Abstract
In southern China, glucose-6-phosphate dehydrogenase (G6PD) deficiency is a significant health problem, and the incidence ranged from 0.5 to 4.08% in different Chinese population. The aims of this study are to investigate the molecular epidemiological characteristic of the G6PD gene among Chinese Hakka in southern Jiangxi province. 2331 unrelated subjects were screened for G6PD deficiency by a fluorescent test. DNA from deficient individuals was analyzed by a gene chip analysis for thirteen common Chinese G6PD mutations. In total, 3.60% (82/2331; 95% CI 2.77-4.27) of the sample were found to be G6PD-deficient. Eight mutations were found from 80 samples. However, mutation(s) for the two remaining samples were unknown. The most common mutations were G6PD Canton (1376 G>T) and G6PD Kaiping (1388 G>A), and the following mutations were 1311 polymorphism (1311 C>T), G6PD Gaohe (95 A>G), G6PD Chinese-5 (1024 C>T), G6PD Maewo (1360 C>T), Shunde (592 C>T), G6PD Viangchan (871 G>A) and Chinese-3 (493 A>G). This is the first report of G6PD deficiency among Chinese Hakka population in Jiangxi province.
Keywords: G6PD deficiency, Chinese Hakka, gene chip, gene mutation
Introduction
G6PD is a key enzyme in the pentose phosphate pathway that plays an important role in the body’s oxidative defenses by governing the formation of nicotinamide adenine dinucleotide phosphate-oxidase (NADPH) from nicotinamide adenine dinucleotide phosphate (NADP) [1]. Many variants of G6PD resulted from point mutations in the G6PD gene, cause deficiency of the enzyme. The situation is more frequently found in the male patients for the X-linked condition. Affected individuals are usually asymptomatic and not aware of their deficiency through their life [2]. However, G6PD deficiency may cause a large spectrum of diseases including neonatal jaundice, acute hemolysis and severe chronic non-spherocytic hemolytic anemia [3]. Worldwide, estimated 400 million people carry a deficient variant of the G6PD gene, with disproportionately higher prevalence observed in tropical Africa, the Middle East, tropical and sub-tropical Asia including Southern China [4]. As previously reported, this deficiency has a prevalence of 0.5-10% in the different population of China [5,6].
Ganzhou (Kanchow) is a large city covering the southern part of Jiangxi Province, with an area of 39,400 km2 and a population 8.96 million. It borders Fujian Province to the east, Guangdong Province to the south, and Hunan Province to the west (Figure 1). More than 95% people lived in Ganzhou are Hakka. Hakka is a intriguing Han Chinese populations that mainly inhabit southern China, which is characteristic of their unique culture and is distinct from the traditional culture of southern Hans (SHs), but show lots of similarities to that of northern Hans (NHs), including some features in dialects, life styles, customs, and habits [7,8]. Many studies of genetic markers, such as autosomal microsatellites/SNPs, and Y chromosome SNP, preferred the northern origins of Hakka [8,9]. Currently, few data are available on the prevalence and molecular characterization of G6PD deficiency for the Hakka population of the Ganzhou region.
Figure 1.
Geographic location of the Ganzhou region, Jiangxi province, P.R. China.
Here, we reported a population screening of 2331 Chinese Hakka in Southern Jiangxi province (Ganzhou), which allows us to document the prevalence and molecular characterization of G6PD deficiency in this region.
Materials and methods
Population samples
We obtained data from health examination surveys. The study population included 2331 unrelated subjects for G6PD deficiency between August 2011 and November 2011 (Male: 642 Female: 1689). The ages of these subjects ranged from 18 to 70-year-old and about 95% were Hakka aborigines, i.e. Information sheets with nationality, sex, age, dialect, aborigines or not and written consent forms were available in Chinese to ensure comprehensive understanding of the study objectives, and informed consent was signed or thumb printed by the participants or their guardians. After obtaining informed consent, peripheral blood sample was collected into a tube with EDTA-K2 by the medical laboratory, First Affiliated Hospital of Gannan Medical University, and stored at 4°C.
