Abstract
DNA methylation has been recently recognized as a novel tumor marker. This study investigated the methylation status of Reprimo and hMLH1 gene in both plasma and tissue samples from gastric cancer patients, in an attempt to investigate their diagnostic implications in gastric cancer. A total of 180 tissue and plasma samples (including 50 cases of gastric cancer, 50 dysplasia, 50 chronic atrophic gastritis with intestinal metaplasia and 30 normal controls) were collected for detecting DNA methylation status of Reprimo and hMLH1 genes using MSP method. Tissue protein expression levels were further tested by immunohistochemical (IHC) staining. The positive rate of DNA methylation rate was, in ascending sequence, gastritis tissue, dysplasia tissue and gastric carcinoma tissue. All those tissues had significantly elevated DNA methylation level compared to normal group (P < 0.05). Expression level of Reprimo and hMLH1 proteins were, however, decreased in pathological tissues compared to normal ones (P < 0.05). A significantly negative relationship existed between protein level and promoter region methylation level. The DNA methylation occurred in promoter regions of both Reprimo and hMLH1 genes depressed the protein expression, and may participate in the occurrence and progression and gastric cancer. The combined assay of serum Reprimo and hMLH1 DNA methylation levels thus had critical importance in the early diagnosis and gastric cancer.
Keywords: DNA methylation, reprimo gene, hMLH1 gene, gastric cancer, early tumor diagnosis
Introduction
As one common malignant tumor in digestive tract, the pathogenesis of gastric cancer involves a continuously pathogenic process from chronic gastritis towards mucosal atrophy, intestinal metaplasia, dysplasia and eventually tumors [1]. The underlying genetic alternation thus owns critical values for the early diagnosis and exploration of pathogenesis.
Besides genetic mutation, epigenetic alternation is also correlated with gastric cancer. DNA methylation is the most common epigenetic mechanism, involving the addition of methyl group to 5’-carbon of cytosine under the direction of DNA transferase [2]. DNA methylation may affect the expression of tumor suppressor gene and mismatch repair gene, both of which participate in processes including cell cycle modulation, DNA repair, oncogenic metabolites, intracellular interaction, apoptosis and angiogenesis [3]. Specifically, the methylation of CpG island in promoter region of tumor suppressor gene and mismatch repair gene may impede the binding between promoter and transcriptional factors, thus promoting the occurrence and progression of tumors [4]. Studies have revealed the existence of CpG island hyper-methylation in almost all human tumor tissues, especially during the early stage [5]. Such tumor-specific DNA methylation can be detected in blood samples, suggesting its potency as one non-invasive tumor marker [6,7].
Located on human chromosome 2, Reprimo gene is one of genes that regulating normal cell growth. The deactivation of Reprimo gene occurs at the early stage of multiple tumors, mainly due to DNA methylation [8]. In one study of gastric cancer patients’ plasma and tissue samples, the positive rate of Reprimo gene methylation was 95.3% and 97.7% m respectively, in sharp contrast to 9.7% in normal people [9]. As the major component of DNA mismatch repair system, hMLH1 gene is also correlated with gastric cancer pathogenesis. In most tumor-adjacent tissues from gastric cancer patients, methylation can be found in gene promoter region, suggesting that hMLH1 gene methylation may be one important molecular event during the early progression of gastric cancer [10,11].
Currently no study has been performed regarding the abnormal expression of both tumor suppressor gene Reprimo and mismatch repair gene hMLH1 in various stages during the occurrence of gastric cancer. This study thus for the first time tested the expression of Reprimo and hMLH1 gene in both peripheral blood and tissue samples from gastric cancer patients, in an attempt to analyze the correlation between those two genes and different stages of gastric cancer, thus providing novel molecular marker for the early diagnosis of gastric carcinoma.
Materials and methods
Study objects
A total of 180 individuals were recruited in the Lin-Yi Cancer Hospital from October 2012 to October 2014. Tissue samples were collected from gastroscopy. Inclusive criteria were: (1) Confirmed pathological diagnosis as chronic atrophy gastritis complicated with intestinal metaplasia, dysplasia or gastric carcinoma; (2) All participants have signed written consents with fully understanding of this study. Exclusive criteria: (1) Complicated with other digestive diseases; (2) With other systemic tumors and/or metastasis to the gastric; (3) Have undergone surgical or chemotherapy; (4) Complicated with other severe systematic disease or organ failure. A total of 50 chronic atrophy gastritis, 50 cases of dysplasia, 50 cases of gastric carcinoma and 30 normal gastric mucosal tissues. The general information of all people involved is listed in Table 1. No significant difference regarding age or sex distribution has been discovered between control and diseases animals.
