao JY, Yang XY, Shi KH, Sun SN, Hou J, Ye ZZ, Wang J, Duan WY, Qiao B, Chen YJ, Shen HB, Huang GY, Jin L, Wang HY. A functional variant in the cystathionine β-synthase gene promoter significantly reduces congenital heart disease susceptibility in a Han Chinese population. Cell Res. 2013;23:242–253. doi: 10.1038/cr.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward C, Stadt H, Hutson M, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol. 2005;284:72–83. doi: 10.1016/j.ydbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Esmer AÇ, Yüksel A, Calı H, Ozsürmeli M, Omeroğlu RE, Kalelioğlu I, Has R. Prenata l Diagnosis of Persistent Left Superior Vena Cava and Clinical Significance. Balkan Med J. 2014;31:50–54. doi: 10.5152/balkanmedj.2014.9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinesh SM, Kusuma L, Smitha R, Savitha MR, Krishnamurthy B, Narayanappa D, Ramachandra NB. Single-nucleotide polymorphisms of NKX2.5 found in congenital heart disease patients of Mysore, South India. Genet Test Mol Biomarkers. 2010;14:873–879. doi: 10.1089/gtmb.2010.0100. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Zou L, Zhong R, Zhu B, Chen W, Shen N, Ke J, Lou J, Song R, Miao XP. Associations between two genetic variants in NKX2-5 and risk of congenital heart disease in Chinese population: a meta-analysis. PLoS One. 2013;8:e70979. doi: 10.1371/journal.pone.0070979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


