Abstract
Background: Expression patterns of microRNAs in serum are involved in potentially biomarkers for various diseases. The aim of the study was to investigate the expression level of miR-21 in diffuse large B cell lymphoma (DLBCL) and its prognostic value. Methods: Real-time quantitative polymerase chain reaction (qRT-PCR) was used to measure miR-21 levels in serum samples from 112 patients with DLBCL as well as in serum samples from 45 healthy controls. The associations between miR-21 expression and clinicopathologic parameters and overall survival of the patients, were analyzed by chi-square test and Kaplan-Meier method. The Cox proportional hazards regression analyses were performed to estimate the prognostic values for patient survival prediction. Results: We found that serum miR-21 expression was markedly upregulated in patients with DLBCL than healthy controls. Increased miR-21 expression was significantly correlated with B symptoms, IPI score, CHOP-like treatment and Rituximab (all Ps<0.05). Moreover, DLBCL patients with miR-21 higher expression have shown significantly worse overall survival than those with lower miR-21 expression. And miR-21 expression was an independent prognostic marker of overall survival in a multivariate analysis (P=0.001, HR: 4.404, 95% CI: 1.770-10.956). Conclusion: The results of the present study suggested miR-21 expression level could be a novel potential biomarker for DLBCL prognosis.
Keywords: MicroRNA-21, diffuse large B-cell lymphoma, prognosis
Introduction
Diffuse large B-cell lymphoma (DLBCL) is one of the most lethal malignancies and is becoming one of most deadly threat to human health and life worldwide [1]. DLBCL is the most common lymphoma worldwide, accounting for nearly 30 to 40% of non-Hodgkin’s lymphoma cases and is highly heterogeneous from both morphological and clinical standpoints [2]. The pathogenic mechanism contributing to the malignant biological characteristics in DLBCL urgently remains to be clarified by the reason of lacking of specific clinical manifestations and responding poor to existing treatment [3,4]. Although there are latest advancements in diagnostic and therapeutic techniques, a large number of DLBCL patients still have an unfavorable prognosis every year. In addition, despite several biomarkers emerged to better classify and predict outcome at diagnosis, there are not yet routinely used in clinical practice [5]. Thus, exploring more molecular biomarkers involved in DLBCL pathogenesis may novel provide effective therapeutic opportunity.
MicroRNAs (miRNAs) are a class of small, naturally occurring, noncoding and single-stranded RNA molecules (18, 22 nucleotides) that function as post-transcriptional regulators by directly cleaving target messenger RNA (mRNA) or translational repression [6]. A growing number of both direct and indirect evidence suggests a relationship between differential miRNA expression and cancer [7,8]. However, some miRNAs were found to act as tumor suppressors, whereas others acted as oncogenes, depending on the targets of the miRNAs, which may provide insights into the functional detection of human malignancies [9].
It is reported that specific miRNAs may be associated with outcome in patients with DLBCL [10]. The microRNA-21 gene (miR-21) is the most commonly over-expressed miRNA in cancers. It has been identified as the only miRNA commonly over-expressed in various solid tumors, including lung, breast, stomach, prostate, colon, brain, head and neck, esophagus, and pancreas, as well as in chronic lymphocytic leukemia, uterine leiomyomas, and malignant hepatocytes [9,11-15]. In addition, a correlation between miR-21 expression and the carcinogenesis of DLBCL has also been reported [16]. However, the underlying mechanism is not completely clear. Thus, further analyses are needed to clarify the role of miR-21 in DLBCL prognosis based on clinicopathologic stage.
In the present study, serum miR-21 expression levels in DLBCL were examined, and the clinicopathologic significance and potential prognostic value for DLBCL were assessed.