Biochemical screeing for G6PD deficiency
As our previous study [10], the G6PD enzyme activity of all subjects was determined by the G6PD mensuration reagent kit (Kerfen, Guangzhou, China) on HCP-7600-020 automatic biochemistry analyzer (Hitachi, Japan) according to the manufacturer’s specification. Quality control was performed using G6PDH controls each day. In normal RBCs, the G6PD activity ranges from 1,300 to 3,600 U/I. For this assay, the cut-off for G6PD deficiency was set at 1,300 U/I according to the manufacturer’s specification. To prevent a reduction of the G6PD activity, all samples were assayed within 72 h [10,11].
Molecular diagnosis of G6PD deficiency
Genomic DNA of subjects with reduction of the G6PD activity was extracted from peripheral blood leukocytes by DNA blood mini kit (QIAGEN China Shanghai Co., Ltd). The DNA concentration was determined by UV spectrophotometer (UNICO Shanghai Instruments Co., Ltd) at the wavelength of 260 nm.
The 13 known G6PD gene mutations most commonly seen in the Chinese population including G6PD Gaohe (95 A>G), 392 (c.482G>T), Chinese-3(493 A>G), Shunde (592 C>T), G6PD Viangchan (871 G>A), G6PD Fushan (1004 C>T), G6PD Chinese-5 (1024 C>T), G6PD Maewo (1360 C>T), G6PD Canton (1376 G>T), G6PD Keelung (1387 C>T), G6PD Kaiping (1388 G>A) and two polymorphism (1311 C>T, 1381 G>A) were analyzed by commercial detection kit. The G6PD gene chip were designed and made by Chaozhou Hybribio Limited Corporation. All probes were immobilized on a nylon membrane. Their localization in the membrane is shown in Figure 2A. The detailed information of the primers and probes are described in Tables 1 and 2. Some of the primers of the kit were biotinylated (Table 1).
Figure 2.
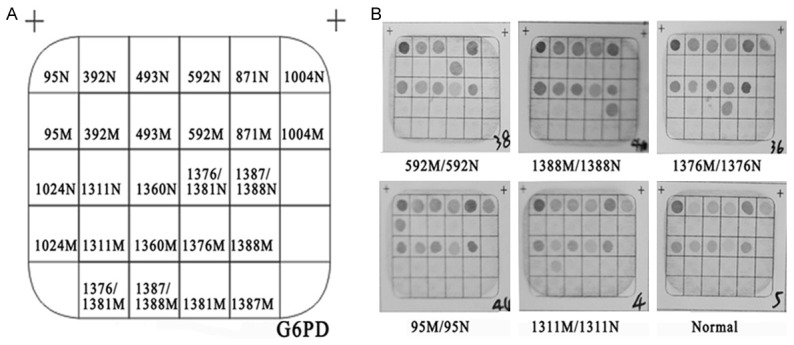
The hybridization results of G6PD gene chip. A: The location of the probes dotted in the gene chip used for the reverse dot blot assay. Locations of wild-type and mutant probes are denoted N and M, respectively. B: The results of G6PD gene in the gene chip.
Table 1.
Detailed information about the primers in the reaction system
| Name | Sequence (5’→3’) | Location GenBank: X55448.1 | Products |
|---|---|---|---|
| Exon2-F | ATTTGGGCAATCAGGTGTCA | 3226-3245 | 348 bp |
| Exon2-R | AACTTAGCAGAGCCTGTGGG | 3554-3573 | 348 bp |
| Exon5-F | AAGCTGGAGGACTTCTTTGCC | 14139-14159 | 273 bp |
| Exon5-R | ACACGCTCATAGAGTGGTGGG | 14391-14411 | 273 bp |
| Exon6-F | CTGGGAGGGCGTCTGAATGA | 14920-14939 | 413 bp |
| Exon6-R | GGGCAAGGTGGAGGAACTGA | 15313-15332 | 413 bp |
| Exon9-F | AACACCCAAGGAGCCCATTC | 16334-16353 | 470 bp |
| Exon9-R | CACTGCTGGTGGAAGATGTCG | 16783-16803 | 470 bp |
| Exon11-12-F | TGGCATCAGCAAGACACTCTCTC | 17010-17032 | 318 bp |
| Exon11-12-R | CCCTTTCCTCACCTGCCATAAA | 17306-17327 | 318 bp |
Table 2.