Table 1.
General information of all patients
| General info | Intestinal metaplasia | Dysplasia | Gastric cancer | Normal | |
|---|---|---|---|---|---|
| Sex | Male | 30 | 27 | 331 | 17 |
| Female | 20 | 23 | 17 | 12 | |
| Age | Average | 55.4±11.2 | 59.4±13.9 | 63.9±16.5 | 58.4±14.7 |
| Range | 32~69 | 37~73 | 42~79 | 35~78 |
DNA extraction and bisulfite modification
Fasting blood samples (5 mL) were collected from veins of all participants and were stored in EDTA-containing tubes. After centrifugation at 1 500 g for 10 min, plasma was saved to extract genomic DNA using DNA purification kit (Zymo Research, US). The purity of DNA was determined by agarose gel electrophoresis and UV spectrometer. After modification using bisulfite, DNA samples were kept at -20°C for further use.
MSP assay of plasma sample
Specific methylation (M) and un-methylation (U) primers were designed based on specific sequences of CpG islands of hMLH1 and Reprimo gene promoter regions [9,10]. Sequences were: Reprimo-M-Forward, 5’-GCGAG TGAGC GTTTA GTTC-3’, Reverse, 5’-TACCT AAAAC CGAA TTCAT CG-3’; Peprimo-U-Forward, 5’-TTGTG AGTGA GTGTT TAGTT TG-3’; Reverse, 5’-TAATT ACCTA AAACCA AATTCA TC-3; hMLH-1-M-Forward, 5’-TTGGT TGGAT ATTTT GTATT TTTTG A-3’; Reverse, 5’-CTCCCT AAAAC AACTA CTACCC; hMLH1-U forward, 5’-ATTGG TTGGA TATTT CGTA TTTTT C-3’; hMLH1-U reverse, 5’-CCTAA AACGA CTCTA CCCGC T-3’. PCR was carried under the following condition: 95°C for 5 min, followed by 30 cycles each containing 95°C denature for 30 sec, 62°C annealing for 60 sec and 72°C elongation for 30 sec. PCR products with methylation activity was employed as a positive control using healthy adjacent tissues without modification. Another cohort of positive control will be used for non-methylation assay. A gel imaging equipment (BIO-RAD, US) was used to capture pictures. The methylation status (Band M: methylated, DNA Bank U: un-methylated DNA).
Protein expression level
All tissue samples were immediately fixed in 10% neutral buffered formaldehyde (NBP). After embedding in paraffin, 4 μm-thickness consecutive slices were made. Protein expression levels of Reprimo and hMLH1 gene. A total of 12 high magnification (× 400) fields were randomly selected from the family. The number of IHC-positive cells was counted to define positive slices (i.e. those with less than 5% of NBF were regarded as negative slices and vice versa).
Statistical analysis
SPSS 16.0 software package was used to process all collected data, of which measurement data was presented as mean ± standard deviation (SD). Student t-test was used to compare means between two groups. Multiple group comparison was done by analysis of variance (ANOVA). Enumeration data, on the other hand, was tested by chi-square approach. A statistical significance was defined when P < 0.05.
Results
MSP amplification of plasma sample
Methylation assay of Reprimo gene showed that, 14 out of 50 samples with chronic atrophy gastritis with intestinal metaplasia showed gene methylation (positive rate = 26%). In all dysplasia samples, 28 cases had methylation (positive rate = 56%). In gastric cancer patients, this number was even as higher as 62% (31 out of 50). All 30 normal controlled people, however, did not have detectable methylation of Reprimo gene (Figure 1). Compared to those in control people, the methylation rates in intestinal metaplasia, dysplasia and gastric carcinoma patients were significantly elevated (P < 0.05). Among those three groups, gastric cancer and dysplasia had highest level of methylation, as compared to intestinal metaplasia and control people (P < 0.05).