Methods and materials
Patients and serum samples
Serum samples were obtained from 112 patients who were diagnosed with DLBCL enrolled at the Tianjin Medical University General Hospital at the time of diagnosis. All of these patients have undergone molecular and phenotypic classification with available clinical data. 45 serum samples from healthy individuals. Blood samples of all patients and healthy control patients were collected. The samples were allowed to stand at room temperature for 30 min and then centrifuged at 3,000 rpm for 15 min at 4°C. To remove cellular contaminants, serum samples were subjected to additional centrifugation at 12,000 rpm for 10 min. The supernatants to be used for RNA extraction were snap-frozen and then stored at -80°C. The study was approved by the Ethics Committee of Tianjin Medical University General Hospital, and ethical permission and informed consent were obtained from all participants.
Isolation of total RNA and real-time quantitative PCR analysis (qRT-PCR)
MiR-21 expression in serum samples from 112 patients with DLBCL and 45 healthy controls was measured by reverse transcription and real-time PCR (RT-PCR). Total RNA was isolated from frozen samples using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The TaqMan microRNA assay and TaqMan universal PCR master mix were used to detect the expression of miR-21, and the U6 gene was used as an internal control to normalize variances. Relative quantification of target miRNA expression was evaluated using the comparative cycle threshold (CT) method. Each sample was examined in triplicate and the raw data were presented as the relative quantity of target miRNA, normalized with respect to U6. RT-PCR primers: miR-21: F: 5’-GCGGGTAGCTTATCAGACTG-3’; R: 5’-GTGCAGGGTCCGAGGT-3’; U6: F: 5’-GCGCGTCGTGAAGCGTTC-3’; R: 5’-GTGCAGGGTCCGAGGT-3.
Statistical analysis
All statistical calculations were performed using SPSS 18.0 for Windows (SPSS Inc, IL, USA) and GraphPad Prism 5 (GraphPad Software Inc., CA, USA). miR-21 expression levels in serum samples were shown by mean and standard deviation (mean ± SD) and compared using Student’s t-test. The two-tailed Chi-squared test was employed to explore the correlation between miR-21 expression and clinical pathological features. Survival rates were calculated according to the Kaplan-Meier method and survival curves were plotted; statistical differences were analyzed using the log-rank test. Multivariate analysis of the prognostic factors was performed with Cox regression model. P<0.05 was considered statistically significant.
Results
Serum miR-21 is significantly up-regulated in patients with DLBCL
We analyzed the expression levels of miR-21 in serum samples from 112 DLBCL patients and 45 healthy individuals. As revealed by qRT-PCR analysis, miR-21 expression was significantly higher in serum samples from DLBCL patients than that from healthy controls (P<0.05, Figure 1).
Figure 1.
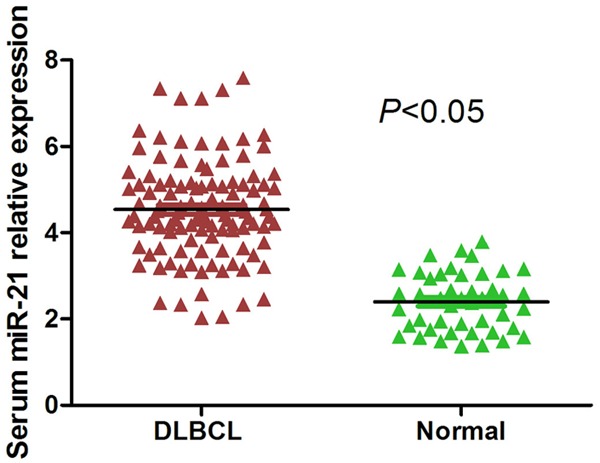
The relative expression level of miR-21 in serum samples from DLBCL patients and healthy individuals. Serum miR-21 expression was significantly higher in DLBCL patients compared with healthy individuals (P<0.05).