Detailed information about the probes in the G6PD gene chip
| Probe | Sequence (5’→3’) |
|---|---|
| 95N-P | CGGATACACACATATTCATC |
| 95M-P | GGATACACGCATATTCATC |
| 392N-P | CCACCTGGGGTCACAG |
| 392M-P | TCCACCTGGTGTCACAG |
| 493N-P | CTGGAACCGCATCATC |
| 493M-P | CTGGGACCGCATCATC |
| 592N-P | CAGATCTACCGCATCGA |
| 592M-P | ATCTACTGCATCGACCA |
| 871N-P | CAGGTCAAGGTGTTGAAA |
| 871M-P | TCAGGTCAAGATGTTGAAA |
| 1004N-P | TCCACCGCCACTTTTG |
| 1004M-P | CACCACCGTCACTTTTG |
| 1024N-P | CCGTCGTCCTCTATGTG |
| 1024M-P | GCCGTCGTCTTCTATGT |
| 1311N-P | CTGACGCCTACGAGCG |
| 1311M-P | CCTGACGCCTATGAGC |
| 1360N-P | CACTTCGTGCGCAGGT |
| 1360M-P | ACTTCGTGTGCAGGTGA |
| 1376/1381N-P | GCTCCGTGAGGCCTGG |
| 1376M-P | CGACGAGCTCCTTGAG |
| 1381M-P | GAGCTCCGTGAGACCTG |
| 1376/1381M-P | AGCTCCTTGAGACCTGG |
| 1387/1388N-P | CCTGGCGTATTTTCACC |
| 1387M-P | CCTGGTGTATTTTCACC |
| 1388M-P | CCTGGCATATTTTCACC |
| 1387/1388M-P | CCTGGTATATTTTCACCC |
The assay was performed according to the manufacturer’s protocol. Briefly, reaction system 1 was performed with a reaction volume of 20 μl containing 100 ng of DNA template, 3.0 μl 10 × buffer, 3.5 mmol/L MgCl2, 0.2 mmol/L of each primer (Exon2-F, Exon2-R, Exon5-F, Exon5-R, Exon6-F, Exon6-R), 0.2 mmol/L of each dNTP, 2.5 units of DNA Taq polymerase in a MJ Mini Personal Thermal Cycler (Bio-RAD Company) with an initial 9 min denaturation at 95°C, 40 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension at 72°C for 15 min. PCR Reaction system 2 was performed in 20 μl reaction mixture containing 100 ng DNA template, 3.0 μl 10 × buffer, 3.5 mmol/L MgCl2, 0.2 mmol/L of each primer (Exon9-F, Exon9-R, Exon11-12-F, Exon11-12-R), 0.2 mmol/L of each dNTP, 2.5 units DNA Taq polymerase. PCR reaction condition was: preheating at 95°C, 40 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension at 72°C for 15 min. The amplicons of the two reaction system (two tubes) were subsequently denatured and subjected to hybridization.
Hybridization reactions were performed using HMM2I (Chaozhou Hybribio Limited Corporation, Chaozhou, China) as previous report [12]. The hybridization process was performed according to the manufacturer’s protocol. The assay utilized a Hybribio’s proprietary Flow-through Hybridization Technology (US Patents 5,741,647 and 6,020,187) by actively directing the targeting molecules toward the immobilized probes within the membrane fibers, with the complementary molecules being retained by the formation of duplexes [12]. After a stringent wash, the hybrids were detected by the addition of the streptavidin-horseradish peroxidase conjugate, which binds to the biotinylated PCR products and a substrate (nitro-blue tetrazolium-5-bromo-4-chloro-3-indolylphosphate) to generate a blue-purple precipitate at the probe dot. The results were interpreted by direct visualization [12].