Figure 1.
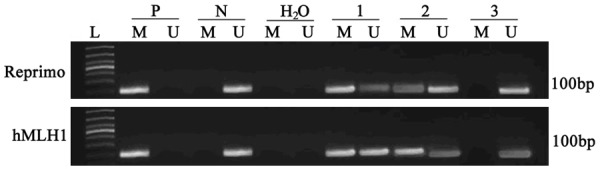
Methylation level of Reprimo and hMLH1 genes. L, DNA ladder; M, methylation; U, un-methylation; P, positive control; N, negative control; H2O, water control. Lane 1, gastric cancer; Lane 2, dysplasia; Lane 3, intestinal metaplasia.
Methylation assay for hMLH1 gene showed that, 10 out of 50 in gastritis with intestinal metaplasia, 22 out of 50 of dysplasia patients, and 24 out of 50 gastric cancer patients had DNA methylation, making the positive rates at 20%, 44% and 48%, respectively. In contrast, healthy people had the methylation rate only 3.3% (1 out of 30 people, Figure 1). These data suggested that dysplasia and gastric cancer were accompanied with significantly higher hMLH1 gene methylation level compared to that in intestinal metaplasia patients (P < 0.05), who further had higher methylation level than that in control group (P < 0.05).
Protein expression level of Reprimo and hMLH1 genes
We further used IHC staining to visualize the protein expression level of Reprimo and hMLH1 genes in tissue samples. As shown in Figure 2, the positive expression rate of Reprimo and hMLH1 genes in intestinal metaplasia, dysplasia, gastric cancer and normal group were 20% (10/50) and 22% (11/50), 44% (22/50) and 40% (20/50), 48% (24/50) and 46% (23/50), and 3.3% (1/30) and 0%, respectively. Therefore, the positive expression rate of both genes were positively correlated with methylation rate in plasma (r = 0.99, P < 0.05).
Figure 2.
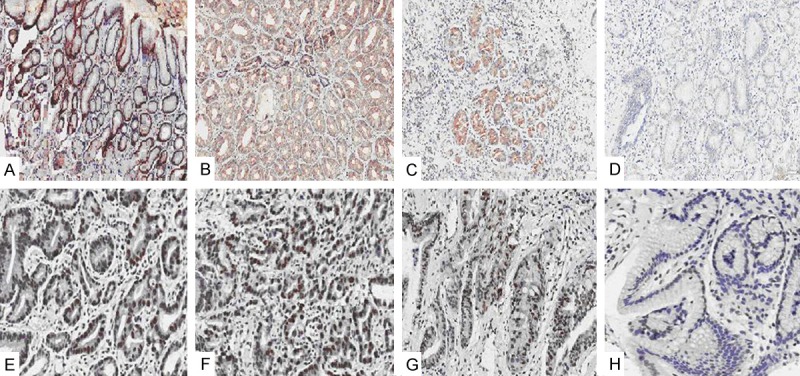
Reprimo and hMLH1 protein expression (× 100). A-D. Reprimo; E-H. hMLH1 protein. A and E. Intestinal metaplasia; B and F. Dysplasia; C and G. Gastric cancer; D and H. Control.
Combined assay of methylation of two genes
As shown in Table 2, the positive rates of combined methylation assay in the plasma of intestinal metaplasia, dysplasia, gastric cancer and normal group were 34%, 76%, 84% and 3.3%, respectively. The combined negative rates in tissue samples were 36%, 78%, 82% and 0%. Therefore, the combined assay of two gene markers can improve the specificity and sensitivity of diagnosis.
Table 2.
