Abstract
Gene promoter methylation may be used a potential biomarker for detecting solid tumor including cervical cancer. Here, we used methylation sensitive-high resolution melting (MS-HRM) analysis to detecting promoter methylation ratios of DAPK1, MGMT and RARB gene in patients with different cervical disease grade. The detection of gene promoter methylation was conducted in two hundred fifty patients’ samples including normal cytology (n=48), cervical intraepithelial neoplasia grade 1 (CIN1, n=54), cervical intraepithelial neoplasia grade 2 (CIN2, n=47), cervical intraepithelial neoplasia grade 3 (CIN3, n=56) and cervical squamous cell carcinomas (SCS, n=45). We found there were a significant positive correlation between the promoter methylation status of DAPK1 and cervical disease grade (P=0.022). In addition, the methylated promoters of DAPK1 combined with MGMT, MGMT combined with RARB, DAPK1 combined with RARB were positive correlated with cervical disease grade (P < 0.05). All three genes promoters methylated were positive correlated with cervical disease grade (P < 0.001). Receiver operating characteristic (ROC) curves was conducted to evaluate whether the three genes methylation could be used to be a potential marker for diagnosing high grade cervical disease (HSIL and SCC). The cutoff values for the methylation rates of all these genes were 0-5%. Regrettably, only the methylation of MGMT combined with DAPK1 gave 43.4% sensitivity and 68.6% specificity. The current results indicated that MS-HRM-based testing for DNA methylations of MGMT plus DAPK1 genes holds some promise for high grade cervical disease screening.
Keywords: Cervical cancer, DNA methylation, MS-HRM
Introduction
Cervical cancer is one of the leading causes of mortality for females, with the worldwide incidence of cervical cancer reaching 454,000 cases per year in 2010 [1]. According to preliminary statistics, there are about 62,000 women diagnosed cervical cancer and about 30,000 died due to cervical cancer in China [2]. Some conventional cancer screening methods have a lower sensitivity or a higher sensitivity but tend to give false positives, such as cytology screening and HPV testing [3-5]. In a clinical setting, some screening methods generally cause excessive diagnosis and unnecessary hurt for patients with both physically and mentally, such as vaginal endoscopy and biopsy [6]. Therefore, a screening test with high sensitivity and specificity and without hurt is urgently needed for early diagnosis and monitoring of cervical cancer.
DNA methylation is a well-known epigenetic marker, as histone modification [7]. In cervical cancer and precancerous lesions, some gene promoter regions existed high methylation status, for the detection of specific gene promoter region methylation status for cervical disease and cervical cancer diagnosis. Recently, more and more researchers focus on methylation detection in the diagnosis of cervical lesions, especially for high level forecast and screening of cervical lesions.
The DNA hypomethylation and hypermethylation of several cancer-related genes have been extensively studied [8-10]. In cervical cancer, DNA methylation also affects the expression of several genes, including CDH1, TIMP-3, p16 (INK4a), FHIT and RASSF1A 9 [11]. DAPK1, MGMT and RARB genes have been shown to be associated with HPV-associated cervical carcinogenesis. Moreover, DAPK1, MGMT and RARB genes have previously been shown to be targets for aberrant DNA methylation in cervical cancer, however the genes have not been comprehensively tested using quantitative methods [12]. In this study, we detected the gene promoter region methylation level of MGMT, DAPK1 and RARB by the methylation sensitivity-high resolution dissolution curve (MS-HRM) analysis. Additionally, we analyzed the relationship between the promoter methylation level with cervical disease and cancer, and evaluated clinical value of the promoter methylation in diagnosis of cervical disease and cancer. The objective of this study was to identify promoter methylation status of MGMT, DAPK1 and RARB in cervical disease patients, and to determine whether the gene promoter methylation could be used as biomarkers to distinguish high grade cervical disease from low grade cervical disease.