Statistical analysis
Statistical analysis was conducted with SPSS 16.0 statistical software. The prevalence of different mutation alleles was calculated from the standard Hardy-Weinberg formula. Data from the Ganzhou region and previous studies were analyzed by Pearson χ2 test with Bonferroni adjustment for multiple testing. P<0.05 was considered statistically different.
Results
Phenotypic screening for G6PD deficiency in healthy individuals showed that the prevalence of the G6PD deficient phenotype in the Hakka population is relatively lower at 3.60% (82/2331; 95% CI 2.77-4.27). Of 642 males and 1689 females tested, 4.83% (31/642; 95% CI 3.17-6.49) and 3.02% (51/1689; 95% CI 2.20-3.84) were found to have G6PD deficiency, respectively. There was significant difference in the incidence between males and females (P=0.034).
To identify the molecular cause of G6PD deficiency in this population, the G6PD gene was screened for mutations by a gene chip. The hybridization results of G6PD gene chip were shown in Figure 2, which included heterozygote, hemizygote and compound homozygote. All normal control dots on the chip would be colored except the homozygous or hemizygous sample with G6PD gene mutations after the hybridization reactions (Figure 2B). Twenty-one kinds of genotypes including 8 kinds of heterozygous genotypes, 7 kinds of hemizygous genotypes, 6 kinds of homozygous genotypes were identified. The results were listed in Table 3. Eight mutation sites were detected from 82 G6PD-deficient cases. The most common mutations were G6PD Canton (1376 G>T) and G6PD Kaiping (1388 G>A), and the following mutations were 1311 polymorphism (1311 C>T), G6PD Gaohe (95 A>G), G6PD Chinese-5 (1024 C>T), G6PD Maewo (1360 C>T), Shunde (592 C>T), G6PD Viangchan (871 G>A) and Chinese-3(493 A>G).
Table 3.
Twenty-one different genotypes and their contributions in Ganzhou
| Heterozygote | n (%) | Hemizygote | n (%) | Homozygote | n (%) |
|---|---|---|---|---|---|
| 95 A>G | 4 | 95 A>G | 2 | 871 G>A/1388 G>A | 1 |
| 493 A>G | 1 | 592 C>T | 1 | 1311 C>T/1376 G>T | 1 |
| 592 C>T | 1 | 1024 C>T | 3 | 1311 C>T/1360 C>T | 2 |
| 871 G>A | 1 | 1311 C>T | 1 | 1311 C>T/1388 G>A | 2 |
| 1024 C>T | 1 | 1360 C>T | 2 | 1376 G>T /1388 G>A | 5 |
| 1311 C>T | 3 | 1376 G>T | 12 | 1376 G>T/1376 G>T | 1 |
| 1376 G>T | 14 | 1388 G>A | 10 | ||
| 1388 G>A | 12 | ||||
| Unknown | 2 | ||||
| 39 | 31 | 12 |
Discussion
The prevalence of G6PD deficiency in Southeast China is highly variable [5,13,14]. Our present study showed that the overall prevalence of G6PD deficiency was 3.60% (95% CI 2.77-4.27) from 2332 subjects of Ganzhou region, 4.83% (95% CI 3.17-6.49) from males and 3.02% (95% CI 2.20-3.84) from females, respectively. Because the technical reason for the missed female heterozygotes may be due to insufficient sensitivity of the assay, the detection rate of G6PD deficiency in male was higher than in female.
The prevalence of G6PD deficiency were lower than that found in Guangxi Zhuang autonomous region (7.43%) [14], middle region (Guangzhou, 4.2%) [14] and Southern region (Zhanjiang, 4.6%) of Guangdong province [15], but higher than that in Fujian province (1.03%) [16], Shanghai (0.78%) [17] and Nanchang region of Northern Jiangxi (1.02%) [18]. Our result was similar to that finding in Hainan province (3.73%) [16]. The distribution of G6PD deficiency frequency was similar to the situation found with β-thalassemia in this region, further confirming the hypothesis that both human G6PD and the β-globin gene was involved in malaria-protective selection factors. With a current annual birth rate of about 113,225 divided equally into males and females, we can estimate that the number of affected newborns in Ganzhou at risk of G6PD-deficiency because of hemizygosity in males or homozygosity in females is 2734 (95% CI, 1795-3674) and 483 (95% CI, 311-543), respectively. The data obtained in our study, the high frequency of G6PD deficiency and the elucidation of mutation patterns enhances the basic knowledge of this disorder and also provides a more rational approach to population screening for prevention of and counselling on this disease in this area.