Reprimo and hMLH1 gene methylation and protein expression
| Group | N | Reprimo methylation rate | Reprimo protein expression | hMLH1 methylation rate | hMLH1 protein expression | Combined rate of methylation | Combined expression rate |
|---|---|---|---|---|---|---|---|
| Intestinal metaplasia | 50 | 14 (28%) | 16 (32%) | 10 (20%) | 11 (22%) | 17 (34%) | 18 (36%) |
| Dysplasia | 50 | 28 (56%) | 30 (60%) | 22 (44%) | 20 (40%) | 38 (76%) | 39 (78%) |
| Gastric cancer | 50 | 31 (62%) | 33 (66%) | 24 (48%) | 23 (46%) | 42 (84%) | 41 (82%) |
| Normal | 30 | 0 (0%) | 0 (0%) | 1 (3.3%) | 0 (0%) | 1 (3.3%) | 0 (0%) |
Discussion
The 5-year survival rate of early-stage gastric cancer can reach up to 95%. However, once the tumor has invaded into muscular or serous layer, the survival rate has dropped to less than 20% [12]. Therefore, the early intervention of gastric cancer can total cure the disease. However, due to the lack of unique symptom, the diagnostic rate of gastric cancer at its early stage was less than 10%. Most patients were already at the advanced stage at the time of first diagnosis, leading to lower survival rate [13]. The effective early diagnostic approach is thus of significant values and has drawn lots of research interests [14-16]. The precancerous lesion of gastric carcinoma includes atrophy gastritis, intestinal metaplasia and intraepithelial neoplasia, all of which are risk factors for gastric cancer and can be used to improve the diagnostic rate at early stage for improving the prognosis. Recently, epigenetic studies including DNA methylation has provided more insights regarding the early diagnosis and prognostic prediction of tumors [17]. The methylation of both tumor suppressor gene and mismatch repair gene at their promoter region is closely correlated with tumor occurrence [18].
This study investigated the methylation status of tumor suppressor gene Reprimo and mismatch repair gene hMLH1 at their promoter regions, for further exploration of the potency as biological indexes for screening and early diagnosis of gastric cancer. As one newly discovered tumor suppressor gene, Reprimo codes a highly glycosylated protein in the cytoplasm for mediating p53-directed cell cycle. It can arrest abnormal cells at G2 phase, inhibit the Cdc2 activity and nuclear translocation of cyclin B1, and prevent hyper-proliferation of cells [19]. Studies have found that the deactivation of Reprimo gene occurred at the early stage of dozens of malignant tumors, mainly as a result of DNA methylation. For example, Reprimo gene methylation has been reported in most of gastric cancer, lymphoma and colorectal carcinoma, in contrast to minimal level in normal tissues [20,21]. The abnormal methylation of Reprimo gene is thus a hallmark at the early stage of certain cancers. Recently, the correlation between mismatch repair system and gastric cancer occurrence has also been investigated [22,23]. The major function of mismatch repair gene is to correct the mismatching of base pairs occurred during DNA replication, thus maintaining genome stability [24]. As one DNA mismatch repair gene, hMLH1 can affect the endogenous repair function of cells and has been suggested to play an important role in the occurrence of gastric cancer [25]. The microsatellite instability (MSI) is related with mismatch repair gene, and is one feature of gastric cancer [26]. Many studies have showing the deactivation of hMLH1 gene due to its hyper-methylation under MSI environment. The absence of MSI, however, helps to depress gene methylation level for facilitating gene expression, suggesting the possible involvement of hyper-methylation of hMLH1 gene promoter in gastric cancer [27].
To investigate the correlation between methylation of tumor suppressor gene Reprimo and mismatch repair gene hMLH1 and the occurrence of gastric cancer, we recruited patients including intestinal metaplasia, dysplasia and gastric cancer at different stages. On those patients, both MSP and IHC methods were used to detect the promoter methylation level and protein expression level in plasma and tissue samples. Our results showed elevated gene methylation level in intestinal metaplasia, dysplasia and gastric cancer patients, with an ascending order. Therefore, during the transformation from intestinal metaplasia towards neoplasia, the significant methylation of promoter region in Reprimo and hMLH1 gene might be an early event during gastric cancer onset.
IHC staining showed an elevated expression rate of Reprimo and hMLH1 proteins in intestinal metaplasia, dysplasia and cancer group (in ascending order). Statistical analysis revealed a positive correlation between methylation rate and tissue protein expression rate. As tumor cell metabolites may enter into the plasma during the process of necrosis and apoptosis, the assay of plasma gene methylation level thus may provide a novel non-invasive method for tumor diagnosis with small sample size and simple procedures.
The abnormal methylation of single gene cannot determine the tumor progression. We thus used a combined assay scenario including Reprimo and hMLH1 genes’ methylation status. This method has been shown to own significant value for early diagnosis of gastric cancer. As in either of dysplasia and tumor tissue samples, both genes have been found to have methylation at various levels. The combined assay for methylation owned the positive rate as high as 76% and 84%. Therefore, the combined assay of tissue and plasma levels of Reprimo and hMLH1 gene, can work as one clinical index for the early diagnosis of gastric cancer.