Materials and methods
Study population
The cervical cytology remaining specimens were collected from the Department of Obstetrics and Gynecology in the Peking Union Medical College Hospital. For this study, we retrospectively selected different level of pathological cases. Pathological diagnosis was identified as the gold standard. The ethics committee approved the relating screening, inspection, and data collection from the patients, and all subjects signed a written informed consent form. Patients name, age, medical history, TCT examination results, Hybrid Capture II (HC2) test results and colposcopy, cervical biopsy, taper cutting or cervical cancer surgical pathologic result were collected in this study. The exclusion criteria included unreceived uterine cervical surgery or radiation and chemotherapy, without cervical physical treatment such as laser and freezing for at least three months.
The study sample comprised exfoliated cervical cell specimens from 250 patients. The average age of the patients was 42.5 (range from 21 to 73), including 157 cases of CIN (54 grade I, 47 grade II, 56 grade III) and 45 cases of cervical squamous cell carcinoma and 48 cases of uterine benign disease as healthy control. According to the 2014 WHO reclassified cervical tumor classification, CIN1 was classified as LSIL (low-grade squamous intraepithelial lesions), CIN2 and CIN3 were classified as HSIL (high-grade squamous intraepithelial lesions). So there were 54 cases of LSIL and 103 cases of HSIL in the study sample.
Thin-prep cytology test (TCT)
The cervical cell specimens were collected according to the TCT standards and stored at room temperature up to six months. The diagnosis reports of TCT were provided based on the Bethesda System (TBS) by cytopathologic experts from the Department of Obstetrics and Gynecology in the Peking Union Medical College Hospital.
HC2-HPV detection
At enrollment, all samples were tested by HR HC2 assay (Qiagen, Gaithersburg, MD), according to the manufacturer’s protocol. All samples were stored at room temperature up to 7 days. HC2-HPV detection reports were provided by physician from the Department of Obstetrics and Gynecology in the Peking Union Medical College Hospital.
Pathological diagnosis
All pathological results were diagnosed by two pathologists from the Department of Obstetrics and Gynecology in the Peking Union Medical College Hospital. Read and review were performed by the two pathologists respectively [13]. Patients with atypical squamous cells of undetermined significance (ASCUS) and HC2 cytology for positive, or simple cytology for ASCUS (including) above, needed detection of Colposcopy with directed biopsy in the study.
DNA isolation and extraction
DNA was extracted from those samples by using the DNeasy Tissue Extraction Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) for bisulphite modification of the DNA samples. CpGenomeTM Universal Methylated DNA (Chemicon, Millipore, Billerica, MA) as a positive/methylated control in our experiments. DNA from peripheral blood mononuclear cells was used as a negative/unmethylated reference.
High resolution melting analysis (HRM) analysis
PCR amplification was carried out in a 20 µL total volume containing: 10 μL LightCycler 480 HRM Master Mix (Roche), 2 µL MgCl2 (25 mM), 200 nM of each primer, 1 µL of bisulphite modified template (theoretical concentration 20 ng/µL) and RNase-free H2O. The amplification consisted of 15 min at 95°C, followed by 50 cycles of 5 s 95°C, 5 s at the primer annealing temperature (Ta) and 10 s at 72°C. Primers sequence as follows: DAPK1, 5’-GGTTGTTTCGGAGTGTGAGGAGG-3’ as forward, 5’-GCCGACCCCAAACCCTACC-3’ as reverse; MGMT, 5’-GCGTTTCGGATATGTTGGGATAGT-3’ as forward, 5’-AACGACCCAAACACTCACCAAA-3’ as reverse; RARB 5’-TTTATGCGAGTTGTTTGAGGAT-3’ as forward, 5’-ATCCCAAATTCTCCTTCCAA-3’ as reverse. High resolution melting analyses were performed at the temperature ramping and florescence acquisition setting recommended by the manufacturer i.e. temperature ramping from 70-95°C, rising by 0.1°C/2 s. All the reactions were performed in triplicate. The melting curves were normalized by calculation of the ‘line of best fit’ in between two normalization regions before and after the major fluorescence decrease representing the melting of the PCR product using software. This algorithm allows the direct comparison of the samples that have different starting fluorescence levels.