Association between miR-21 expression and the clinicopathological features of DLBCL
For better understanding of the clinical relevance of miR-21 expression in DLBCL, we divided the 112 DLBCL patients into a high expression group (n=53) and a low expression group (n=59), according to the expression level of miR-21 in all samples. And the relationship of the miR-21 with various clinical features of DLBCL was analyzed and is summarized in Table 1. As shown in the results, miR-21 expression was closely associated with B symptoms, IPI score, CHOP-like treatment and Rituximab (all Ps<0.05). However, but there was no relationship with other characteristics, such as age, gender, Ann Arbor stages, extra nodal status, and serum CRP (all Ps>0.05).
Table 1.
miR-21 expression and clinicopathological features
| Characteristics | No. (n=112) | miR-21 expression levels | P values | |
|---|---|---|---|---|
|
| ||||
| Low (n=53) | High (n=59) | |||
| Age (years) | ||||
| <60 | 55 | 27 | 28 | 0.713 |
| ≥60 | 57 | 26 | 31 | |
| Gender | ||||
| Male | 58 | 28 | 30 | 0.834 |
| Female | 54 | 25 | 29 | |
| Ann Arbor stages | ||||
| I-II | 47 | 25 | 22 | 0.290 |
| III-IV | 65 | 28 | 37 | |
| B symptoms | ||||
| Absent | 67 | 37 | 30 | 0.041 |
| Present | 45 | 16 | 29 | |
| Extra nodal status | ||||
| <2 | 41 | 21 | 20 | 0.530 |
| ≥2 | 71 | 32 | 39 | |
| Serum CRP | ||||
| Normal | 52 | 26 | 26 | 0.597 |
| High | 60 | 27 | 33 | |
| IPI score | ||||
| 0-2 | 42 | 25 | 17 | 0.045 |
| 3-5 | 70 | 28 | 42 | |
| CHOP-like treatment | ||||
| No | 65 | 25 | 40 | 0.027 |
| Yes | 47 | 28 | 19 | |
| Rituximab | ||||
| No | 55 | 20 | 35 | 0.023 |
| Yes | 57 | 33 | 24 | |
IPI: International Prognostic Index; CHOP-like refers to CHOP (cyclophosphamide, adriamycin, vincristine and prednisone) or a CHOP-like regimen.
Prognostic values of miR-21 expression in serum samples from DLBCL patients
The correlation between miR-21 level and survival time of the patients with DLBCL was evaluated by Kaplan-Meier survival analysis. As determined by the log-rank test in Figure 2, the survival rates of the patients with high miR-21 level were significantly lower than those with low miR-21 level (P=0.000). Multivariate analysis using the Cox proportional hazards model for all variables suggested that high miR-21 expression was an independent prognostic factor for patients with DLBCL (Table 2) (P=0.001, HR: 4.404, 95% CI: 1.770-10.956).
Figure 2.
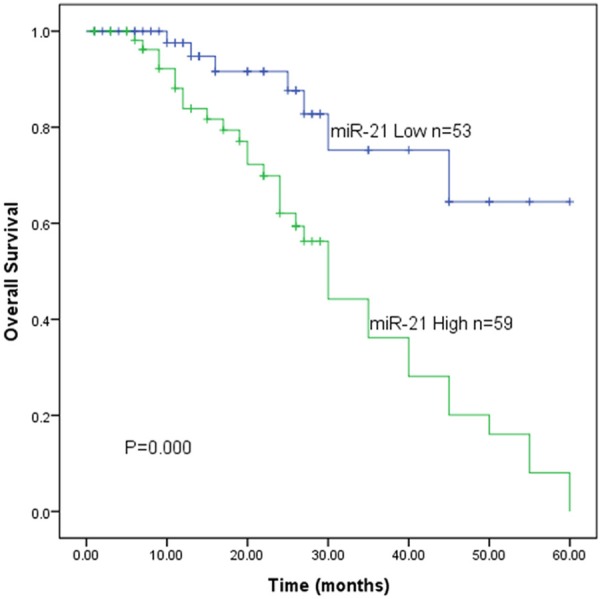
Overexpression of miR-21 is associated with poor overall survival in patients with DLBCL. Kaplan-Meier analysis of overall survival was analyzed according to miR-21 expression levels (P=0.000).