Disclosure of conflict of interest
None.
References
- 1.Dirnu R, Secureanu FA, Neamţu C, Totolici BD, Pop OT, Mitruţ P, Mălăescu DG, Mogoantă L. Chronic gastritis with intestinal metaplasia: clinico-statistical, histological and immunohistochemical study. Rom J Morphol Embryol. 2012;53:293–7. [PubMed] [Google Scholar]
- 2.Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, Amarantidis K, Chelis L, Trypsianis G, Chatzaki E, Lianidou ES, Kakolyris S. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res. 2015;778:46–51. doi: 10.1016/j.mrfmmm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Kulis M, Queirós AC, Beekman R, Martín-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161–74. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Gilsbach R, Preissl S, Grüning BA, Schnick T, Burger L, Benes V, Würch A, Bönisch U, Günther S, Backofen R, Fleischmann BK, Schübeler D, Hein L. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun. 2014;5:5288. doi: 10.1038/ncomms6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto E, Suzuki H, Takamaru H, Yamamoto H, Toyota M, Shinomura Y. Role of DNA methylation in the development of diffuse-type gastrie cancer. Digestion. 2011;83:241–249. doi: 10.1159/000320453. [DOI] [PubMed] [Google Scholar]
- 6.Ye T, Chen Y, Fang J. DNA methylation biomarkers in serum for gastric cancer screening. Mini Rev Med Chem. 2010;10:1034–8. doi: 10.2174/1389557511009011034. [DOI] [PubMed] [Google Scholar]
- 7.Ye T, Chen Y, Fang J. DNA methylation based biomarkers in no-invasive cancer screening. Mini Rev Med Chem. 2010;10:1034–1038. doi: 10.2174/1389557511009011034. [DOI] [PubMed] [Google Scholar]
- 8.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in no-invasive cancer screening. Curr Mol Med. 2010;10:123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernal C, Aguayo F, Villarroel C, Vargas M, Díaz I, Ossandon FJ, Santibáñez E, Palma M, Aravena E, Barrientos C, Corvalan AH. Reprimo as a potential biomarker for early detection in gastric cancer. Clin Cancer Res. 2008;14:6264–9. doi: 10.1158/1078-0432.CCR-07-4522. [DOI] [PubMed] [Google Scholar]
- 10.Mir MR, Shabir N, Wani KA, Shaff S, Hussain I, Banday MA, Chikan NA, Bilal S, Aejaz S. Association between p16, hMLH1 and Ecadherin promoter hypermethylation and intake of local hot salted tea and sun-dried foods in Kashmiris with gastric tumors. Asian Pac J Cancer Prev. 2012;13:181–6. doi: 10.7314/apjcp.2012.13.1.181. [DOI] [PubMed] [Google Scholar]
- 11.Xiao XQ, Gong WD, Wang SZ, Zhang ZD, Rui XP, Wu GZ, Ren F. Polymorphisms of mismatch repair gene hMLH1 and hMSH2 and risk of gastric cancer in a Chinese population. Oncol Lett. 2012;3:591–598. doi: 10.3892/ol.2011.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao K, Doyama H, Gotoda T, Ishikawa H, Nagahama T, Yokoi C, Oda I, Machida H, Uchita K, Tabuchi M. Diagnostic performance and limitations of magnifying narrow-band imaging in screening endoscopy of early gastric cancer: a prospective multicenter feasibility study. Gastric Cancer. 2014;17:669–79. doi: 10.1007/s10120-013-0332-0. [DOI] [PubMed] [Google Scholar]
- 13.Badgwell B, Roy-Chowdhuri S, Chiang YJ, Matamoros A, Blum M, Fournier K, Mansfield P, Ajani J. Long-term survival in patients with metastatic gastric and gastroesophageal cancer treated with surgery. J Surg Oncol. 2015;111:875–81. doi: 10.1002/jso.23907. [DOI] [PubMed] [Google Scholar]