Statistical analysis
Statistical analysis was performed by SPSS (version 20.0). Measurement results (e.g age) were expressed in mean ± standard deviation (SD). Hierarchical data were analyzed by using the Mann-Whitney Test and Kruskal Wallis Test. Pearson’s correlation coefficient was calculated to analyze the correlation between the genes promoter methylation and different grade cervical disease. Receiver operating characteristic (ROC) curves were generated to confirm the accuracy of diagnosis of each gene, and sensitivity and specificity were computed for each combination. A P-value of less than 0.05 was considered statistically significant.
Results
A total of 250 tissue specimens were included in this study. The age characteristics and clinicopathological features of this study population were presented in Figure 1. The correlations between age and clinicopathological features were not significantly (P > 0.05). In addition, we also showed that the age was not associated with cervical disease grade by Thin-prep cytology test (TCT) test (P=0.47) and by pathological diagnosis (P=0.46).
Figure 1.
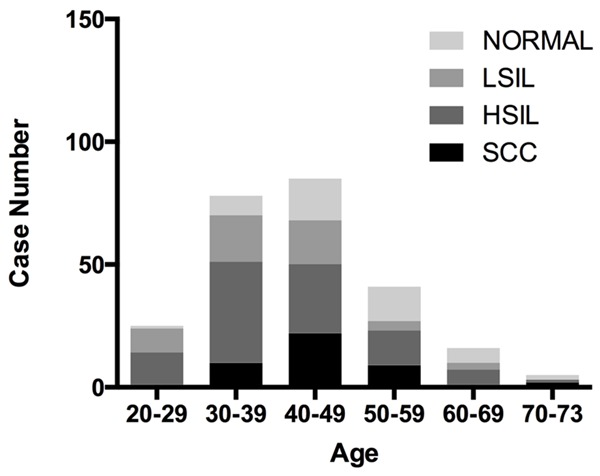
Demographics of patients with different grade cervical disease and age.
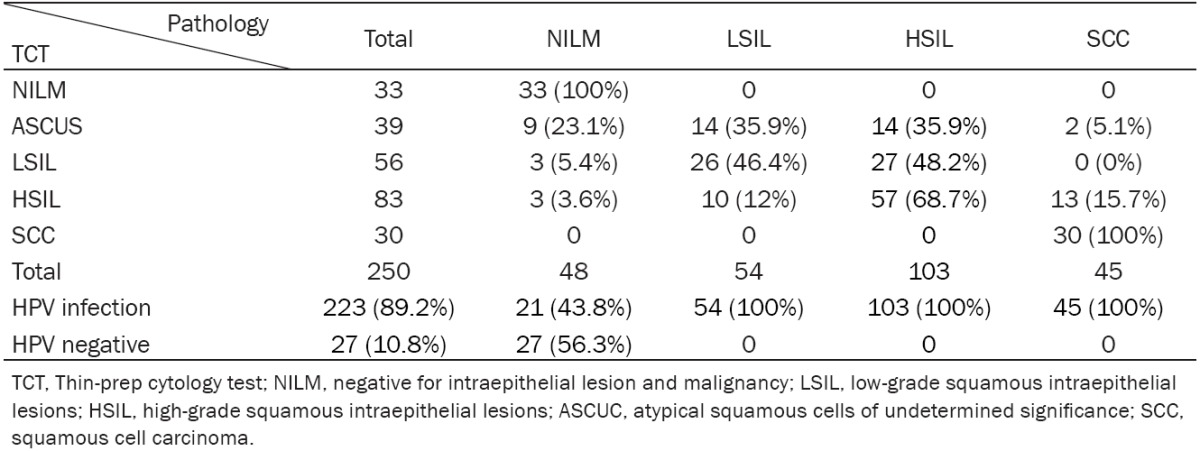
By pathological diagnosis, the 250 tissue specimens included 48 samples with normal cytology, 54 samples with cervical intraepithelial neoplasia grade 1 (CIN1), 47 samples with cervical intraepithelial neoplasia grade 2 (CIN2), 56 samples with cervical intraepithelial neoplasia grade 3 (CIN3) and 45 samples with cervical squamous cell carcinomas (SCS). In addition, by TCT test, the above tissue specimens included negative for intraepithelial lesion and malignancy (NILM, n=33), atypical squamous cells of undetermined significance (ASCUS, n=39), ASCUS-H (n=9), low-grade squamous intraepithelial lesion of cervix (LSIL, n=56), high-grade squamous intraepithelial lesions (HSIL, n=83), squamous cell carcinoma (SCC, n=30). As shown in Table 1, the results from pathological diagnosis were basically consistent with the data obtained by TCT test. Additionally, by HC2 test, Table 1 indicated that 250 tissue specimens included 223 samples with HPV positive (HPV > 1 pg/ml) and 27 samples with HPV negative (HPV < 1 pg/ml). The positive rate of HPV was 89.2%.