Table 2.
Multivariate analyses of prognostic variables of overall survival in DLBCL patients
| Variables | HR | 95% CI | P Values |
|---|---|---|---|
| MiR-21 expression | 4.404 | 1.770-10.956 | 0.001 |
| B symptoms | 1.137 | 0.551-2.348 | 0.729 |
| IPI score | 1.128 | 0.498-2.556 | 0.773 |
| CHOP-like treatment | 1.036 | 0.484-2.217 | 0.928 |
| Rituximab | 1.709 | 0.831-3.514 | 0.145 |
Discussion
Recently, miRNAs have been demonstrated to play a key role in tumorigenesis. miRNAs may offer a new regulatory model of gene expression, and miRNA expression levels correlate closely with specific clinical features of cancer, so that they can be used to classify normal and cancerous tissues, as well as for prognosis [17-19]. It has been reported that miR-21 plays a role in the development of tumor via regulating the expression of the tumor suppressor, such as PDCD4, PTEN, and TPM1. Suppression of miR-21 can inhibit tumor growth, which could indicate miR-21 functions as an oncogene [20-22]. It has been reported that the dysregulation of miR-21 performed an important function in different types of cancer [9,11-15]. However, few studies are available on its expression and functions in DLBCL.
In this study, we investigated the expression of miR-21 in serum samples from DLBCL patients and healthy controls by qRT-PCR for the first time. Based on the relative expression level analysis, it was investigated that the association of miR-21 with clinicopathological factors and prognosis of patients with DLBCL. Results showed that the serum level of miR-21 was up-regulation in DLBCL patients compared with that in healthy controls (P<0.05). The expression pattern of miR-21 found in our study is in line with previous findings that the expression of miR-21 was increased in DLBCL cells and was used as an independent prognostic indicator for DLBCL patients by Lawrie’s studies [23]. Fang et al. found that miR-21 were significantly elevated in DLBCL serum when compared with normal controls [24]. Wang et al. reported that miR-21 expression was significantly higher in hepatocellular carcinoma tissues compared with normal adjacent liver tissues [25]. Alexander et al. showed that miR-21 showed significantly increased levels in the cerebrospinal fluid of patients with primary central nervous system lymphoma compared with the cerebrospinal fluid of control patients [26]. Charles et al. revealed that miR-21 levels were higher in DLBCL patient than healthy control serum [27]. Chen et al. reported that expression of miR-21 was increased in DLBCL cell lines (OCI-Ly1, OCI-Ly3, OCI-Ly4, OCI-Ly7, OCI-Ly8, OCI-Ly10, OCI-Ly18, OCI-Ly19, and HBL) [28].
In addition, it was also proved that the relative expression level of miR-21 was closely associated with B symptoms, IPI score, CHOP-like treatment and Rituximab. However, miR-21 expression was not associated with age, gender, Ann Arbor stages, extra nodal status, and serum CRP.
More importantly, we proved that miR-21 expression was significantly associated with overall survival by Kaplan-Meier analysis and log-rank test. Patients with high levels of miR-21 expression had worse overall survival compared with those with low levels of miR-21 expression. By a Cox proportional hazards model adjusted for factors related to survival of DLBCL, miR-21 up-regulation could be an independent prognostic marker in patients with DLBCL. These data indicated that miR-21 expression play a crucial role in tumorigenesis, and progression of DLBCL. Mao et al. revealed that serum miR-21 was used as an independent and powerful predictor of overall survival for primary central nervous system lymphoma [29]. Go et al. reported that overexpression of miR-21 was significantly associated with shorter progression-free survival and overall survival and was an independent prognostic factor in DLBCL patients treated with rituximab-combined chemotherapy [16]. MG Narducci et al. reported that miR-21 was upregulated in cutaneous T-cell lymphoma and could discriminate patients with unfavorable and favorable outcome [30].