Eight mutation sites were detected from 82 G6PD-deficient cases in our study. G6PD Canton (1376 G>T), G6PD Kaiping (1388 G>A) and G6PD Gaohe (95 A>G) account for 78.05% (64/82) mutations. The finding was similar to Guangdong and Guangxi, Yunan and Taiwan [14,20,21]. The 1311 polymorphism (1311 C>T) was identified from 10.98% (9/82) samples of enzymatic activity deficient. The recently report indicated that there was strong association between haplotype 1311T/93C and rs1050757G located inside the 3’-UTR, which could effect on the downregulation of mRNA and consequently G6PD deficiency either by affecting mRNA stability and translation or mirRNA regulation process [22].
In a conclusion, this study firstly provides information about the molecular epidemiological character of G6PD deficiency in southern area of Jiangxi province. It could be useful for future prevention and control of G6PD deficiency aimed at Chinese Hakka population.
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (contract/grant number 81360265 to Tian-Yu Zhong) and Science and Technology Projects of Jiangxi Educational Committee (contract/grant number GJJ13690 to Rong Hu). Reagent discounts and reagent gifts was from Hybribio Biotechnology Limited Corp. The funders had no role in study design and data analysis, decision to publish, or reparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Stanton RC. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB. Life. 2012;64:362–369. doi: 10.1002/iub.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martini G, Toniolo D, Vulliamy T, Luzzatto L, Dono R, Viglietto G, Paonessa G, D’Urso M, Persico MG. Structural analysis of the X-linked gene encoding human glucose 6-phosphate dehydrogenase. EMBO J. 1986;5:1849–1855. doi: 10.1002/j.1460-2075.1986.tb04436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler E. G6PD deficiency. Blood. 1984;84:3613–3636. [PubMed] [Google Scholar]
- 4.Nkhoma ET, Poole C, Vannappagari V, Hall SA, Beutler E. The global prevalence of glucose-6-phosphate dehydrogenase deficiency: a systematic review and meta-analysis. Blood Cells Mol Dis. 2009;42:267–278. doi: 10.1016/j.bcmd.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Du C. Studies on glucose-6-phosphate dehydrogenase (G6PD) deficiency in China: forty years retrospection and perspective. Zhonghua Xue Ye Xue Za Zhi. 2000;21:174–175. [PubMed] [Google Scholar]
- 6.Beutler E. Glucose-6-phosphate dehydrogenase deficiency: a historical perspective. Blood. 2008;111:16–24. doi: 10.1182/blood-2007-04-077412. [DOI] [PubMed] [Google Scholar]
- 7.Wang WZ, Wang CY, Cheng YT, Xu AL, Zhu CL, Wu SF, Kong QP, Zhang YP. Tracing the origins of Hakka and Chaoshanese by mitochondrial DNA analysis. Am J Phys Anthropol. 2010;141:124–130. doi: 10.1002/ajpa.21124. [DOI] [PubMed] [Google Scholar]
- 8.Hu SP, Luan JA, Li B, Chen JX, Cai KL, Huang LQ, Xu XY. Genetic link between Chaoshan and other Chinese Han populations: Evidence from HLA-A and HLA-B allele frequency distribution. Am J Phys Anthropol. 2007;132:140–150. doi: 10.1002/ajpa.20460. [DOI] [PubMed] [Google Scholar]