Table 1.
Correlation between pathological diagnosis and TCT test in patients
|
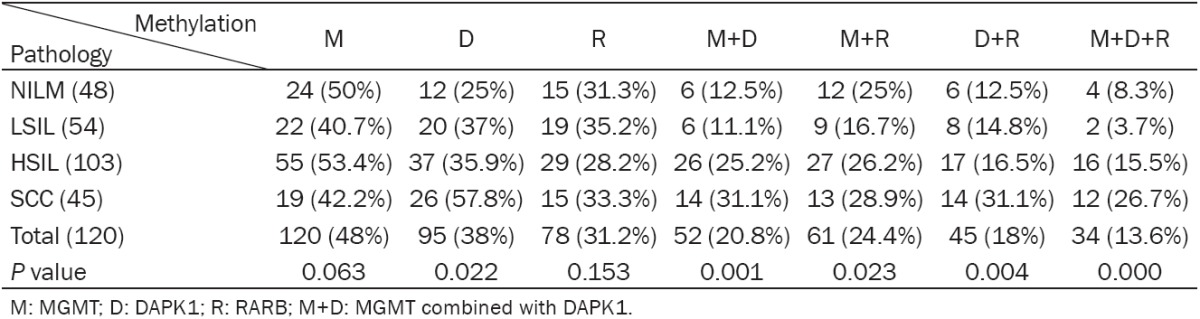
By MS-HRM analysis, we determined the frequencies of methylation at the promoter of the DAPK1, MGMT and RARB gene respectively. When dichotomized as methylation negative (the frequencies of methylation=0), and methylation positive (the frequencies of methylation > 0), the difference in the frequencies of methylation was not statistically significant for each gene. As shown in Table 2, for MGMT gene, we found that the 130 tissue specimens with methylation positive (frequency > 0%) and 130 samples with methylation negative (frequency=0%). The positive rate of the promoter methylation of MGMT was 48% (n=120). In addition, the positive rates of the promoters’ methylation of DAPK1, RARB were 38% and 31.2% respectively. Moreover, there were not significant associations between the promoters’ methylation of all three genes and patient age (data not shown).
Table 2.
The percentage of methylation for each gene in patients
| MGMT | DAPK1 | RARB | |
|---|---|---|---|
| Total | 250 | 250 | 250 |
| Negative | 130 (52.0%) | 155 (62.0%) | 172 (68.8%) |
| Positive | 120 (48.0%) | 95 (38.0%) | 78 (31.2%) |
| 0-5% | 69 (27.6%) | 72 (28.8%) | 66 (26.4%) |
| 5-25% | 11 (4.4%) | 9 (3.6%) | 12 (4.8%) |
| 25-50% | 16 (6.4%) | 8 (3.2%) | 0 (0%) |
| 50-75% | 9 (3.6%) | 5 (2.0%) | 0 (0%) |
| 75-100% | 15 (6.0%) | 1 (0.4%) | 0 (0%) |
For analysis of the correlations between methylation status of the MGMT, DAPK1 and RARB gene promoter and cervical disease grade, there was a significant association between the promoter methylation status of DAPK1 and cervical disease grade (P=0.022) (Table 3). Further analysis revealed that the correlation was positive. In addition, DAPK1 combined with MGMT promoters methylated, MGMT combined with RARB, DAPK1 combined with RARB were positive correlated with cervical disease grade (P < 0.05, Table 3). All three genes promoters methylated (34/250) were associated with cervical disease grade (P < 0.001). The correlation was positive (Table 4). However, the differences were not significant between the promoter methylation status of single MGMT gene (P=0.063) and single RARB gene (P=0.153) with cervical disease grade respectively (Table 3).