- 14.Jin Z, Jiang W, Wang L. Biomarkers for gastric cancer: Progression in early diagnosis and prognosis (Review) Oncol Lett. 2015;9:1502–1508. doi: 10.3892/ol.2015.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang GD, Konda VJ. Early diagnosis and management of esophageal and gastric cancer. Minerva Gastroenterol Dietol. 2013;59:357–76. [PubMed] [Google Scholar]
- 16.Saragoni L, Morgagni P, Gardini A, Marfisi C, Vittimberga G, Garcea D, Scarpi E. Early gastric cancer: diagnosis, staging, and clinical impact. Evaluation of 530 patients. New elements for an updated definition and classification. Gastric Cancer. 2013;16:549–54. doi: 10.1007/s10120-013-0233-2. [DOI] [PubMed] [Google Scholar]
- 17.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzese DM, Hoon DS. Emerging technologies for studying DNA methylation for the molecular diagnosis of cancer. Expert Rev Mol Diagn. 2015;15:647–64. doi: 10.1586/14737159.2015.1027194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saavedra K, Valbuena J, Olivares W, Marchant MJ, Rodríguez A, Torres-Estay V, Carrasco-Avino G, Guzmán L, Aguayo F, Roa JC, Corvalán AH. Loss of Expression of Reprimo, a p53-induced Cell Cycle Arrest Gene, Correlates with Invasive Stage of Tumor Progression and p73 Expression in Gastric Cancer. PLoS One. 2015;10:e0125834. doi: 10.1371/journal.pone.0125834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooki A, Yamashita K, Yamaguchi K, Mondal A, Nishimiya H, Watanabe M. DNA damage-inducible gene, reprimo functions as a tumor suppressor and is suppressed by promoter methylation in gastric cancer. Mol Cancer Res. 2013;11:1362–74. doi: 10.1158/1541-7786.MCR-13-0091. [DOI] [PubMed] [Google Scholar]
- 21.Xu M, Knox AJ, Michaelis KA, Kiseljak-Vassiliades K, Kleinschmidt-DeMasters BK, Lillehei KO, Wierman ME. Reprimo (RPRM) is a novel tumor suppressor in pituitary tumors and regulates survival, proliferation, and tumorigenicity. Endocrinology. 2012;153:2963–73. doi: 10.1210/en.2011-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HJ, Jang YJ, Lee EJ, Kim JH, Park SS, Park SH, Kim CS, Mok YJ. The significance of mismatch repair genes in gastric cancer. J Cancer Res Ther. 2013;9:80–3. doi: 10.4103/0973-1482.110382. [DOI] [PubMed] [Google Scholar]
- 23.van Grieken NC, Aoyama T, Chambers PA, Bottomley D, Ward LC, Inam I, Buffart TE, Das K, Lim T, Pang B, Zhang SL, Tan IB, Carvalho B, Heideman DA, Miyagi Y, Kameda Y, Arai T, Meijer GA, Tsuburaya A, Tan P, Yoshikawa T, Grabsch HI. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the East and the West: results from a large international multicentre study. Br J Cancer. 2013;108:1495–501. doi: 10.1038/bjc.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillotin D, Martin SA. Exploiting DNA mismatch repair deficiency as a therapeutic strategy. Exp Cell Res. 2014;329:110–5. doi: 10.1016/j.yexcr.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Wang D, Song L, Li S, Ding J, Chen S, Li J, Ma G, Zhang X. A new familial gastric cancerrelated gene polymorphism: T1151A in the mismatch repair gene hMLH1. Mol Biol Rep. 2011;38:3181–7. doi: 10.1007/s11033-010-9989-1. [DOI] [PubMed] [Google Scholar]
- 26.Ling ZQ, Tanaka A, Li P, Nakayama T, Fujiyama Y, Hattori T, Sugihara H. Microsatellite instability with promoter methylation and silencing of hMLH1 can regionally occur during progression of gastric carcinoma. Cancer Lett. 2010;297:244–51. doi: 10.1016/j.canlet.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Vogelsang M, Komel R. Non-truncating hMLH1 variants identified in Slovenian gastric cancer patients are not associated with Lynch Syndrome: a functional analysis report. Fam Cancer. 2011;10:255–63. doi: 10.1007/s10689-010-9409-7. [DOI] [PubMed] [Google Scholar]