In conclusion, the results suggest that miR-21 is a novel biomarker and a prognostic target for DLBCL in future. Because there are only a few studies on the relationship between miR-21 expression and the prognosis of DLBCL, further study is required to elucidate the exact molecular mechanisms to verify our conclusions.
Disclosure of conflict of interest
None.
References
- 1.Hunt KE, Reichard KK. Diffuse large B-cell lymphoma. Arch Pathol Lab Med. 2008;132:118–124. doi: 10.5858/2008-132-118-DLBL. [DOI] [PubMed] [Google Scholar]
- 2.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, et al. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994;84:1361–1392. [PubMed] [Google Scholar]
- 3.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 5.Berglund M, Thunberg U, Amini RM, Book M, Roos G, Erlanson M, Linderoth J, Dictor M, Jerkeman M, Cavallin-Stahl E, Sundstrom C, Rehn-Eriksson S, Backlin C, Hagberg H, Rosenquist R, Enblad G. Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod Pathol. 2005;18:1113–1120. doi: 10.1038/modpathol.3800396. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, Thiere M, Loeffler M, Klapper W, Pfreundschuh M, Matolcsy A, Bernd HW, Reiniger L, Merz H, Feller AC. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 11.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 12.Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, Su F, Yao H, Song E. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617–1626. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak HJ, Kim YJ, Chun KR, Woo YM, Park SJ, Jeong JA, Jo SH, Kim TH, Min HS, Chae JS, Choi EJ, Kim G, Shin SH, Gwak HS, Kim SK, Hong EK, Lee GK, Choi KH, Kim JH, Yoo H, Park JB, Lee SH. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 2011;30:2433–2442. doi: 10.1038/onc.2010.620. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 16.Go H, Jang JY, Kim PJ, Kim YG, Nam SJ, Paik JH, Kim TM, Heo DS, Kim CW, Jeon YK. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6:15035–49. doi: 10.18632/oncotarget.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 18.Perera RJ, Ray A. MicroRNAs in the search for understanding human diseases. BioDrugs. 2007;21:97–104. doi: 10.2165/00063030-200721020-00004. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 22.Lou Y, Yang X, Wang F, Cui Z, Huang Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med. 2010;26:819–827. doi: 10.3892/ijmm_00000530. [DOI] [PubMed] [Google Scholar]
- 23.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, Wainscoat JS, Hatton CS. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 24.Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, Fan L, Miao KR, Liu P, Xu W, Li JY. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol. 2012;91:553–559. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang WY, Zhang HF, Wang L, Ma YP, Gao F, Zhang SJ, Wang LC. miR-21 expression predicts prognosis in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2014;38:715–719. doi: 10.1016/j.clinre.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, Maghnouj A, Zollner H, Reinacher-Schick A, Schmiegel W, Hahn SA, Schroers R. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–3146. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 27.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumourassociated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen W, Wang H, Chen H, Liu S, Lu H, Kong D, Huang X, Kong Q, Lu Z. Clinical significance and detection of microRNA-21 in serum of patients with diffuse large B-cell lymphoma in Chinese population. Eur J Haematol. 2014;92:407–412. doi: 10.1111/ejh.12263. [DOI] [PubMed] [Google Scholar]
- 29.Mao X, Sun Y, Tang J. Serum miR-21 is a diagnostic and prognostic marker of primary central nervous system lymphoma. Neurol Sci. 2014;35:233–238. doi: 10.1007/s10072-013-1491-9. [DOI] [PubMed] [Google Scholar]
- 30.Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, Scala E, Fadda P, Cristofoletti C, Facchiano A, Frontani M, Monopoli A, Ferracin M, Negrini M, Lombardo GA, Caprini E, Russo G. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]