Table 3.
The percentage of methylation for each gene in patients with different cervical disease grade
|
Table 4.
Correlation between pathological diagnosis, TCT test and promoter methylation of three genes in patients
| Pathology | TCT | MGMT | DAPK1 | RARB | HC2 | |
|---|---|---|---|---|---|---|
| Pathology | 1 | |||||
| TCT | r=0.679 | 1 | ||||
| (P=0.000) | ||||||
| MGMT | r=0.009 | r=0.146 | 1 | |||
| (P=0.871) | (P=0.006) | |||||
| DAPK1 | r=0.188 | r=0.173 | r=0.142 | 1 | ||
| (P=0.001) | (P=0.002) | (P=0.012) | ||||
| RARB | r=-0.001 | r=0.139 | r=0.375 | r=0.264 | 1 | |
| (P=0.981) | (P=0.013) | (P=0.000) | (P=0.000) | |||
| HC2 | r=0.199 | r=0.268 | r=0.158 | r=0.129 | r=0.172 | 1 |
| (P=0.000) | (P=0.000) | (P=0.001) | (P=0.01) | (P=0.001) |
We analyzed the correlations between histological diagnosis and all that TCT, HC2 and the methylation status of DAPK1, MGMT and RARB. As shown in Table 4, there was a strong positive correlation between histological diagnosis and TCT (r=0.679, P < 0.001). Additionally, Table 4 indicated a weak association between histological diagnosis and both HC2 and the methylation of DAPK1 (r=0.199 and r=0.188 respectively, P < 0.001). Moreover, both TCT and HC2 were weakly associated with the methylation status of DAPK1, MGMT and RARB (all r < 0.3, P < 0.05).
When dichotomized as low grade cervical disease (including normal and LSIL) and high grade cervical disease (including HSIL and SCC), the differences between methylation negative and methylation positive were statistically significant for DAPK1 and MGMT gene in combination (P=0.004, data not shown) and all three gene in combination (P=0.003, data not shown).
ROC curve analysis was conducted to evaluate whether the three genes methylation could be used to be a potential marker for diagnosing high grade cervical disease (HSIL and SCC). The area under the curve (AUC) of ROC plots for MGMT, DAPK1 and RARB in the detection of HSIL and SCC were 0.53, 0.57 and 0.49, respectively (Table 5). The cutoff values for the methylation rates of all these genes were 0-5%. The methylation of DAPK1 combined with MGMT gave 43.4% sensitivity and 68.6% specificity. At a meth-index cutoff value of 2.6, the HC2 measure achieved 92.4% sensitivity and 35.3% specificity (Figure 2). In addition, the TCT conferred 67.6% sensitivity and 78.8% specificity (Figure 2).
Table 5.
The AUC of ROC curve of HC2, TCT test and promoter methylation of three genes
| AUC | 95% CI | P | |
|---|---|---|---|
| MGMT | 0.53 | 0.46-0.6 | 0.445 |
| DAPK1 | 0.57 | 0.5-0.64 | 0.052 |
| RARB | 0.49 | 0.42-0.56 | 0.754 |
| M+D | 0.58 | 0.51-0.65 | 0.034 |
| M+R | 0.54 | 0.46-0.61 | 0.349 |
| D+R | 0.54 | 0.47-0.61 | 0.306 |
| M+D+R | 0.57 | 0.5-0.64 | 0.072 |
| TCT | 0.84 | 0.8-0.89 | 0.000 |
| HC2 | 0.62 | 0.55-0.7 | 0.001 |
AUC: area under the curve. The AUC of ROC curve was calculated for the diagnosis of HSIL and SCC. M: MGMT; D: DAPK1; R: RARB; M+D: MGMT combined with DAPK.
Figure 2.