- 9.Lin M, Wen YF, Wu JR, Wang Q, Zheng L, Liu GR, Huang Y, Yang H, Lin F, Zhan XF, Lin CP, Yang HT, Weng QQ, Huang FT, Wang Y, Yao MQ, Chen HZ, Wu DH, Zeng JB, Zeng RX, Yang H, Li GC, Lu M, Zhu JJ, Xie LX, Wang JL, Yang LY. Hemoglobinopathy: molecular epidemiological characteristics and health effects on Hakka people in the Meizhou region, southern China. PLoS One. 2013;8:e55024. doi: 10.1371/journal.pone.0055024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan M, Lin M, Yang L, Wu J, Zhan X, Zhao Y, Wen Y, Liu G, Yang L, Cai Y. Glucose-6-phosphate dehydrogenase (G6PD) gene mutations detection by improved high-resolution DNA melting assay. Mol Biol Rep. 2013;40:3073–3082. doi: 10.1007/s11033-012-2381-6. [DOI] [PubMed] [Google Scholar]
- 11.Castro SM, Weber R, Dadalt V, Santos VF, Reclos GJ, Pass KA, Giugliani R. Evaluation of glucose-6-phosphate dehydrogenase stability in blood samples under different collection and storage conditions. Clin Chem. 2005;51:1080–1081. doi: 10.1373/clinchem.2005.048520. [DOI] [PubMed] [Google Scholar]
- 12.Lin M, Zhu JJ, Wang Q, Xie LX, Lu M, Wang JL, Wang CF, Zhong TY, Zheng L, Pan MC, Wu JR, Wen YF, Liu GR, Zhan XF, Lin F, Yang LY. Development and evaluation of a reverse dot blot assay for the simultaneous detection of common alpha and beta thalassemia in Chinese. Blood Cells Mol Dis. 2012;48:86–90. doi: 10.1016/j.bcmd.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Zhou YQ, Xiao QZ, Yan TZ, Xu XM, et al. Development and evaluation of a reverse dot blot assay for the simultaneous detection of six common Chinese G6PD mutations and one polymorphism. Blood Cells Mol Dis. 2008;41:17–21. doi: 10.1016/j.bcmd.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Yan T, Cai R, Mo O, Zhu D, Ouyang H, Huang L, Zhao M, Huang F, Li L, Liang X, Xu X. Incidence and complete molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Guangxi Zhuang autonomous region of southern China: description of four novel mutations. Haematologica. 2006;91:1321–1328. [PubMed] [Google Scholar]
- 15.Pei XY, Cao ZY, Cao XY, et al. The neonatal screening of glucose-6-phosphate dehydrogenase in Zhangjiang, Guangdong province. Chinese Journal of Birth Health & Heredity. 2002;10:86–89. [Google Scholar]
- 16.Su YQ, Zhu WB, Wang Q. Survey on the glucose-6-phosphate dehydrogenase (G6PD) deficiency incidence in Fujian province. Data compilation of the Eighth National Laboratory Medicine Conference of Chinese Medical Association and the 30th Anniversary Celebration of its Laboratory Medicine Branch. 2009 [Google Scholar]
- 17.Xu HP, Wang YM, Tiang GL. Neonate screening for G6PD deficiency in Shanghai. Shanghai Journal of preventive Medicine. 2008;20:285–286. [Google Scholar]
- 18.Xu XL, Wang F, Yang R, et al. Analysis of the screening results of G6PD deficiency among 10792 neonates in Nanchang region. Chinese Journal of Birth Health & Heredity. 2005;13 85-85-92. Chinese. [Google Scholar]
- 19.Huang QD, Wang J, Zhao ZD, et al. Analysis of results in screening of neonatal G6PD in Hainan province. China Tropical Medicine. 2007;7:2145–2146. [Google Scholar]
- 20.Yang Z, Chu J, Ban G, et al. The genotype analysis of glucose-6-phosphate dehydrogenase deficiency in Yunnan province. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2001;18:259–263. [PubMed] [Google Scholar]
- 21.Tseng CP, Huang CL, Chong KY, Hung IJ, Chiu DT. Rapid detection of glucose-6-phosphate dehydrogenase gene mutations by denaturing high-performance liquid chromatography. Clin Biochem. 2005;38:973–980. doi: 10.1016/j.clinbiochem.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Amini F, Ismail E. 3’-UTR variations and G6PD deficiency. J Hum Genet. 2013;58:189–194. doi: 10.1038/jhg.2012.155. [DOI] [PubMed] [Google Scholar]