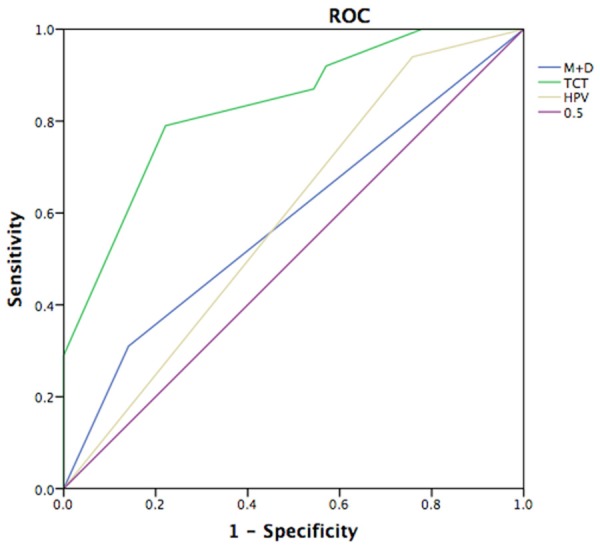
Receiver Operator Curve (ROC) analysis of TCT, HPV, methylated MGMT and DAPK1. ROC analysis of predicted sensitivity and 1-specificity was performed using SPSS 20.0, when TCT, HPV, methylated MGMT and DAPK1 were added to the model.
Discussion
Methylation of DNA is a common regulatory process that regulates gene transcription in cells. DNA methylation is a well-balanced process in normal cells. However, aberrant CpG methylation changes may occur in cancer cells [10]. Low methylation of oncogene promoters and high methylation of tumor suppressor gene promoters are pivotal alterations in tumorigenesis [14]. Several investigators have found that DNA methylation occurs frequently in CIN3 and cervical cancer cells, but rarely in CIN1 and normal cells [15], suggesting that aberrant methylation may indicate an increased risk of developing cervical cancer.
Several investigators have found that DAPK1, MGMT and RARB showed elevated methylation in cervical cancers [16]. DAPK1 can induce cell apoptosis and inhibit cell survival and proliferation [17]. Previous investigations have revealed that the level of DAPK1 was low in many malignant tumors [18]. The RARB gene encodes a nuclear receptor that binds retinoic acid, and is important in differentiation of stratified squamous epithelium, including cervical epithelium [19]. Furthermore, RARB silencing was found in both cervical dysplastic and carcinoma cells, suggesting this protein can suppress cervical carcinogenesis. MGMT is a DNA methyltransferase. High methylation rate in MGMT gene promoter was detected in some cancer tissue including cervical cancer and non-small cell lung cancer [20]. MGMT also can inhibit cell survival and proliferation [20]. Thereafter, we selected these three genes to assess the association between ratio of methylation and high grade cervical disease. DNA methylation was assessed by semi-quantitative methods tend to overestimate methylation prevalence causing a high number of low specificity or false positives. Therefore, the use of quantitative MS-HRM analysis to determine DNA methylation levels is critical to accurately classify methylation status. In this study, we used MS-HRM analysis to quantitate the level of DNA methylation within DAPK1, MGMT and RARB gene promoters in normal cervical and cervical cancer specimens.
Our findings demonstrated that DAPK gene methylation was associated with HSIL and SCC patients. Therefore, DAPK was a novel potential biomarker for high grade cervical disease. In addition, we also evaluated the association between HC2, TCT and high grade cervical disease. However, our results revealed that TCT and HC2 were significantly more sensitive and specific than DAPK methylation for the diagnosis of HSIL and SCC. In conclusion, a correlation between the DAPK1 methylation and high grade cervical disease including HSIL and SCC was observed in this study. However, whether methylation of DAPK1 confers a poorer prognosis of high grade cervical disease patients deserves further study.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 81101977).
Disclosure of conflict of interest
None.
References
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.Setoodeh R, Hakam A, Shan Y. Cerebral metastasis of cervical cancer, report of two cases and review of the literature. Int J Clin Exp Pathol. 2012;5:710–4. [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman ME, Solomon D, Schiffman M. Qualification of ASCUS. A comparison of equivocal LSIL and equivocal HSIL cervical cytology in the ASCUS LSIL Triage Study. Am J Clin Pathol. 2001;116:386–394. doi: 10.1309/JM3V-U4HP-W8HJ-68XV. [DOI] [PubMed] [Google Scholar]
- 4.Layfield LJ, Qureshi MN. HPV DNA testing in the triage of atypical squamous cells of undetermined significance (ASCUS): cost comparison of two methods. Diagn Cytopathol. 2005;33:138–143. doi: 10.1002/dc.20316. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Romero R, Iglesias-Chiesa C, Alatorre B, Vazquez K, Pina-Sanchez P, Alvarado I, Lazos M, Peralta R, Gonzalez-Yebra B, Romero A, Salcedo M. HPV frequency in penile carcinoma of Mexican patients: important contribution of HPV16 European variant. Int J Clin Exp Pathol. 2013;6:1409–1415. [PMC free article] [PubMed] [Google Scholar]
- 6.Solomon D, Breen N, McNeel T. Cervical cancer screening rates in the United States and the potential impact of implementation of screening guidelines. CA Cancer J Clin. 2007;57:105–111. doi: 10.3322/canjclin.57.2.105. [DOI] [PubMed] [Google Scholar]
- 7.Dong Y, Tan Q, Tao L, Pan X, Pang L, Liang W, Liu W, Zhang W, Li F, Jia W. Hypermethylation of TFPI2 correlates with cervical cancer incidence in the Uygur and Han populations of Xinjiang, China. Int J Clin Exp Pathol. 2015;8:1844–1854. [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 9.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 10.Silva TD, Vidigal VM, Felipe AV, DE Lima JM, Neto RA, Saad SS, Forones NM. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol Lett. 2013;6:1687–1692. doi: 10.3892/ol.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong DH, Youm MY, Kim YN, Lee KB, Sung MS, Yoon HK, Kim KT. Promoter methylation of p16, DAPK, CDH1, and TIMP-3 genes in cervical cancer: correlation with clinicopathologic characteristics. Int J Gynecol Cancer. 2006;16:1234–1240. doi: 10.1111/j.1525-1438.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 12.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Z, Yuan Z, Zhang Q, Long Z, Chen J, Tang Z, Zhu Y, Chen S, Xu J, Yan M, Wang J, Liu Q. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy. 2012;8:1798–1810. doi: 10.4161/auto.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polakova VV, Vannucci L, Korenkova V, Prochazka P, Slyskova J, Vodickova L, Rusnakova V, Bielik L, Burocziova M, Rossmann P, Vodicka P. Evaluation of tumor suppressor gene expressions and aberrant methylation in the colon of cancer-induced rats: a pilot study. Mol Biol Rep. 2013;40:5921–5929. doi: 10.1007/s11033-013-2699-8. [DOI] [PubMed] [Google Scholar]
- 15.Wang KH, Lin CJ, Liu CJ, Liu DW, Huang RL, Ding DC, Weng CF, Chu TY. Global methylation silencing of clustered proto-cadherin genes in cervical cancer: serving as diagnostic markers comparable to HPV. Cancer Med. 2015;4:43–55. doi: 10.1002/cam4.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geering B. Death-associated protein kinase 2: Regulator of apoptosis, autophagy and inflammation. Int J Biochem Cell Biol. 2015;65:151–154. doi: 10.1016/j.biocel.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Martins CR, Fansler ZB, Roemer KL, Kincaid EA, Gustafson KS, Heitjan DF, Clark DP. DNA methylation in anal intraepithelial lesions and anal squamous cell carcinoma. Clin Cancer Res. 2005;11:6544–6549. doi: 10.1158/1078-0432.CCR-05-0374. [DOI] [PubMed] [Google Scholar]
- 19.Dolle P. Developmental expression of retinoic acid receptors (RARs) Nucl Recept Signal. 2009;7:e006. doi: 10.1621/nrs.07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb) 2015;25:161–176. doi: 10.11613/BM.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


